Viruses have multifaceted approaches to ensure that viral genome amplification can be achieved in an efficient and, in some instances, a cell-type-specific manner. To accomplish this task, some viruses encode their own polymerases which selectively amplify the viral genomes; other viruses have evolved a variety of ways to compete directly with the host cell for factors that are needed for viral gene replication and packaging (5). However, there is one piece of macromolecular machinery in the host cell for which all viruses have to compete: the ribosome. Early in infection, the viral mRNAs have to compete with the host, not so much for ribosomes, but for the limited pool of eukaryotic initiation factors (eIFs) that mediate the recruitment of ribosomes to both viral and cellular mRNAs (10). To circumvent this competition, viruses often modify certain eIFs within infected cells so that ribosomes can be recruited selectively to viral mRNAs even though only a limited repertoire of eIFs is present (2). Of course, this strategy implies that such viral mRNAs have structural features that are distinct from most polymerase II-derived host mRNAs.
For example, it was a long-standing puzzle how poliovirus, a human picornavirus, can inhibit the translation of capped host cell mRNAs when translation of its own uncapped mRNA remained uninhibited. More than a decade ago, it was discovered that poliovirus, and all other picornaviruses, contain internal ribosome entry site elements, commonly abbreviated as IRES elements, in their 5′ noncoding regions that can directly recruit ribosomal 40S subunits with a reduced set of eIFs (13, 24). The cap binding protein eIF-4E is especially dispensable for IRES activity in most viral IRES-containing mRNAs. Since then, IRES elements have been detected in many positive-stranded viral RNA genomes (9). More recently, IRESs have also been identified in Kaposi's sarcoma-associated herpesvirus, which contains a DNA genome. Specifically, a polycistronic transcript, found in all latently infected cells, is used to express the v-FLIP (FLICE-inhibitory protein) protein whose function is to counteract fatty acid synthase-induced apoptosis (1, 7). These findings have provided ample evidence that IRES elements have important functions in the viral life cycle, mostly to ensure efficient viral translation when components of the host translation machinery are limited due to virus-induced modification or host-induced antiviral responses, such as the phosphorylation of eIF-2 (9).
In this minireview, I will discuss the surprising structural information we have obtained from studies on binary hepatitis C virus (HCV) IRES-40S complexes and the roles of specific canonical initiation and IRES-transacting factors (ITAFs) in translation initiation and in viral pathogenesis. I apologize for not mentioning and citing the many important contributions of other investigators who have made contributions in the viral IRES field. Due to space constraints in this minireview, I needed to focus on a few selected topics.
ANATOMY OF A RIBOSOME-IRES INTERACTION: THE BINARY HCV IRES-40S COMPLEX
Although the eIFs which are modified in picornavirus-infected cells have been identified and the elements in the viral IRES which are important for the recruitment of ribosomes have been delineated, the exact mechanism by which 40S subunits are recruited onto IRES elements has been an enigma. This has been mostly due to the complexity of the eukaryotic translation machinery that is involved in the assembly of 40S mRNA complexes. However, with the discoveries that the IRES elements in the flavivirus HCV (26) and cricket paralysis virus (CrPV) (36) can recruit 40S subunits as binary complexes without the aid of known canonical eIFs, structural and kinetic analyses of 40S-IRES complexes became feasible. As a consequence, several structural biologists have entered the IRES field and their studies have provided exciting insights into structural features of the HCV IRES and into the anatomy of the HCV IRES-40S complex.
Initially, a structural model of the HCV IRES (Fig. 1) was obtained from both the phylogenetic and chemical enzymatic RNA structure probing approaches (11, 34, 35). Kieft and coworkers provided the first evidence that the IRES folds into a distinct three-dimensional structure at physiological salt concentrations in the presence of magnesium, as judged by the migration of a single band in native polyacrylamide gels; importantly, the folded IRES was found to bind 40S subunits with a high affinity of 2 nM (17).
