Abstract
The herpes simplex virus type 1 (HSV-1) UL51 gene products are virion-associated phosphoproteins with apparent molecular masses of 27, 29, and 30 kDa in HSV-1-infected cells. In this study, we have investigated the intracellular localization and distribution of UL51 protein both in infected cells and in transfected cells expressing only UL51. We found that this protein colocalized closely with Golgi marker proteins such as the Golgi-58K protein and GM130 in transfected cells expressing only UL51. However, in infected cells, the UL51 protein localized to the juxtanuclear region but only partially colocalized with the Golgi maker proteins. Mutant protein analysis revealed that the N-terminal 15 amino acid residues of the UL51 protein sufficed for this Golgi localization property. The UL51 protein redistributed on addition of brefeldin A. This was prevented by pretreatment with 2-deoxyglucose and sodium azide, which results in ATP depletion, but not by pretreatment with NaF and AlCl3, which activates heterotrimeric G proteins. Moreover, we found that palmitoylation of the UL51 protein through the N-terminal cysteine at position 9 was necessary for its Golgi localization. Protease digestion analysis suggested that the UL51 protein localized on the cytoplasmic face of the membrane in UL51-transfected cells, while in infected cells it localized mainly to the inside of cytoplasmic vesicles and/or the viral envelope. Transmission immunoelectron microscopy revealed an association of UL51 protein-specific labeling with cytoplasmic virions and also with some membranous structure. We infer from these observations that internalization of UL51 protein into the cytoplasmic vesicle and/or virion may occur in association with viral envelopment in HSV-infected cells.
Herpes simplex virus (HSV) is a large, enveloped DNA virus, the genome of which contains at least 74 different protein-coding genes (16, 39, 53). Approximately half of these genes are not essential for virus replication in cell cultures (52). The UL51 gene of HSV type 1 (HSV-1) is one of the dispensable genes that is located at the right end of the unique long (UL) region of the virus genome (6, 39), and its homologs are conserved throughout the herpesvirus family (1, 3, 7, 11, 14, 24, 26, 33, 59); these homologs, except in human cytomegalovirus (HCMV), have similar molecular sizes ranging from 200 to 259 amino acids and contain some highly conserved motifs. In a previous study, we raised a rabbit polyclonal antiserum against an HSV-1 UL51 fusion protein and characterized the UL51 protein in HSV-1-infected cells. It is present as a series of 27-, 29-, and 30-kDa phosphoproteins that are expressed at the late stage of infection and are localized to the perinuclear region of the cytoplasm in infected cells. Additionally, analysis of extracellular virions revealed that the UL51 protein is a component of the virion (13).
Little is known about localization signals for membrane-associated proteins which contain neither an N-terminal signal sequence nor a stretch of hydrophobic transmembrane domains (15, 35, 44, 60). A number of proteins are associated with the cytoplasmic face of the Golgi apparatus, and some of them are modified by covalent attachment of fatty acid (48). Such fatty acylated proteins play key roles in regulating cellular structure and function, membrane attachment, Golgi localization, and plasma membrane targeting (21, 40, 43, 44, 48). The two most common forms of protein fatty acylation are modification with myristate, a 14-carbon saturated fatty acid, and with palmitate, a 16-carbon saturated fatty acid (48). Myristic acid is cotranslationally attached N terminally via amide bonds to glycine residues and immediately after the start methionine is removed by methionine aminopeptidase. N myristoylation is catalyzed by N-myristoyl transferase (48). In contrast, palmitoylation of membrane proteins occurs posttranslationally, and nearly all palmitoylated proteins are acylated by attachment of palmitate through a thioester linkage to the sulfhydryl group of cysteine, although the precise mechanism of palmitoylation reactions is poorly understood (21, 43, 48).
Covalent attachment of fatty acids to membrane proteins was first described for the viral glycoproteins of Sindbis and vesicular stomatitis viruses (57, 58). Since then, an increasing number of viral proteins have been found to be acylated; these include proteins of ortho- and paramyxoviruses (56), retroviruses (67, 68), filoviruses (22), adenoviruses (28), vaccinia viruses (25), and coronaviruses (12). In herpesviruses, the UL11 gene product of HSV-1 has been shown to be a myristoylated protein (36, 37). Recently, Loomis et al. reported that the UL11 protein also undergoes palmitoylation, which is required for its targeting to the Golgi apparatus and for strong membrane binding (34).
In the present study, we have examined the intracellular localization of the UL51 protein in greater detail and found that it has the intrinsic property of localizing to the Golgi apparatus. The UL51 protein redistributed into the cytoplasm on addition of brefeldin A (BFA), but this was prevented by pretreatment with 2-deoxyglucose and sodium azide. Moreover, mutational analyses revealed that the Golgi localization of the UL51 protein was highly dependent on its N-terminal position 9 cysteine, which is required for palmitoylation of this protein.
MATERIALS AND METHODS
Viruses and cells.
U251 cells, a stable line of human glioma cells, were grown in Dulbecco's minimal essential medium supplemented with 10% fetal bovine serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml and were used throughout this study. ST51 cells, stably expressing the UL51 protein, were grown in Dulbecco's minimal essential medium supplemented with 10% fetal bovine serum, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 1 mg of Geneticin (Life Technologies Inc.) per ml. U251 cells transfected with pcDNA-UL51w/t were grown in maintenance medium supplemented with Geneticin. Some Geneticin-resistant stable transformants were isolated, and one of these clones was used in subsequent experiments. HSV-1 strain KOS, derived from single plaques, was propagated in Vero cells by infection at a low multiplicity (0.01 PFU per cell). Infected cells were harvested when almost all cells exhibited cytopathic effects. After freezing and thawing three times and elimination of cell debris at 3,000 rpm for 10 min, virus was stored at −80°C.
Plasmids and PCR.
The plasmid pcDNA-UL51 w/t (13) which expresses the HSV-1 UL51 gene under the control of the HCMV immediate-early promoter, was used for expression of the UL51 protein and for construction of N- or C-terminal deletion mutants of the UL51 protein. The following PCR conditions were used throughout the present study: an initial 2-min denaturation step at 96°C, followed by 35 cycles of denaturation (96°C, 30 s), annealing (56°C, 1 min), and extension (72°C, 2 min) and a final extension at 72°C for 5 min. The PCR product was digested with EcoRI and NotI and then ligated into the multicloning site of pcDNA3.1(+) (Invitrogen).
The expression plasmid UL51N15-GFP was constructed for expression of the UL51 N-terminal 15 amino acid residues tagged upstream with green fluorescent protein (GFP). The DNA fragment encoding the UL51 N-terminal 15 amino acid residues was first synthesized as UL51N15Fxho (5′-TCGAGatggcttctcttctcggggctatatgtggctggggagcgcgccccG-3′) and UL51N15Rbam (5′-GATCCggggcgcgctccccagccacatatagccccgagaagagaagccatC3′), and mixtures of these were incubated for 1 min at 96°C and then for 1 h at room temperature. The annealed DNA fragment was then ligated into the XhoI-BamHI restriction site of the multicloning site of pEGFP-N3 (Clontech) in frame to give UL51N15-GFP.
The expression plasmids pcDNA-UL51C9S and -UL51C9A, which express versions of the UL51 protein with a single amino acid substitution in which cysteine at N-terminal position 9 was replaced by serine and alanine, respectively, were constructed. The UL51C9S and UL51C9A coding sequences were made by PCR amplification from pcDNA-UL51w/t by using UL51C9Seco (5′-gGAATTCatggcttctcttctcggggctataTCTggctgg-3′/EcoRI) and UL51C9Aeco (5′-gGAATTCatggcttctcttctcggggctataGCTggctgg-3′/EcoRI), respectively, as the forward primer. UL51Rnot (5′-caagcGCGGCCGCttattgacccaaaacacacgg-3′/NotI) was used for the reverse primer. Each PCR product was digested with EcoRI and NotI and ligated into the multicloning site of pcDNA3.1(+).
Construction of deletion mutants of the UL51 protein.
Plasmids pcDNA3.1-UL51DN15, pcDNA3.1-UL51DN44, pcDNA3.1-UL51DN75, pcDNA3.1-UL51DN88, pcDNA3.1-UL51DC111, pcDNA3.1-UL51DC132, pcDNA3.1-UL51DC164, pcDNA3.1-UL51DC183, and pcDNA3.1-UL51DC208 were constructed for expression of N- and C-terminal deletion mutants of the UL51 protein. The forward primers for the N-terminal deletion mutants were UL51delN15 (5′-ggaGAATTCatggaggaacaatatgagatgatccg-3′/EcoRI), UL51delN44 (5′-gtcGAATTCatgctacttcccgcccccatcacg-3′/EcoRI), UL51delN75 (5′-gtgGAATTCatgcacgcctgcatggtgaacctag-3′/EcoRI), and UL51delN88 (5′-gtgGAATTCatgcaccccgggttcgaggccccc-3′/EcoRI). The reverse primer was UL51Rnot (5′-caagcGCGGCCGCttattgacccaaaacacacgg-3′/NotI). The forward PCR primer for the C-terminal deletion mutants was UL51Feco (5′-gtgGAATTCatggcttctcttctcggggctatatg-3′/EcoRI). The reverse primers and the incorporated restriction enzyme sites were UL51delC111 (5′-cacGCGGCCGCttagcgccgcatcttgtcctgatgggc-3′/NotI), UL51delC132 (5′-gagcGCGGCCGCttacttgtcagccgcccccacggac-3′/NotI), UL51delC164 (5′-ccGCGGCCGCttagaggccaagggcccgttcggcgatg-3′/NotI), UL51delC183 (5′-tgcGCGGCCGCttacgcctcggtcaccccaatccc-3′/NotI), and UL51delC208 (5′-gtGCGGCCGCttagagggcgtcgccgtttcgggc-3′/NotI). The PCR products were ligated into pcDNA3.1(+).