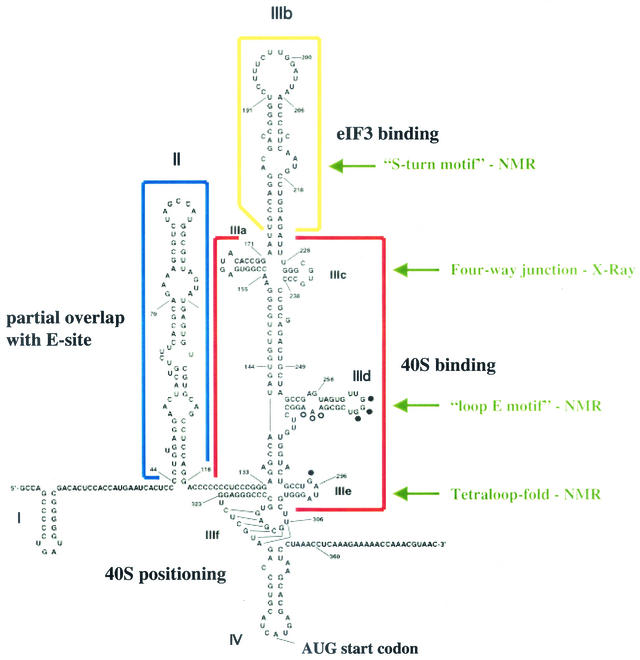
FIG. 1.
Sequence and structure of the HCV IRES (modified from references 11 and 21). Individual domains in the IRES are highlighted, and their known functions in the translation initiation step are indicated. Solved structural motifs in various IRES domains are indicated by the green arrows. See the text for details.
Armed with a structural model, several laboratories tested the effects on translation of defined mutations in predicted conserved elements of the HCV IRES. Overall, the results suggested that the HCV IRES is modular and contains domains that can have both distinct and overlapping functions in 40S binding, eIF-3 binding, and the induction of conformational changes in the bound 40S subunit (Fig. 1).
The heart of the HCV IRES, which recruits 40S subunits, is located in the basal part of domain III, comprising subdomains IIIa, IIIc, IIId, and IIIe (Fig. 1). Subdomain IIIabc forms a four-way junction which is essential for binding both the 40S subunit and eIF-3 (16). X-ray crystal structure showed that this junction forms two helical stacks. Stem IIIa stacks on stem IIIb, and stem IIIc forms a coaxial stack with the basal part of stem III in an A-form duplex. However, the single-stranded junction nucleotides (A154, A155, and U228) influence the overall stacking geometry; such distorted RNA backbone structures often facilitate RNA-protein interactions (16). More importantly, the IIIabc junction resembles a structure formed between four helices in the prokaryotic 50S ribosomal subunit, where the rRNA junction interacts with ribosomal proteins (15). Using nuclear magnetic resonance (NMR), it was found that stem-loop IIId, which contains a helical stem followed by an internal loop and a hexanucleotide loop region, folds into a loop E motif, which contains rich hydrogen-bonding potential for the formation of RNA-protein or RNA-RNA interactions (21). Loop E motifs have been detected both in prokaryotic 5S rRNA and in the sarcin-ricin loop of eukaryotic 28S rRNA, suggesting that IIId interacts with conserved components of the ribosome. Finally, stem-loop IIIe folds into a tetraloop structure, suspected to have a role in the initial assembly of IRES-40S complexes (21).