Immunofluorescence microscopy.
Cells were fixed with cold acetone or with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. The cells were labeled with anti-UL51 rabbit polyclonal antibodies (13) alone or together with mouse monoclonal antibodies for β-COP, Golgi-58K protein (Sigma), GM130, clathrin heavy chain, and γ-adaptin (BD Transduction Laboratories). Goat anti-rabbit antibodies and goat anti-mouse antibodies conjugated to fluorescein isothiocyanate (FITC) or tetramethyl rhodamine isocyanate (TRITC) were used to visualize the primary antibodies. Fluorescent images were examined under the Bio-Rad MRC 1024 imaging system. For HSV-infected cells, cells were treated with normal goat serum before being labeled with primary antibodies to avoid nonspecific reactions.
Western blotting.
Cells were lysed in sodium dodecyl sulfate (SDS) sample buffer, electrophoretically separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred to polyvinylidene difluoride membranes. Nonspecific protein binding was blocked by treating membranes at 4°C overnight with phosphate-buffered saline (PBS) containing 5% skim milk. The membranes were washed with PBS containing 0.05% Tween 20 (PBST) and incubated with appropriately diluted primary antibodies in PBS at 37°C for 1 h. After being washed with PBST, the membranes were incubated with the goat anti-rabbit or anti-mouse peroxidase-labeled secondary antibodies at 37°C for 1 h. The membranes were then washed with PBST, treated with ECL Western blotting detection system reagents (Amersham Pharmacia), and exposed to Hyperfilm-ECL (Amersham Pharmacia).
Subcellular fractionation.
Subconfluent cells were washed twice with PBS, scraped, and resuspended in cold buffer A (10 mM Tris-HCl [pH 7.4] and 5 mM sucrose containing 1 mM phenylmethylsulfonyl fluoride and a cocktail of protease inhibitors [Sigma]). Subsequent procedures were carried out at 4°C. The cells were homogenized with a Dounce homogenizer. Unbroken cells and nuclei were removed by centrifugation at 700 × g for 10 min. Postnuclear supernatants (PNS) were mixed with the same quantity of buffer A or 2% Triton X-100 in buffer A (final concentration, 1%). After incubation on ice for 30 min, PNS mixtures were centrifuged at 45,000 rpm (120,000 × g) in a TLS 55 rotor for 1 h in a Beckman Optima tabletop ultracentrifuge. The resulting supernatants and membrane pellets were collected and analyzed.
Drug treatment.
To examine the effect of BFA on the distribution of the UL51 protein, ST51 cells were treated with 5 μg of BFA per ml for 0, 10, and 30 min at 37°C. The cells were then processed for immunofluorescence analysis. To examine the effect of aluminum fluoride or ATP depletion on BFA-induced redistribution of the UL51 protein, cells were pretreated with AlF (30 mM NaF and 50 μM AlCl3) or with 50 mM 2-deoxyglucose and 0.05% sodium azide (DOG-Az) for 10 min at 37°C and then treated with 5 μg of BFA per ml for 0, 10, and 30 min at 37°C.
Immunoprecipitation and electrophoresis.
Subconfluent U251 cells were separately transfected with each plasmid expressing wild-type UL51 protein (UL51w/t) or the N- or C-terminal deletion mutants of the UL51 protein by using Lipofectamine reagent, according to the protocols recommended by the supplier (GibcoBRL). After 24 h, the transfected cells were washed twice with methionine-free minimal essential medium, incubated for 30 min at 37°C, and then labeled for 30 min with 100 μCi of [35S]methionine (Amersham Pharmacia) per ml. The labeled cells were washed three times with cold PBS and solubilized in buffer A containing 1% Triton X-100. The samples were incubated 4°C for 1 h and centrifuged at 14,000× g at 4°C for 30 min to remove the cell debris. After preabsorption of the supernatants with preimmune rabbit serum and protein A-agarose (Roche), they were incubated with an appropriate amount of anti-UL51 rabbit polyclonal antibody and the immune complexes were precipitated by incubation with protein A-agarose. The immunoprecipitates were washed five times with buffer A containing 1% Triton X-100 to remove nonspecifically adsorbed proteins, eluted from the agarose beads by boiling in 2× SDS sample buffer (125 mM Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, and 0.06% bromophenol blue), and centrifuged at 12,000 rpm at 4°C for 10 min. The supernatants were collected and analyzed by SDS-PAGE. After electrophoresis, the gels were fixed with 10% acetic acid and 10% methanol, dried, and autoradiographed with the Fujix Bioimaging Analyzer BAS 2000 system (Fuji Photo Film).
To detect palmitoylation of the UL51 protein, transfected cells expressing one of the mutant proteins were metabolically labeled with [3H]palmitic acid (400 μCi/60-mm-diameter dish) (Amersham Pharmacia) for 30 min at 37°C. The labeled cells were washed three times with cold PBS and lysed in buffer A containing 1% Triton X-100, and the UL51w/t or mutant UL51 proteins were immunoprecipitated with an appropriate amount of anti-UL51 rabbit polyclonal antibody as described above. Immunoprecipitated proteins were then mixed with 2× SDS-sample buffer (β-mercaptoethanol was omitted to prevent release of palmitate) (28, 34) and separated by SDS-PAGE. Gels were treated with EN3HANCE (NEN Life Science Products, Inc.) for 1 h prior to drying and autoradiographed. The film was exposed to the gels for 3 weeks. HSV-1-infected U251 cells were also labeled with [35S]methionine or [3H]palmitic acid, and samples were prepared and analyzed as described above.
Protease digestion assay.
HSV-1-infected cells or pcDNA-UL51w/t-transfected cells were scraped into reticulocyte standard buffer supplemented with a protease inhibitor cocktail to prevent proteolysis by cellular proteases. After the cells were homogenized by 10 strokes with a glass homogenizer, proteinase K (TaKaRa) was added to cell homogenates in the presence or absence of 1% Triton X-100. After incubation for 30 min at room temperature, phenylmethylsulfonyl fluoride was added to each sample to a final concentration of 2 mM to inhibit further proteolysis. Samples were then separated by SDS-PAGE and analyzed by Western blotting.
Transmission immunoelectron microscopy (TIEM).
U251 cells were grown in 60-mm-diameter culture dishes and were either mock infected or infected with HSV-1 at a multiplicity of 5 PFU per cell. At 24 h after infection, the cells were fixed with modified PLP fixative (10 mM NaIO4, 75 mM lysine, 37.5 mM phosphate buffer [PB] [pH 7.4], 4% paraformaldehyde, 0.1% glutaraldehyde) for 17 h and then washed with 0.1 M PB (46). The cells were then harvested from the culture dish by scraping, resuspended in PB, and pelleted by low-speed centrifugation. The cell pellet was washed with PB, dehydrated in graded ethanol solutions with progressive lowering of the temperature, embedded in the acrylic resin Lowicryl K4M (Polysciences), and polymerized by UV light at −30°C for 24 h. Ultrathin sections were collected onto Formvar-coated nickel grids. The sections were incubated with 5% normal goat serum in PBS containing 1% bovine serum albumin for 1 h at room temperature, washed five times in PBS, and incubated with preimmune rabbit serum or anti-UL51 rabbit polyclonal antibody adequately diluted in PBS-1% bovine serum albumin for 2 h. After five washes in PBS, the sections were incubated for 1 h with the secondary antibody goat anti-rabbit immunoglobulin conjugated with 10-nm-diameter gold particles (British BioCell International) and then washed five times in PBS and twice in double-distilled water. The sections were double stained with 4% uranyl acetate for 30 min followed by Reynold's lead citrate solution for 2 min. Carbon-coated sections were examined with a Hitachi H7100 transmission electron microscope at 75 to 100 kV.
RESULTS
The UL51 protein colocalizes with Golgi marker proteins in singly expressing cells.
We have previously shown that the UL51 protein localizes mainly to the cytoplasm both in HSV-1-infected Vero cells and in transfected cells expressing only this viral gene (13). In order to further examine the intrinsic properties of this protein, we established a U251-derived cell line which stably expressed UL51 protein (ST51) and examined the intracellular localization of the protein. As shown in Fig. 1a, d, and g, the UL51-specific fluorescence was detected in the juxtanuclear region. Because the juxtanuclear staining pattern was reminiscent of a Golgi localization pattern, we examined whether the UL51 protein colocalized with Golgi-associated proteins by double-staining analysis. We indeed found that the UL51 protein colocalized well with the Golgi-58K protein (Fig. 1a to c), which is a peripheral membrane protein exposed to the cytoplasmic side of the Golgi complex (8, 23). Colocalization of the UL51 protein with other Golgi-associated proteins, including GM130 (Fig. 1d to f), β-COP (Fig. 1g to i), and γ-adaptin (data not shown), was also examined, and UL51 was found to colocalize with all of them at the juxtanuclear region, although β-COP was apparently more broadly distributed than the Golgi-58K protein and GM130. These results indicate that the UL51 protein localized predominantly to the Golgi apparatus in singly expressing cells.