The role of domain II in HCV IRES activity has been puzzling. Mutational analysis has shown that stem-loop II is essential in IRES activity; however, these mutated RNAs bound 40S subunits with wild-type affinity (23). Insights into the functional roles of domain II came from the remarkable cryoelectron microscopy structure of HCV IRES-40S complexes reported by Spahn and colleagues (31). This study showed that the IRES binds through multiple contacts to the solvent side of the 40S subunit. Domain II nestles around the head of the 40S subunit and points down into the ribosomal E site, normally occupied by the deacylated tRNA before it is expelled from the ribosome. The presence of domain II induces or stabilizes drastic conformational changes in the 40S subunit. These changes were not observed in IRES-40S complexes containing deletions of domain II, even though their binding affinities were similar. Apparently, the additional energy derived from binding to stem-loop II is used to stabilize this altered conformation. Very recently, an additional important feature of domain II has been obtained from NMR studies: domain II can fold independently in the context of the entire HCV IRES. NMR spectroscopy is usually restricted, because of resonance overlap and broadening, to the analysis of 25-kDa molecules (approximately 70 nucleotides); thus, the full-length 100-kDa HCV IRES is out of limits. However, Kim and coworkers (18) have recently developed an elegant approach by which the structure of a small RNA segment can be monitored in the context of surrounding RNA sequences. Briefly, HCV stem-loop II sequences 40 to 104 (Fig. 1) were synthesized by T7 RNA polymerase in the presence of 15N-labeled nucleotide triphosphates. The 5′ ends of these RNAs were generated after cleavage by a cis-acting hammerhead ribozyme, resulting in an RNA species that contained 5′ and 3′ OH groups. Next, stem-loop II sequences 105 to 354 were synthesized in the presence of GMP and a 3′-end-located hammerhead ribozyme, resulting in RNAs with monophosphates at their 5′ and 3′ ends. Purified RNAs were ligated by T4 RNA ligase, yielding segmentally labeled full-length HCV IRES RNAs without intramolecular ligation by-products. Comparison of full-length IRES with a labeled domain II segment revealed imino resonances similar to those of domain II in solution, suggesting that domain II of the HCV IRES forms an independent structure and does not engage in long-range interactions with other parts of the IRES (18). Thus, this novel technology should be very useful in studying the global domains of small RNA segments in the context of a higher-ordered structure.
Once binary IRES-40S complexes are formed and domain II-induced changes in the 40S subunits have taken place, 60S subunits join to form an 80S ribosome. Binding of eIF-3 to the apical domain IIIb (Kd = 35 nM) is essential for 60S subunit joining (26). However, binding of eIF-3 to domain IIIb is not necessary for 40S-IRES assembly. This finding is supported by the cryoelectron microscopy study which shows that domain IIIb projects into solution (31). NMR spectroscopy has revealed that the internal loop sequence in IIIb contains mismatched RNA held in an S-like motif flanked by two helices. The more apical of these helices contains two unprotonated cytosine residues stacked inside the helix (4). So far, the exact role of this structure in the IRES-eIF-3 complex is unknown.
The above discussion shows that much information has been obtained on structural features in the HCV IRES in quite a short time. However, very little is known about the components of the ribosome that contact the IRES. Initial UV cross-linking experiments have shown that ribosomal proteins S9 and S5 could be cross-linked to the HCV IRES (6, 26). More recently, Otto and coworkers (23) have examined whether rRNA or ribosomal proteins could be cross-linked to HCV IRES molecules that contained 4-thiouridine residues. Curiously, no cross-links between rRNA and the IRES could be detected. In contrast, a series of ribosomal proteins S2, S3, S10, S15, S27, and S16 or S18 could be readily cross-linked to the IRES. Comparisons with their prokaryotic homologs provide some clues to their functional roles in the eukaryotic ribosome (23). For example, S2 and S3 could aid in the unwinding of domain IV, which contains the start-site AUG codon, because their bacterial counterparts are suspected to have RNA helicase activity (23). Based upon the ribosomal locations of their prokaryotic homologs, S2, S16/S18, and S15 are suspected to regulate IRES activities in the ribosomal A, P, and E sites, respectively (23).