FIG. 1.
Intracellular localization of the UL51 protein in ST51 cells. ST51 cells were fixed; double stained with the anti-UL51 rabbit polyclonal antibody (a, d, and g) and mouse monoclonal antibodies against Golgi-58K protein (b), GM130 (e), or β-COP (h); and then reacted with anti-rabbit IgG-conjugated FITC or anti-mouse IgG-conjugated TRITC. The merged images are shown in panels c, f, and i. The insets show high magnification of the juxtanuclear region.
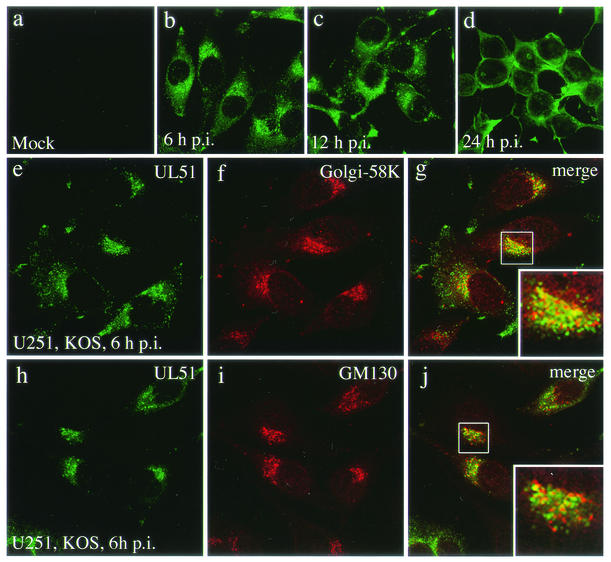
Next, the intracellular localization of the UL51 protein was examined in HSV-1-infected U251 cells. At various times after infection, HSV-1-infected cells were fixed, treated with normal goat serum to block nonspecific binding, and stained with anti-UL51 rabbit polyclonal antibody. As shown in Fig. 2b, the UL51 protein-specific fluorescence became detectable in the cytoplasm by 6 h postinfection (p.i.) and was marked in the juxtanuclear region of infected cells (Fig. 2b). At 12 h p.i., UL51-specific fluorescence was found more widely distributed in the cytoplasm, although the fluorescence was stronger in the juxtanuclear region (Fig. 2c). The pattern of juxtanuclear staining dispersed at later times of infection. At 24 h p.i., UL51-specific fluorescence was detected mainly in the vicinity of the plasma membrane of infected cells (Fig. 2d). No specific fluorescence was observed in mock-infected cells stained with the anti-UL51 rabbit polyclonal antibody (Fig. 2a). Interestingly, the juxtanuclear UL51-specific fluorescence in infected cells was somewhat different from the pattern observed in singly expressing cells. Representative images double stained for the Golgi-58K protein or GM130 are shown in Fig. 2e to g and h to j. The merged images show that the UL51 protein only partially colocalized with the Golgi-58K protein and GM130 (Fig. 2g and j, insets).
FIG. 2.
Intracellular localization of the UL51 protein in HSV-1-infected U251 cells. Mock-infected (a) and HSV-1-infected (b to j) U251 cells were fixed at 6 (b and e to j), 12 (c), and 24 (d) h p.i. as described in Materials and Methods. The samples were double stained with the anti-UL51 rabbit polyclonal antibody (a to e and h) and anti-Golgi-58K protein mouse monoclonal antibody (f) or anti-GM130 mouse monoclonal antibody (i) and then reacted with anti-rabbit IgG-conjugated FITC or anti-mouse IgG-conjugated TRITC. The merged images are shown in panels g and j. Infected cells were pretreated with normal goat serum to block nonspecific antibody reaction. The insets show high magnification of the juxtanuclear region.
The UL51 protein is distributed in both membrane and cytosolic fractions.
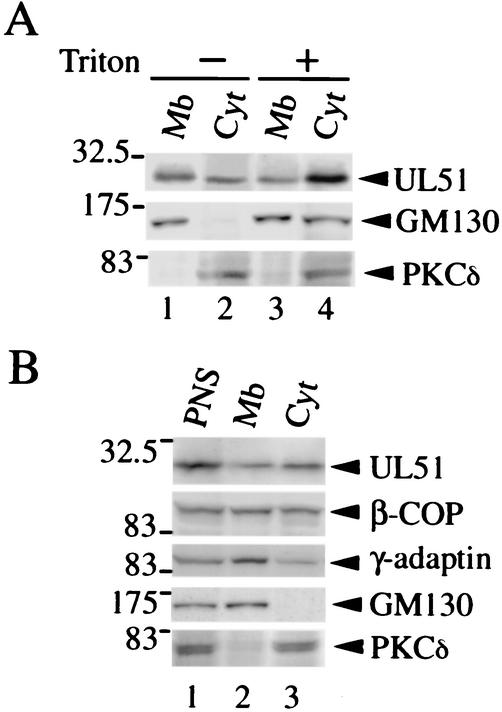
In immunofluorescence staining analysis, it was shown that the UL51 protein localized to the Golgi apparatus but was also detectable in the cytoplasm (Fig. 1a, d, and g). In order to biochemically specify the localization of the UL51 protein, U251 cells transfected with pcDNA-UL51w/t were separated into the membranous pellet and the cytosolic supernatant fractions by ultracentrifugation (120,000 × g) after treatment of the cells with or without 1% Triton X-100 for 30 min on ice. The resulting supernatants and pellets were analyzed by Western blotting with the anti-UL51 rabbit polyclonal antibody (13). The UL51 protein was detected in both pellet and supernatant fractions when the cells were fractionated in the absence of Triton X-100 (Fig. 3A, upper panel, lanes 1 and 2). The relative amount of the UL51 protein detected in each fraction varied to some extent, but the amount of UL51 protein in the supernatant fractions was always markedly increased by treatment with the detergent (Fig. 3A, upper panel, lanes 3 and 4). As a control, the same fractions were subjected to Western blotting with monoclonal antibodies against GM130 and protein kinase C delta (PKCδ). In the absence of Triton X-100, GM130, as expected, was detected predominantly in the pellet fraction, while PKCδ was detected in the supernatant fraction (Fig. 3A, middle and lower panels, lanes 1 and 2). A considerable amount of GM130 was shifted to the supernatant fractions by the addition of the detergent (Fig. 3A, middle panel, lanes 3 and 4).
FIG. 3.
Subcellular distribution of the UL51 protein. (A) Lysates of U251 cells transiently expressing the UL51w/t protein were separated into the membranous pellet (Mb) (lanes 1 and 3) and the cytosolic supernatant (Cyt) (lanes 2 and 4) by ultracentrifugation (120,000 × g, 1 h) in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of 1% Triton X-100 as described in Materials and Methods. The samples were subjected to SDS-PAGE and analyzed by Western blotting with the anti-UL51 rabbit polyclonal antibody (upper panel) and mouse monoclonal antibodies against GM130 (middle panel) or PKCδ (lower panel). (B) ST51 cells were homogenized, the resulting PNS (lane 1) was ultracentrifuged (120,000 × g, 1 h), and the membranous pellets (lane 2) and the cytosolic supernatants (lane 3) were collected. The samples were separated by SDS-PAGE and analyzed by Western blotting with the anti-UL51 rabbit polyclonal antibody and mouse monoclonal antibodies against β-COP, γ-adaptin, GM130, or PKCδ.
The subcellular distribution of UL51 was also examined in ST51 cells. PNS (Fig. 3B, lane 1) were subjected to centrifugation at 120,000 × g, and both membranous pellet (Fig. 3B, lane 2) and cytosolic (Fig. 3B, lane 3) fractions were subjected to SDS-PAGE and then analyzed by Western blotting with the anti-UL51 antiserum. As shown in Fig. 3B, the UL51 protein was detected in both the membranous pellet and cytosolic fractions. When the same fractions were probed with anti-β-COP or anti-γ-adaptin monoclonal antibodies, similar results were obtained, although the relative amounts of the proteins detected in the pellet and supernatant fractions were different (Fig. 3B). On the other hand, GM130 and PKCδ were found predominantly in the membranous pellet and cytosolic fractions, respectively, demonstrating that the fractionation protocol was effective (Fig. 3B, lower panels). Taking these observations together, it is suggested that the UL51 protein is present in at least two different forms, namely, membrane-associated and cytosolic soluble forms.
Redistribution of the UL51 protein in response to BFA treatment.
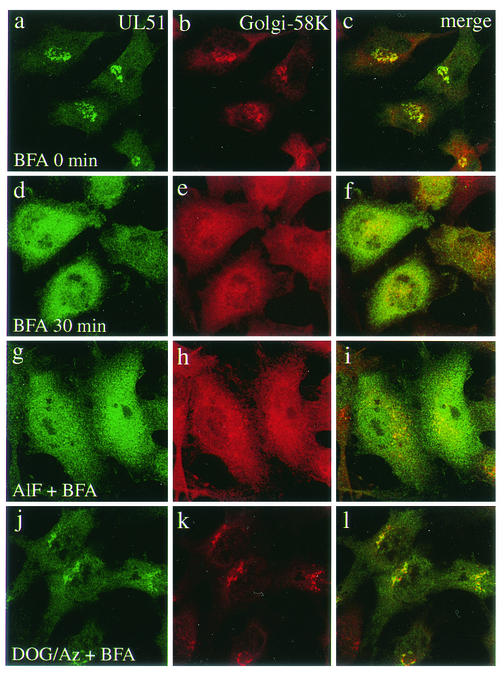
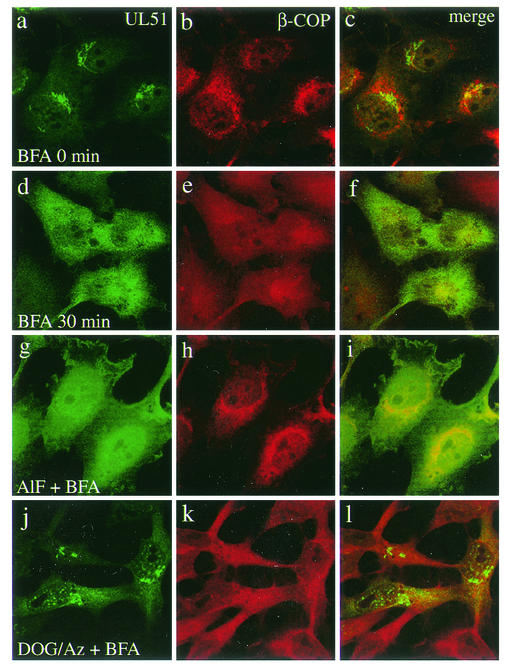
Previous studies have shown that many Golgi-associated proteins are redistributed into the cytosol after BFA treatment. We therefore examined the effects of BFA on the distribution of UL51 protein. ST51 cells were incubated with BFA at a concentration of 5 μg/ml for various lengths of time and then examined by immunofluorescence microscopy. Cells were stained with the rabbit antiserum against the UL51 protein and mouse monoclonal antibodies against Golgi-58K protein (Fig. 4) or β-COP (Fig. 5). Figure 4c illustrates an obvious overlap between the UL51 protein (Fig. 4a) and the Golgi-58K protein (Fig. 4b) in the juxtanuclear region. At 10 min after the addition of BFA, the UL51 protein was completely redistributed into the cytoplasm (data not shown), while the Golgi-58K protein was present both in a diffuse cellular pattern and in small punctuate structures clustered in the juxtanuclear region (data not shown). After 30 min, the Golgi-58K protein was more widely redistributed (Fig. 4e). β-COP, like the UL51 protein, was already redistributed at 10 min after BFA addition (data not shown), as reported previously (18), and thereafter no significant change in its distribution pattern was observed (Fig. 5e). The BFA-induced change of the UL51 protein and Golgi-associated proteins was completely reversible when BFA was removed (data not shown).
FIG. 4.
Effect of BFA on the distribution of UL51 protein and Golgi-58K protein. ST51 cells were pretreated with either AlF (g to i) or DOG-Az (j to l) for 10 min at 37°C and mock treated (a to c) or treated with BFA (5 μg/ml) (d to l) for 30 min at 37°C. Cells were then fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. The cells were double stained with anti-UL51 rabbit polyclonal antibody and anti-Golgi-58K protein mouse monoclonal antibody and then reacted with anti-rabbit IgG-conjugated FITC and anti-mouse IgG-conjugated TRITC. Fluorescence images were obtained with the Bio-Rad MRC 1024 confocal imaging system.
FIG. 5.
Effect of BFA on the distribution of UL51 protein and β-COP. ST51 cells were pretreated with either AlF (g to i) or DOG-Az (j to l) for 10 min at 37°C and mock treated (a to c) or treated with BFA (5 μg/ml) (d to l) for 30 min at 37°C. Cells were then fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. The cells were double stained and fluorescence images were obtained as described in the legend to Fig. 4.
Aluminum fluoride cannot inhibit BFA-induced redistribution of the UL51 protein.
Aluminum fluoride (AlF) has been shown to exert a protective effect on dissociation of β-COP and mannosidase II, but not ADP-ribosylation factor (ARF), induced by BFA treatment (17, 19). Similar to the situation with β-COP, the BFA-induced redistribution of γ-adaptin, a component of clathrin-coated vesicles, was prevented by pretreatment with AlF (51). We therefore examined whether AlF has such a protective effect on dissociation of the UL51 protein caused by BFA treatment. It was found that pretreatment with AlF had no protective effect on BFA-induced redistribution of the UL51 protein (Fig. 4g and 5g). Redistribution of both β-COP (Fig. 5h) and γ-adaptin (data not shown) was significantly reduced by pretreatment with AlF, as shown by previous studies (51). On the other hand, neither the Golgi-58K protein (Fig. 4 h) nor GM130 (data not shown) was protected from BFA-induced redistribution by pretreatment with AlF.
ATP is required for BFA-induced redistribution of the UL51 protein from the Golgi apparatus.
It has been reported that treatment of cells with DOG-Az results in ATP depletion and thereby disrupts the association of β-COP and ARF with the Golgi apparatus, whereas mannosidase II is not affected (18). It has also been shown that pretreatment with DOG-Az impairs BFA-induced retrograde movement of CTR 433, a resident protein of the medial-Golgi cisternae (30), and Rab 6 (49) but not β-COP (49). We therefore studied the effect of DOG-Az on the distribution of the UL51 protein. DOG-Az alone had no obvious effect on the redistribution of UL51 protein and other Golgi-associated proteins except β-COP (data not shown). DOG-Az markedly impaired the redistribution of UL51 protein induced by BFA (Fig. 4j and 5j), whereas β-COP (Fig. 5k) and γ-adaptin (data not shown) were not affected. The redistribution of both the Golgi-58K protein (Fig. 4k) and GM130 (data not shown) was impaired to the same extent as that of the UL51 protein. These results suggest that ATP is required for the redistribution of UL51 protein caused by BFA.
Golgi targeting information is contained within the first 15 N-terminal amino acid residues of UL51.
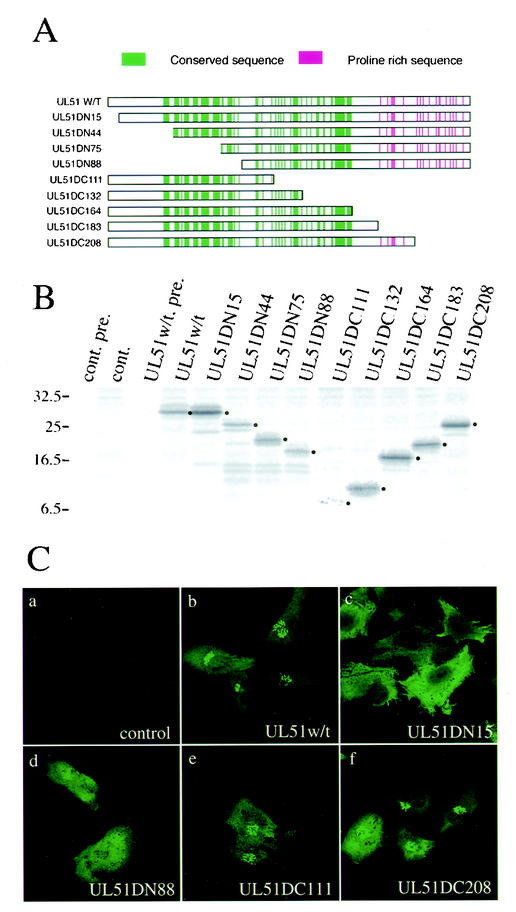
Analysis of the predicted sequence of the UL51 protein suggests that it has no obvious hydrophobic transmembrane domain, although a proline-rich domain is predicted in the C-terminal region (13). To better understand the relationship between localization properties and the structure of the UL51 protein, plasmids pcDNA-UL51w/t, -UL51DN15, -UL51DN44, -UL51DN75, -UL51DN88, -UL51DC111, -UL51DC132, -UL51DC164, -UL51DC183, and -UL51DC208 were constructed (Fig. 6A). These plasmids produce N- and C-terminal deletion mutants of the UL51 protein under the control of the HCMV promoter. U251 cells were transfected with each plasmid, harvested at 24 h posttransfection, and analyzed by immunoprecipitation (Fig. 6B). The yield of UL51DC111 protein was very poor, resulting in only a faint band. However, the results showed that these deletion mutants were expressed as proteins of the expected sizes. Next, the localization of these mutant proteins in transfected cells was analyzed by immunofluorescence staining. At 24 h posttransfection, UL51w/t was observed predominantly as a mass or a cluster of dots in the juxtanuclear region (Fig. 6C, panel b). The localization of all C-terminal deletion mutants was similar to that of UL51w/t (Fig. 6C, panels e and f), whereas marked changes were observed in cells transfected with N-terminal deletion mutants. As shown in Fig. 6C, panels c and d, N-terminal deletion mutant proteins did not show the Golgi localization pattern. No specific fluorescence was observed in control cells that were stained with the anti-UL51 rabbit polyclonal antibody (Fig. 6C, panel a).
FIG. 6.