The recently discovered HCV-like IRES elements in the CrPV-like viruses (30, 36) share many features of the HCV IRES. For example, the CrPV-like IRESs fold into a higher-ordered tertiary structure that contain three pseudoknot-like motifs (12, 14) with distinct functions in both the formation of binary 40S complexes and the positioning of the 40S subunit in the ribosomal P site (12). In contrast to the HCV IRES, however, the binary CrPV IRES-40S complex can assemble 60S subunits without any eIF to form 80S ribosomes (36) that are very stable in solution. Clearly, we should soon be learning more exciting information about IRES-ribosome complexes from studies with divergent viral IRES elements that lack requirements for different eIFs.
ROLES OF IRES ELEMENTS IN VIRAL PATHOGENESIS
Unlike HCV, human picornaviruses can be grown in culture and in animals. However, the host genomes can still not be manipulated genetically in easy ways. Of course, with the advent of RNA interference (20), the expression of host factor genes suspected of being involved in the formation of IRES-40S complexes can be reduced and subsequent effects on the assembly of viral mRNA-ribosome complexes can be studied. However, studies performed with a mouse picornavirus, Theiler's murine encephalomyelitis virus (TMEV), have recently pointed to important roles for the viral IRES in the regulation of viral pathogenesis in an organism.
It has been known for a long time that viral IRES elements can bind certain canonical translation and tissue-specific host factors (9). Studies with the poliovirus IRES, for example, have shown that mutations within the IRES elements of attenuated strains lead to decreased translational efficiencies in neuronal cells (8, 19). Similar observations were made in extracts (8, 32), opening the possibility to study the mechanism by which IRES-mediated translation from virulent and attenuated picornaviruses is regulated. It became immediately evident that this was not a simple task, due to the complexity of the eukaryotic translation apparatus. However, landmark findings by Pestova et al. (25, 27) showed that IRES-48S ribosomal complexes, which contain mRNA associated with 40S subunits with the initiator tRNA in the ribosomal P site, can be assembled from purified eIFs and 40S subunits. Specifically, it was found that all picornavirus IRES elements can bind intact or truncated forms of eIF-4G which aid in the recruitment of 40S subunits (27).
Analyzing the IRES from the virulent GDVII strain of TMEV, Pilipenko and colleagues showed that the pyrimidine tract-binding protein PTB greatly stimulated the formation of 48S-GDVII IRES complexes that were assembled from purified eIF-2, eIF-3, eIF-4A, eIF-4B, and eIF-4F, initiator tRNA, 40S ribosomal subunits, and viral mRNA (28). Because each monomer of PTB contains four RNA recognition motifs and the functional form is a homodimer, it was proposed that PTB functions as an RNA chaperone, modulating IRES conformation by a concerted interaction with several RNA binding sites. This finding raised the question of whether the effects of PTB on the GDVII IRES observed in the cell-free assembly system have roles in the pathogenesis of TMEV.
The virulent GDVII strain of TMEV infects and replicates in cells of the central nervous system, including motor neurons. However, motor neurons lack PTB; instead, these cells contain a neural homolog of PTB, termed nPTB. Consequently, Pilipenko and coworkers examined whether nPTB can stimulate the assembly of 48S-GDVII IRES complexes. It was found that, like PTB, nPTB binds to the IRES at several sites. However, the conformation of the IRES changed markedly upon binding of nPTB, which correlated with enhanced assembly of 48S-IRES complexes (29). Next, they tested whether GDVII viruses containing IRES mutations that abolished binding of nPTB were as neurovirulent as wild-type virus. Indeed, it was found that such mutant viruses were less virulent in mice (29). As a control, the authors noted that the mutant viruses grew to similar titers in nonneuronal cultured cells, suggesting that the observed effects were not simply due to the infection with an amplification-defective virus. Virulent revertants could be isolated; inspection of the revertant genomes revealed that a second-site mutation was acquired that generated a binding site for nPTB (29). That multiple nPTB binding sites in the IRES were required to elicit a virulent phenotype strongly supports the hypothesis that nPTB functions like the fabric of an umbrella to keep the spokes of the IRES in an ordered conformation, facilitating the recruitment of 40S subunits.