Effect of N- and C-terminal deletions on the Golgi targeting of UL51 protein. (A) Construction of the UL51 deletion mutant proteins. All of the constructs were inserted into pcDNA3.1(+) as described in Materials and Methods. Highly conserved amino acid sequences in alphaherpesviruses are shown by vertical lines in green. Proline-rich region are shown in red. (B) Expression of the UL51 deletion mutant proteins. U251 cells were transfected with pcDNA-UL51w/t or each plasmid and labeled with [35S]methionine as described in Materials and Methods. Cell lysates were subjected to immunoprecipitation with anti-UL51 rabbit polyclonal antibody, and the labeled proteins were separated by SDS-PAGE and detected by autoradiography with the Fujix Bioimaging Analyser BAS 2000 system. The main bands of each deletion protein are shown by black dots. Molecular mass markers (in kilodaltons) are shown on the left. (C) Intracellular localization of the UL51 deletion mutant proteins. U251 cells were transfected with each plasmid. After 24 h, cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. The samples were stained with the anti-UL51 rabbit polyclonal antibody and then reacted with anti-rabbit IgG-conjugated FITC. Fluorescence images were obtained with the Bio-Rad MRC 1024 confocal imaging system.
As described above, the UL51 protein has no obvious hydrophobic transmembrane domain, and so experiments were performed to demonstrate the ability of the first 15 N-terminal amino acid residues to localize the UL51 protein to the Golgi apparatus. U251 cells were transfected with the plasmid UL51N15GFP, which was designed to express the first 15 UL51 N-terminal amino acid residues tagged upstream with GFP under control of the HCMV immediate-early promoter (Fig. 7A), and the translocation of the GFP-specific fluorescence was investigated by confocal laser-scanning microscopy. Although pEGFP-N3, used as a control, was diffusely distributed throughout the cytoplasm and nucleus (Fig. 7A, panel a), UL51N15GFP-specific fluorescence was observed predominantly in the Golgi region (Fig. 7A, panel d). Double-staining analysis revealed that the specific fluorescence of UL51N15GFP colocalized with the Golgi-58K protein (Fig. 7A, panel f) and with GM130 (data not shown).
FIG. 7.
Mutational analysis of the first 15 amino acid residues of the UL51 protein. (A) The first 15 amino acids of UL51w/t were fused to the N terminus of GFP in frame (UL51N15GFP). U251 cells were transfected with pEGFP-N3 (panels a to c) or UL51N15GFP (d to f). At 24 h posttransfection, the cells were fixed and permeabilized as described in Materials and Methods. The samples were reacted with anti-Golgi-58K protein mouse monoclonal antibody (panels b and e). The inset shows high magnification of the juxtanuclear region. (B) The cysteine at position 9 of UL51w/t was replaced by serine (UL51C9S) or alanine (UL51C9A). U251 cells were transfected with UL51w/t (panels a to c), UL51C9S (panels d to f), or UL51C9A (panels g to i). The samples were double stained with anti-UL51 rabbit polyclonal antibody (panels a, d, and g) and anti-Golgi-58K protein mouse monoclonal antibody (panels b, e, and h). Fluorescence images were obtained with the Bio-Rad MRC 1024 confocal imaging system.
These results suggest that the N-terminal 15 amino acid residues, but not the C-terminal proline-rich domain, are crucial for the localization of the UL51 protein to the Golgi apparatus. Moreover, it is suggested that the first 15 N-terminal amino acid residues alone are sufficient to translocate the GFP into the Golgi apparatus.
Golgi targeting is dependent on palmitoylation at N-terminal cysteine.
In order to specify an amino acid residue(s) that is responsible for Golgi targeting within the N-terminal region of the UL51 protein, we carried out amino acid substitution analysis. U251 cells were transfected with the plasmids pcDNA-UL51C9S and -UL51C9A, which were designed to express mutants of the UL51 protein with single amino acid substitutions in which the cysteine residue at N-terminal position 9 was replaced with serine and alanine, respectively (Fig. 7B). The mutated proteins did not colocalize with the Golgi-58K protein (Fig. 7B, panels d to i) and GM130 (data not shown) but showed a more cytosolic distribution pattern than the wild type (Fig. 7B, panel a). These results suggest that cysteine at position 9 is involved in directing the UL51 protein specifically to the Golgi apparatus.
Several proteins peripherally associated with Golgi membranes have N-terminal sequences that include sites for modification with palmitic acid (15, 34, 35, 44). It is also known that nearly all palmitoylated proteins are acylated by attachment of palmitate through a thioester linkage to the sulfhydryl group of cysteine (48). To determine whether the UL51 protein is palmitoylated in vivo, transfected cells were metabolically labeled with [3H]palmitic acid and immunoprecipitated with the anti-UL51 rabbit polyclonal antibody. As shown in Fig. 8B, neither of the cysteine mutants could be labeled with [3H]palmitic acid, while only UL51w/t was labeled efficiently (Fig. 8B, lane 3), indicating that these mutants can no longer be acylated by palmitic acid. Similar to the case for the cysteine mutants, UL51DN15, which is the version of the UL51 protein with the first 15 N-terminal amino acids truncated, also was not labeled to any detectable level (Fig. 8B, lane 4). As controls, these different UL51 mutant proteins were labeled with [35S]methionine (Fig. 8A). All constructs were found to be expressed at levels equal to or greater than that for the wild type (Fig. 8A). The acylation of UL51 protein by palmitic acid was also detected at 9 h p.i. in HSV-1-infected cells (Fig. 8C). These data indicate that the attachment of palmitic acid to UL51 protein occurred through a thioester linkage to the N-terminal cysteine at position 9 and that the UL51 protein is palmitoylated both in transfected and in HSV-1-infected cells.
FIG. 8.
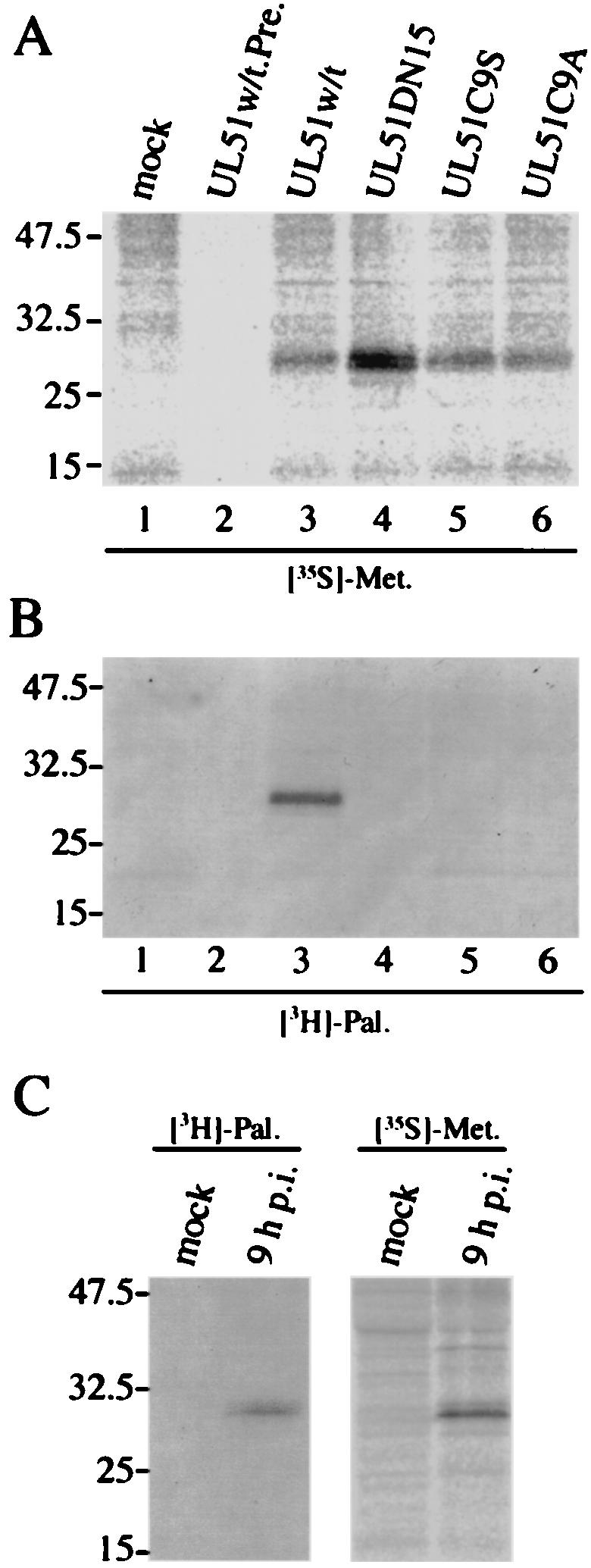
Palmitoylation of UL51 protein. (A and B) Transfected U251 cells were labeled with [35S]methionine (A) or [3H]palmitic acid (B) as described in Materials and Methods. Cell lysates were immunoprecipitated with anti-UL51 rabbit polyclonal antibody (lanes 1 and 3 to 6) or preimmune rabbit serum (lane 2), and the labeled proteins were mixed with the sample buffer (without β-mercaptoethanol). The samples were separated by SDS-PAGE and detected by autoradiography. (C) HSV-1-infected U251 cells were labeled with [35S]methionine or [3H]palmitic acid. At 9 h p.i., cells were harvested and the lysates were immunoprecipitated and analyzed as described in Materials and Methods.
As shown in Fig. 7B, the nonpalmitoylated UL51 mutant proteins showed no predominant localization to the Golgi apparatus. We therefore performed subfractionation analysis to determine whether palmitoylation affects the ability of the UL51 protein to bind to membranes. Figure 9 shows that all of the nonpalmitoylated UL51 mutant proteins, unlike the wild-type protein, were detected predominantly in the cytosolic fractions. As a control, the same cell fractions were also probed with monoclonal antibodies against GM130, β-COP, and PKCδ. These data suggest that the palmitoylation of UL51 protein plays an important role in its membrane binding ability.