In contrast to the virulent GDVII virus, attenuated DA and BeAn strains of TMEV initially infect the brain and subsequently move to the spinal cord where they persist in microglia and oligodendrocytes. Because viral persistence is associated with inflammation and demyelination, attenuated TMEV strains have been used as a model to study multiple sclerosis. Interestingly, the ability to cause a persistent infection correlates with expression of the viral L* protein (3). Mutations that abolish synthesis of L* resulted in mutant viruses that failed to persist in mice, suggesting that L* can interfere with the antiviral cytotoxic-T-lymphocyte response of the infected host cell (3).
The AUG start codon for L* is 13 nucleotides downstream of the polyprotein AUG start codon. Therefore, synthesis of the 18-kDa L* protein is in the +1 reading frame with respect to the polyprotein. It was postulated that the IRES elements of persistent strains recruit ribosomes that can start translation at the AUG codons for both the polyprotein and the L* protein. In the latter case, ribosomes were postulated to scan from the polyprotein start codon to the AUG start codon of L* (37).
However, very recent observations seem to allow another interpretation. Van Eyll and Michiels changed the AUG start codon for L* in the persistent DA virus to an ACG codon and observed that the mutant virus persisted like wild-type virus in macrophages (33). It was noted that the ACG mutant virus could still direct the synthesis of small amounts of the 18-kDa L*; however, a more-prominent 15-kDa L* protein was synthesized, with AUG codon 41 of the L* open reading frame as the start codon (33).
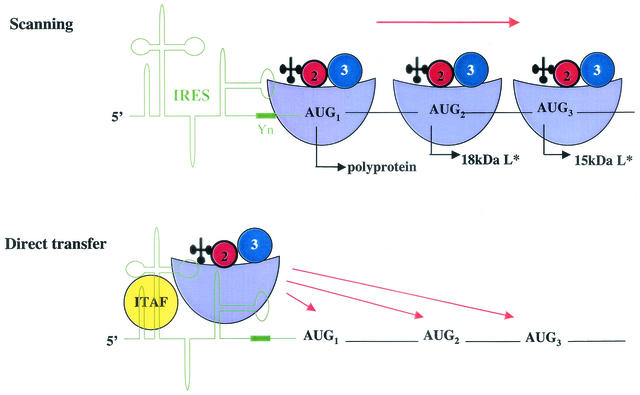
Is the AUG-to-ACG change the mutation that allows persistence? While this is not known, an interesting mechanistic question arises from these findings. How are the start-site codons for the polyprotein (AUG1) and for the 18-kDa (AUG2) and the 15-kDa (AUG3) L* proteins being recognized? One scenario suggests that the IRES recruits ribosomes to AUG1; such ribosomes could start polyprotein synthesis at this AUG or could move to the downstream AUG or ACG codons by a linear scanning mechanism (Fig. 2). Alternatively, ribosomes have been proposed to be directly transferred from the IRES to AUG2 and AUG3, termed dropping by the authors (33) (Fig. 2). Why would ribosomes be transferred directly to different positions, sometimes ignoring perfectly good AUG codons? It is possible that ITAFs, such as nPTB, modulate start-site recognition in subsets of RNA molecules. For example, cell-specific factors could facilitate an IRES conformation which allows the transfer of 48S subunits predominantly to AUG1. This may be the case for the GDVII IRES in motor neurons. In the absence of nPTB or the presence of other ITAFs, the IRES may have more flexibility and position 48S subunits downstream of AUG2 and AUG3 codons by direct transfer (Fig. 2). One can also envisage that viral factors manipulate IRES elements, the translation apparatus, or both in a cell-specific manner. The outcome of such scenarios could lead to the accumulation of heterogeneous pools of ribosomes that could affect the rates of translation initiation differently.
FIG. 2.