FIG. 9.
Membrane binding of nonpalmitoylated UL51 mutant proteins. Transfected U251 cells were harvested at 24 h posttransfection. The cells were homogenized, the resulting PNS was ultracentrifuged (120,000 × g, 1 h), and then the membranous pellets (Mb) and the cytosolic supernatants (Cyt) were collected as described in Materials and Methods. The samples were separated by SDS-PAGE and analyzed by Western blotting with the anti-UL51 rabbit polyclonal antibody and mouse monoclonal antibodies against GM130, β-COP, or PKCδ.
The UL51 protein is incorporated into virions in the process of envelopment.
To learn more about the properties of membrane association of UL51 protein in transfected and infected cells, a protease digestion assay was performed. U251 cells were transfected with pcDNA-UL51w/t and harvested at 24 h posttransfection. After the cells were homogenized by 10 strokes with a glass homogenizer, proteinase K was added to cell homogenates in the presence or absence of Triton X-100. In transfected cells, the UL51 protein was sensitive to proteinase K in either the presence or absence of Triton X-100 (Fig. 10A). Golgi-associated marker proteins, which face the cytoplasm, were also sensitive to the proteinase even in absence of Triton X-100 (Fig. 10A), while a significant part of the transferrin receptor (TfR), which is present within cytoplasmic vesicles and on the cell surface, was significantly resistant to proteinase K in the absence of Triton X-100. Since TfR has an N-terminal cytoplasmic region (32), the decrease in its apparent molecular weight may be due to the digestion of that region (Fig. 10A). Thus, it is likely that the UL51 protein was localized on the outer or cytoplasmic surface of the Golgi membrane in singly expressing cells.
FIG. 10.
Sensitivity of UL51 protein to proteinase K. U251 cells expressing only the UL51w/t protein (A) and HSV-1-infected U251 cells (B) were treated with proteinase K in the absence (−) or presence (+) of 1% Triton X-100. All samples were then separated by SDS-PAGE and analyzed by Western blotting with the anti-UL51 rabbit polyclonal antibody and with mouse monoclonal antibodies against the Golgi-58K protein, GM130, γ-adaptin, β-COP, or TfR. Positions of molecular mass markers (in kilodaltons) are indicated on the left.
We next examined the sensitivity of the UL51 protein in infected cells. As shown in Fig. 10B, the UL51 protein was resistant to 100 μg of proteinase K per ml in the absence of Triton X-100, whereas it was highly sensitive to the proteinase in the presence of the detergent. The sensitivities of Golgi-associated marker proteins and TfR to proteinase K in infected cells and transfected cells were similar.
The association of UL51 protein with intracellular capsids in the infected cells was examined by TIEM. HSV-1-infected U251 cells were fixed, harvested at 24 h p.i., and embedded in acrylic resin. Ultrathin sections were prepared and treated with 5% normal goat serum to reduce nonspecific reactions before incubation with preimmune rabbit serum or anti-UL51 rabbit polyclonal antibody followed by treatment with anti-rabbit goat immunoglobulin G (IgG) conjugated to 10-nm-diameter gold particles. No specific immunolabeling was seen in the control cells reacted with preimmune serum (Fig. 11A). Intense labeling of the UL51 protein was found around intracellular virions (Fig. 11B). Some labeling was also found in the vicinity of L-particle-like structures and with some membranous structure that was observed near the UL51-labeled intracellular virion (Fig. 11B).
FIG. 11.
TIEM of HSV-1-infected U251 cells. Thin sections were prepared as described in Materials and Methods and incubated with preimmune rabbit serum (A) or the anti-UL51 rabbit polyclonal antibody (B) after treatment with 20% normal goat serum to block nonspecific antibody reactions. Samples were then incubated with anti-rabbit IgG-conjugated 10-nm-diameter gold particles. After extensive rinsing, sections were stained with uranyl acetate and lead citrate and examined with a Hitachi H7100 transmission electron microscope at 100 kV. Some of the enveloped capsids (arrows) were labeled. Labeling was also found in L-particle-like structures (large arrowheads) and along with membranous structures (small arrowheads). The inset shows an enveloped intracellular capsid labeled for UL51 proteins. Bars, 200 nm.
DISCUSSION
The present study demonstrates that the UL51 protein colocalizes closely with Golgi marker proteins such as Golgi-58K and GM130 in UL51-transfected cells but does so only partially in HSV-1-infected cells. The protease digestion assay suggests that the UL51 protein localizes on the cytoplasmic face of the Golgi membrane in transfected cells, whereas in HSV-1-infected cells it localizes mainly to the inner side of cytoplasmic vesicles and/or the viral envelope. Furthermore, it is suggested that palmitoylation of the N-terminal cysteine at position 9 of the UL51 protein is required for its Golgi localization.
We have previously shown that the UL51 protein localizes to the perinuclear region of the cytoplasm both in HSV-1-infected Vero cells and in transfected COS-1 cells expressing only UL51 (13). In that study, we exclusively used cold acetone for fixation of cells and failed to specify the subcellular localization of the UL51 protein. We now provide evidence that the UL51 protein localizes to the Golgi region in both HSV-1-infected and transfected cells, although the precise localizations were different. The Golgi localization of the UL51 protein in transfected cells was confirmed by double staining for Golgi-associated marker proteins, including the Golgi-58K protein, GM130, β-COP, and γ-adaptin. Both the Golgi-58K protein and GM130 are peripheral membrane-associated proteins exposed to the cytoplasmic side of the Golgi apparatus (8, 23), while β-COP and γ-adaptin are coat proteins of the non-clathrin-coated vesicles and clathrin-coated vesicles, respectively (19, 20, 38, 54, 55, 63). In contrast to the distribution patterns of the Golgi-58K protein and GM130, β-COP and γ-adaptin were widespread in both infected and transfected cells, as expected from the distribution of these vesicles. The juxtanuclear localization pattern of UL51 protein in infected cells predominated until 12 h p.i., but even at 6 h p.i. a considerable part of the UL51 protein did not colocalize with the Golgi marker proteins. Although our data clearly show the intrinsic nature of UL51 protein to be associated with the Golgi apparatus, the process of HSV replication appears to affect its interaction with the Golgi apparatus. In HSV-1-infected Vero and Hep-2 cells, the Golgi apparatus is known to be fragmented and dispersed (2, 62). This may partly explain the difference in the localization patterns of the UL51 protein and Golgi marker proteins in transfected and infected cells. At 24 h p.i., UL51 protein-specific fluorescence was detected in the vicinity of the plasma membrane, whereas Golgi marker proteins were detected in the cytoplasm as a faint granulated fluorescence. TIEM revealed that a large number of extracellular virions were adjacent to the plasma membrane, especially at the margin of cell-to-cell junctions (data not shown). The UL51-specific fluorescence observed in the vicinity of the plasma membrane at 24 h p.i. may be attributed to virion-associated UL51 protein.
Although analysis of the predicted sequence of the UL51 protein revealed no obvious hydrophobic transmembrane domain, the UL51w/t protein was detected in both membranous pellet and cytoplasmic supernatant fractions. The results are not due to contamination of each fraction, because GM130 and PKCδ, which were used as controls, were detected only in the membranous pellet and cytosolic fractions, respectively (Fig. 3B). It is likely that the UL51 protein has two different forms, a membrane-associated form and a cytosolic soluble one. We found that UL51 protein was labeled with [3H]palmitic acid and that replacement of the cysteine with serine or alanine abolished the Golgi localization (Fig. 7B, panels d and g). These observations suggest that palmitoylation contributes to membrane attachment and targeting of the UL51 protein to the Golgi apparatus. This is further supported by the results of subfractionation of cells expressing nonpalmitoylated UL51 mutant proteins (Fig. 9), all of which were associated with the membranous fractions to a much lesser extent than the palmitoylated UL51w/t. We thus conclude that modification of the cysteine residue at position 9 by palmitic acid is most important for its membrane association and Golgi targeting, although we cannot rule out the possibility that the UL51 protein has another signal for its intracellular localization.
Previous studies have shown that Golgi-associated proteins, including β-COP, γ-adaptin, the Golgi-58K protein, and GM130, are rapidly redistributed into the cytosol after BFA treatment (17, 23, 45, 51). The addition of BFA disperses β-COP throughout the cytoplasm as early as 2 min after exposure (49, 51). Gao et al. also reported that β-COP can be dispersed into the cytosol after only 50 s of BFA treatment in NRK cells (23). In our case, β-COP was completely redistributed into the cytoplasm by 10 min after BFA addition, consistent with previous observations. BFA-induced redistribution of the Golgi-58K protein and GM130 was slightly slower than that of β-COP. Under these conditions, UL51 protein was redistributed into the cytoplasm as rapidly as β-COP. Although BFA has dramatic effects on the intracellular localization of Golgi-associated proteins, these perturbations are fully reversible within minutes after drug removal (17, 23, 45, 51). The reassociation of UL51 protein with the reforming Golgi apparatus was already detected at 30 min after BFA washout, as seen with Golgi marker proteins (data not shown). Thus, the effect of BFA on the UL51 protein is fully reversible.
The BFA-induced redistribution of UL51 protein was prevented by pretreatment of cells with DOG-Az but not with AlF. The reagent AlF, the active species of NaF and AlCl3, interacts with the GDP-bound form of the Gα subunit by mimicking the γ-phosphate of GTP, thereby activating heterotrimeric G proteins (31). AlF stabilizes β-COP binding to the Golgi apparatus even in the presence of BFA (17). In addition to β-COP, AlF can also stabilize the association of γ-adaptin, Rab 6, and mannosidase II with the Golgi apparatus (49, 51). On the other hand, the combination DOG-Az inhibits BFA-induced redistribution of the Golgi apparatus because of ATP depletion. Previous reports have shown that ATP depletion alone disrupts the association of β-COP with the Golgi apparatus (17, 49). We found that the BFA-induced redistribution of the UL51 protein, like that of the Golgi-58K protein and GM130, was prevented by ATP depletion. Our observations thus suggest that at least heterotrimeric G proteins are not involved in regulating the Golgi association of the UL51 protein and that ATP is required for BFA-induced redistribution of the UL51 protein.
The UL51 protein in HSV-1-infected cells, unlike that in transfected cells, was highly resistant to protease in the absence of Triton X-100, suggesting that the majority of the UL51 protein in infected cells was present within the cytoplasmic vesicles or the viral envelope. In the process of HSV replication, progeny nucleocapsids exit from the nucleus by a budding-fusion event on the inner and outer nuclear membranes and then acquire the final envelope in a cytoplasmic area. Previous studies reported that the Golgi apparatus, especially the trans-Golgi network (TGN), is an important organelle with regard to envelopment (10, 27, 41, 66). In addition, Harley and colleagues reported that enveloped infectious virions accumulate in organelles with biochemical properties similar to those of the TGN and endosomes and that nonenveloped capsids are tethered to the cytoplasmic face of these organelles (27). Our TIEM analysis showed that UL51 labeling was detected along with some membranous structures observed near the intracellular virion. Taking these results together with the previous results that the Golgi apparatus is fragmented and dispersed in HSV-1-infected cells (2, 62), it is possible that the membranous structures labeled with the UL51 protein in HSV-infected U251 cells may be such fragmented vesicles derived from the TGN.
A number of viruses encode palmitoylated proteins that play important roles in the process of virus assembly and release. Vaccinia virus encodes six palmitoylated proteins, four of which have been shown to be important in either egress or cell-to-cell spreading (25). Sindbis virus also encodes a palmitoylated protein E2 which functions in virus assembly and envelopment (29). HSV-1 UL11 protein is acylated both with myristic acid (34, 36, 37) and palmitic acid (34), binds to the cytoplasmic face of internal membranes (4, 36), and plays a role in the envelopment and egress of viral nucleocapsids (5, 37). The UL51 protein, unlike the UL11 protein, has no amino-terminal myristoylation motif MGXXXS/T/A/C/N, and in fact [3H]myristic acid labeling of the UL51 protein was not detectable (data not shown), suggesting that it is a type III palmitoylated protein (48).
Palmitoylation involves the posttranslational attachment of the C16 saturated fatty acid palmitic acid to the thiol group of a specific cysteine residue of soluble and transmembrane proteins in the endoplasmic reticulum, the Golgi complex, or the plasma membrane (21, 42, 47). In contrast to the chemical stability of the acyl-amide bond in N myristoylation (9), the chemically labile acyl-thioester bond allows regulated cycles of palmitoylation and depalmitoylation that are thought to be important in controlling a protein's subcellular localization, conformation, protein-protein interactions, and activity (50, 61, 64, 65). Our results indicate that the UL51 protein can take two different forms, a membrane-binding form and a cytosolic soluble form. Thus, it is possible that the specific acyl modifications of the UL51 protein define not only its susceptibility to palmitoylation-depalmitoylation cycling but also its specific targeting to the Golgi membrane or intracellular trafficking pathway of progeny capsids, as in the case of other viral palmitoylated proteins. It has been reported that an HSV-1 mutant lacking the BamHI D′ fragment plus the C-terminal 202 amino acid residues of UL51 forms very small plaques although the plaque size of the recombinant lacking only the BamHI D′ fragment is similar to that of the parent virus (6). From our results, we hypothesize that the UL51 protein may participate in the targeting of viral capsids to the final envelopment site and/or virus particle trafficking associated with the Golgi apparatus. UL51 null mutants will be very useful for further understanding the role of this protein in the HSV replication cycle. Such studies are now in process.
Acknowledgments
We thank E. Iwata and T. Tsuruguchi for technical assistance. We also thank the members of our laboratory for helpful discussions and are grateful to A. Nozawa for support.
This work was supported by a grant from the Japan Society for the Promotion of Science (JSPS-RFTF97L00703) and also by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, and Culture of Japan.
REFERENCES
- 1.Albrecht, J. C., J. Nicholas, D. Biller, K. R. Cameron, B. Biesinger, C. Newman, S. Wittmann, M. A. Craxton, H. Coleman, B. Fleckenstein, and R. W. Honess. 1992. Primary structure of the herpesvirus saimiri genome. J. Virol. 66:5047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avitabile, E., S. D. Gaeta, M. R. Torrisi, P. L. Ward, B. Roizman, and G. Campadelli-Fiume. 1995. Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J. Virol. 69:7472-7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baer, R. J., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. Tuffnell, and B. G. Barrell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature (London) 310:207-211. [DOI] [PubMed] [Google Scholar]
- 4.Baines, J. D., R. J. Jacob, L. Simmerman, and B. Roizman. 1995. The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J. Virol. 69:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baines, J. D., and B. Roizman. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 66:5168-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker, D. E., and B. Roizman. 1990. Identification of three genes nonessential for growth in cell culture near the right terminus of the unique sequences of long component of herpes simplex virus 1. Virology 177:684-691. [DOI] [PubMed] [Google Scholar]
- 7.Baumeister, J., B. G. Klupp, and T. C. Mettenleiter. 1995. Pseudorabies virus and equine herpesvirus 1 share a nonessential gene which is absent in other herpesviruses and located adjacent to a highly conserved gene cluster. J. Virol. 69:5560-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloom, G. S., and T. A. Brashear. 1989. A novel 58-kDa protein associates with the Golgi apparatus and microtubules. J. Biol. Chem. 264:16083-16092. [PubMed] [Google Scholar]
- 9.Boutin, J. A. 1997. Myristoylation. Cell Signal. 9:15-35. [DOI] [PubMed] [Google Scholar]
- 10.Browne, H., S. Bell, T. Minson, and D. W. Wilson. 1996. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J. Virol. 70:4311-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchinson, T. Kouzarides, J. A. Martignetti, E. Preddie, S. Satchwell, P. Tomlinson, K. Weston, and B. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 12.Corse, E., and C. E. Machamer. 2002. The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting. J. Virol. 76:1273-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daikoku, T., K. Ikenoya, H. Yamada, F. Goshima, and Y. Nishiyama. 1998. Identification and characterization of the herpes simplex virus type 1 UL51 gene product. J. Gen. Virol. 79:3027-3031. [DOI] [PubMed] [Google Scholar]
- 14.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 15.Di Paolo, G., R. Lutjens, V. Pellier, S. A. Stimpson, M. H. Beuchat, S. Catsicas, and G. Grenningloh. 1997. Targeting of SCG10 to the area of the Golgi complex is mediated by its NH2-terminal region. J. Biol. Chem. 272:5175-5182. [DOI] [PubMed] [Google Scholar]
- 16.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donaldson, J. G., R. A. Kahn, J. Lippincott-Schwartz, and R. D. Klausner. 1991. Binding of ARF and β-COP to Golgi membranes: possible regulation by a trimeric G protein. Science 254:1197-1199. [DOI] [PubMed] [Google Scholar]
- 18.Donaldson, J. G., J. Lippincott-Schwartz, G. S. Bloom, T. E. Kreis, and R. D. Klausner. 1990. Dissociation of a 100kDa peripheral membrane protein from the Golgi apparatus is an early event in brefeldin A action. J. Cell Biol. 111:2295-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donaldson, J. G., J. Lippincott-Schwartz, and R. D. Klausner. 1991. Guanine nucleotides modulate the effects of brefeldin A in semipermeable cells: regulation of the association of a 110-kD peripheral membrane protein with the Golgi apparatus. J. Cell Biol. 112:579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duden, R., G. Griffiths, R. Frank, P. Argos, and T. E. Kreis. 1991. Beta-COP, a 110 kd protein associated with non-clathrin-coated vesicles and the Golgi complex, shows homology to beta-adaptin. Cell 64:649-665. [DOI] [PubMed] [Google Scholar]