Scanning and direct transfer models by which the TMEV IRES can use alternate start codons. The IRES and a conserved oligopyrimidine-rich sequence motif (Yn) are shown in green. The ribosomal 48S complex is depicted. 40S subunits are light blue; 40S-associated eIF-2 and eIF-3 are red and blue, respectively; initiator tRNA is black. The ITAF is shown in yellow. The start-site AUG codons for the viral polyprotein and various L* proteins are shown. See the text for details.
Such ideas have recently been discussed by Mauro and Edelman in the ribosome filter hypothesis (22). While it is generally assumed that translational control is primarily exerted at the initiation step of translation, Mauro and Edelman hypothesize that the ribosome itself has regulatory roles in the assembly of ribosome-mRNA complexes. Evidence is cited for heterogeneity in rRNA and ribosomal proteins which could lead to heterogeneous pools of ribosomes that may have different activities in protein synthesis (20). In support of the ribosome filter hypothesis, Otto and coworkers (23) noted that 40S subunits from Saccharomyces cerevisiae failed to form binary complexes with the HCV IRES. Alignment of human and yeast S2, S5, and S10 proteins revealed large differences which could account for the failure to assemble 40S-IRES complexes. Thus, it seems very likely that future findings will point to exciting regulatory roles of heterogeneous ribosome populations on the rate of translation.
Acknowledgments
I thank Karla Kirkegaard for critical reading of the manuscript.
Work in my laboratory was supported by grants from the NIH (AI47365, GM55979), the Hutchison Foundation, and Eli Lilly, Inc.
REFERENCES
- 1.Bieleski, L., and S. J. Talbot. 2001. Kaposi's sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. J. Virol. 75:1864-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bushell, M., and P. Sarnow. 2002. Hijacking the translation apparatus by RNA viruses. J. Cell Biol. 158:395-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, H. H., W. P. Kong, L. Zhang, P. L. Ward, and R. P. Roos. 1995. A picornaviral protein synthesized out of frame with the polyprotein plays a key role in a virus-induced immune-mediated demyelinating disease. Nat. Med. 1:927-931. [DOI] [PubMed] [Google Scholar]
- 4.Collier, A. J., J. Gallego, R. Klinck, P. T. Cole, S. J. Harris, G. P. Harrison, F. Aboul-Ela, G. Varani, and S. Walker. 2002. A conserved RNA structure within the HCV IRES eIF3-binding site. Nat. Struct. Biol. 9:375-380. [DOI] [PubMed] [Google Scholar]
- 5.Flint, S. J., L. W. Enquist, R. M. Krug, V. R. Racaniello, and A. M. Skalka. 2000. Principles of virology: molecular biology, pathogenesis, and control. ASM Press, Washington, D.C.
- 6.Fukushi, S., M. Okada, J. Stahl, T. Kageyama, F. B. Hoshino, and K. Katayama. 2001. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J. Biol. Chem. 276:20824-20826. [DOI] [PubMed] [Google Scholar]
- 7.Grundhoff, A., and D. Ganem. 2001. Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:1857-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haller, A. A., S. R. Stewart, and B. L. Semler. 1996. Attenuation stem-loop lesions in the 5′ noncoding region of poliovirus RNA: neuronal cell-specific translation defects. J. Virol. 70:1467-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hellen, C. U., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593-1612. [DOI] [PubMed] [Google Scholar]
- 10.Hershey, J. W. B., and W. C. Merrick. 2000. The pathway and mechanism of initiation of protein synthesis, p. 33-88. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 11.Honda, M., M. R. Beard, L. H. Ping, and S. M. Lemon. 1999. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 73:1165-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jan, E., and P. Sarnow. 2002. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 324:889-902. [DOI] [PubMed] [Google Scholar]