- 21.Dunphy, J. T., and M. E. Linder. 1998. Signalling functions of protein palmitoylation. Biochim. Biophys. Acta 1436:245-261. [DOI] [PubMed] [Google Scholar]
- 22.Funke, C., S. Becker, H. Dartsch, H.-D. Klenk, and E. Muhlberger. 1995. Acylation of the Marburg virus glycoprotein. Virology 208:289-297. [DOI] [PubMed] [Google Scholar]
- 23.Gao, Y. S., C. Alvarez, D. S. Nelson, and E. Sztul. 1998. Molecular cloning, characterization, and dynamics of rat formiminotransferase cyclodeaminase, a Golgi-associated 58-kDa protein. J. Biol. Chem. 273:33825-33834. [DOI] [PubMed] [Google Scholar]
- 24.Gompels, U., J. Nicholas, G. Lawrence, M. Jones, B. Thomson, M. Martin, S. Efstathiou, M. Craxton, and H. Macaulay. 1995. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 209:29-51. [DOI] [PubMed] [Google Scholar]
- 25.Grosenbach, D. W., S. G. Hansen, and D. E. Hruby. 2000. Identification and analysis of vaccinia virus palmitylproteins. Virology 275:193-206. [DOI] [PubMed] [Google Scholar]
- 26.Hamel, F., H. Boucher, and C. Simard. 2002. Transcriptional and translational expression kinetics of the bovine herpesvirus 1 UL51 homologue gene. Virus Res. 84:125-134. [DOI] [PubMed] [Google Scholar]
- 27.Harley, C. A., A. Dasgupta, and D. W. Wilson. 2001. Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles. J. Virol. 75:1236-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hausmann, J., D. Ortmann, E. Witt, M. Veit, and W. Seidel. 1998. Adenovirus death protein, a transmembrane protein encoded in the E3 region, is palmitoylated at the cytoplasmic tail. Virology 244:343-351. [DOI] [PubMed] [Google Scholar]
- 29.Ivanova, L., and M. J. Schlesinger. 1993. Site-directed mutations in the Sindbis virus E2 glycoprotein identify palmitoylation sites and affect virus budding. J. Virol. 67:2546-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jasmin, B. J., J. Cartaud, M. Bornens, and J. P. Changeux. 1989. Golgi apparatus in chick skeletal muscle: changes in its distribution during end plate development and after denervation. Proc. Natl. Acad. Sci. USA 86:7218-7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kahn, R. A. 1991. Fluoride is not an activator of the smaller (20-25kDa) GTP-binding proteins. J. Biol. Chem. 266:15595-15597. [PubMed] [Google Scholar]
- 32.Lawrence, C. M., S. Ray, M. Babyonyshev, R. Galluser, D. W. Borhani, and S. C. Harrison. 1999. Crystal structure of the ectodomain of human transferrin receptor. Science 286:779-782. [DOI] [PubMed] [Google Scholar]
- 33.Lenk, M., N. Visser, and T. C. Mettenleiter. 1997. The pseudorabies virus UL51 gene product is a 30-kilodalton virion component. J. Virol. 71:5635-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loomis, J. S., J. B. Bowzard, R. J. Courtney, and J. W. Wills. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 75:12209-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lutjens, R., M. Igarashi, V. Pellier, H. Blasey, G. Di Paolo, E. Ruchti, C. Pfulg, J. K. Staple, S. Catsicas, and G. Grenningloh. 2000. Localization and targeting of SCG10 to the trans-Golgi apparatus and growth cone vesicles. Eur. J. Neurosci. 12:2224-2234. [DOI] [PubMed] [Google Scholar]
- 36.MacLean, C. A., B. Clark, and D. J. McGeoch. 1989. Gene UL11 of herpes simplex virus type 1 encodes a virion protein which is myristylated. J. Gen. Virol. 70:3147-3157. [DOI] [PubMed] [Google Scholar]
- 37.MacLean, C. A., A. Dolan, F. E. Jamieson, and D. J. McGeoch. 1992. The myristylated virion proteins of herpes simplex virus type 1: investigation of their role in the virus life cycle. J. Gen. Virol. 73:539-547. [DOI] [PubMed] [Google Scholar]
- 38.Matter, K., and I. Mellman. 1994. Mechanisms of cell polarity: sorting and transport in epithelial cells. Curr. Opin. Cell Biol. 6:545-554. [DOI] [PubMed] [Google Scholar]
- 39.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 40.McLaughlin, R. E., and J. B. Denny. 1999. Palmitoylation of GAP-43 by the ER-Golgi intermediate compartment and Golgi apparatus. Biochim. Biophys. Acta 1451:82-92. [DOI] [PubMed] [Google Scholar]
- 41.McMillan, T. N., and D. C. Johnson. 2001. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J. Virol. 75:1928-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milligan, G., M. Parenti, and A. I. Magee. 1995. The dynamic role of palmitoylation in signal transduction. Trends Biochem. Sci. 20:181-187. [DOI] [PubMed] [Google Scholar]
- 43.Mumby, S. M. 1997. Reversible palmitoylation of signaling proteins. Curr. Opin. Cell Biol. 9:148-154. [DOI] [PubMed] [Google Scholar]
- 44.Munro, S. 1998. Localization of proteins to the Golgi apparatus. Trends Cell Biol. 8:11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakamura, N., C. Rabouille, R. Watson, T. Nilsson, N. Hui, P. Slusarewicz, T. E. Kreis, and G. Warren. 1995. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 131:1715-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nozawa, N., T. Daikoku, Y. Yamauchi, H. Takakuwa, F. Goshima, T. Yoshikawa, and Y. Nishiyama. 2002. Identification and characterization of the UL7 gene product of herpes simplex virus type 2. Virus Genes 24:257-266. [DOI] [PubMed] [Google Scholar]
- 47.Olson, E. N., and G. Spizz. 1986. Fatty acylation of cellular proteins. Temporal and subcellular differences between palmitate and myristate acylation. J. Biol. Chem. 261:2458-2466. [PubMed] [Google Scholar]
- 48.Resh, M. D. 1999. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451:1-16. [DOI] [PubMed] [Google Scholar]
- 49.Roa, M., V. Cornet, C. Z. Yang, and B. Goud. 1993. The small GTP-binding protein rab6p is redistributed in the cytosol by brefeldin A. J. Cell Sci. 106:789-802. [DOI] [PubMed] [Google Scholar]
- 50.Robinson, L. J., L. Busconi, and T. Michel. 1995. Agonist-modulated palmitoylation of endothelial nitric oxide synthase. J. Biol. Chem. 270:995-998. [DOI] [PubMed] [Google Scholar]
- 51.Robinson, M. S., and T. E. Kreis. 1992. Recruitment of coat proteins onto Golgi membranes in intact and permeabilized cells: effects of brefeldin A and G protein activators. Cell 69:129-138. [DOI] [PubMed] [Google Scholar]
- 52.Roizman, B. 1996. The function of herpes simplex virus genes: a primer for genetic engineering of novel vectors. Proc. Natl. Acad. Sci. USA 93:11307-11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roizman, B., and A. E. Sears. 1996. Herpes simplex virus and their replication, p. 1043-1107. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 54.Rothman, J. E. 1994. Mechanisms of intracellular protein transport. Nature (London) 372:55-63. [DOI] [PubMed] [Google Scholar]
- 55.Schekman, R., and L. Orci. 1996. Coat proteins and vesicle budding. Science 271:1526-1533. [DOI] [PubMed] [Google Scholar]
- 56.Schmidt, M. F. G. 1989. Fatty acylation of proteins. Biochim. Biophys. Acta 988:411-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmidt, M. F. G., M. Bracha, and M. J. Schlesinger. 1979. Evidence for covalent attachment of fatty acids to Sindbis virus glycoproteins. Proc. Natl. Acad. Sci. USA 76:1687-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidt, M. F. G., and M. J. Schlesinger. 1979. Fatty acid binding to vesicular stomatitis virus glycoprotein—a new type of posttranslational modification of the viral glycoprotein. Cell 17:813-819. [DOI] [PubMed] [Google Scholar]
- 59.Telford, E. A., M. S. Watson, K. McBride, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus-1. Virology 189:304-316. [DOI] [PubMed] [Google Scholar]
- 60.Ugur, O., and T. L. Z. Jones. 2000. A proline-rich region and nearby cysteine residues target Xlαs to the Golgi complex region. Mol. Biol. Cell 11:1421-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van de Loo, J. W., M. Teuchert, I. Pauli, E. Plets, W. J. Van de Ven, and J. W. Creemers. 2000. Dynamic palmitoylation of lymphoma proprotein convertase prolongs its half-life, but is not essential for trans-Golgi network localization. Biochem. J. 352:827-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ward, P. L., E. Avitabile, G. Campadelli-Fiume, and B. Roizman. 1998. Conservation of the architecture of the Golgi apparatus related to a differential organization of microtubules in polykaryocytes induced by syn− mutants of herpes simplex virus 1. Virology 241:189-199. [DOI] [PubMed] [Google Scholar]
- 63.Waters, M. G., T. Serafini, and J. E. Rothman. 1991. ′Coatomer': a cytosolic protein complex containing subunits of non-clathrin-coated Golgi transport vesicles. Nature (London) 349:248-251. [DOI] [PubMed] [Google Scholar]
- 64.Wedegaertner, P. B., and H. R. Bourne. 1994. Activation and depalmitoylation of Gs alpha. Cell 77:1063-1070. [DOI] [PubMed] [Google Scholar]
- 65.Wedegaertner, P. B., P. T. Wilson, and H. R. Bourne. 1995. Lipid modifications of trimeric G proteins. J. Biol. Chem. 270:503-506. [DOI] [PubMed] [Google Scholar]
- 66.Whiteley, A., B. Bruun, T. Minson, and H. Browne. 1999. Effects of targeting herpes simplex virus type 1 gD to the endoplasmic reticulum and trans-Golgi network. J. Virol. 73:9515-9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang, C., and R. W. Compans. 1996. Palmitoylation of the murine leukemia virus envelope glycoprotein transmembrane subunits. Virology 221:87-97. [DOI] [PubMed] [Google Scholar]
- 68.Yang. C., C. P. Spies, and R. W. Compans. 1995. The human and simian immunodeficiency virus envelope glycoprotein transmembrane subunits are palmitoylated. Proc. Natl. Acad. Sci. USA 92:9871-9875. [DOI] [PMC free article] [PubMed] [Google Scholar]