- 13.Jang, S. K., H. G. Krausslich, M. J. H. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanamori, Y., and N. Nakashima. 2001. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA 7:266-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieft, J. S., K. Zhou, A. Grech, R. Jubin, and J. A. Doudna. 2002. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat. Struct. Biol. 9:370-374. [DOI] [PubMed] [Google Scholar]
- 16.Kieft, J. S., K. Zhou, R. Jubin, and J. A. Doudna. 2001. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7:194-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kieft, J. S., K. Zhou, R. Jubin, M. G. Murray, J. Y. Lau, and J. A. Doudna. 1999. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol 292:513-529. [DOI] [PubMed] [Google Scholar]
- 18.Kim, I., P. J. Lukavsky, and J. D. Puglisi. 2002. NMR study of 100 kDa HCV IRES RNA using segmental isotope labeling. J. Am. Chem. Soc. 124:9338-9339. [DOI] [PubMed] [Google Scholar]
- 19.La Monica, N., and V. R. Racaniello. 1989. Differences in replication of attenuated and neurovirulent polioviruses in human neuroblastoma cell line SH-SY5Y. J. Virol. 63:2357-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindenbach, B. D., and C. M. Rice. 2002. RNAi targeting an animal virus: news from the front. Mol. Cell 9:925-927. [DOI] [PubMed] [Google Scholar]
- 21.Lukavsky, P. J., G. A. Otto, A. M. Lancaster, P. Sarnow, and J. D. Puglisi. 2000. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 7:1105-1110. [DOI] [PubMed] [Google Scholar]
- 22.Mauro, V. P., and G. M. Edelman. 2002. The ribosome filter hypothesis. Proc. Natl. Acad. Sci. USA 99:12031-12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otto, G. A., P. J. Lukavsky, A. M. Lancaster, P. Sarnow, and J. D. Puglisi. 2002. Ribosomal proteins mediate the hepatitis C virus IRES-HeLa 40S interaction. RNA 8:913-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 25.Pestova, T. V., C. U. Hellen, and I. N. Shatsky. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pestova, T. V., I. N. Shatsky, and C. U. Hellen. 1996. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 16:6870-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pilipenko, E. V., T. V. Pestova, V. G. Kolupaeva, E. V. Khitrina, A. N. Poperechnaya, V. I. Agol, and C. U. Hellen. 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 14:2028-2045. [PMC free article] [PubMed] [Google Scholar]
- 29.Pilipenko, E. V., E. G. Viktorova, S. T. Guest, V. I. Agol, and R. P. Roos. 2001. Cell-specific proteins regulate viral RNA translation and virus-induced disease. EMBO J. 20:6899-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasaki, J., and N. Nakashima. 2000. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc. Natl. Acad. Sci. USA 97:1325-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spahn, C. M., J. S. Kieft, R. A. Grassucci, P. A. Penczek, K. Zhou, J. A. Doudna, and J. Frank. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291:1959-1962. [DOI] [PubMed] [Google Scholar]
- 32.Svitkin, Y. V., T. V. Pestova, S. V. Maslova, and V. I. Agol. 1988. Point mutations modify the response of poliovirus RNA to a translation initiation factor: a comparison of neurovirulent and attenuated strains. Virology 166:394-404. [DOI] [PubMed] [Google Scholar]
- 33.van Eyll, O., and T. Michiels. 2002. Non-AUG-initiated internal translation of the L* protein of Theiler's virus and importance of this protein for viral persistence. J. Virol. 76:10665-10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, C., S. Y. Le, N. Ali, and A. Siddiqui. 1995. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA 1:526-537. [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, C., P. Sarnow, and A. Siddiqui. 1994. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J. Virol. 68:7301-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson, J. E., T. V. Pestova, C. U. Hellen, and P. Sarnow. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 102:511-520. [DOI] [PubMed] [Google Scholar]
- 37.Yamasaki, K., C. C. Weihl, and R. P. Roos. 1999. Alternative translation initiation of Theiler's murine encephalomyelitis virus. J. Virol. 73:8519-8526. [DOI] [PMC free article] [PubMed] [Google Scholar]