Abstract
Although considerable progress has been made towards characterizing virus assembly processes, assignment of the site of tegumentation and envelopment for human cytomegalovirus (HCMV) is still not clear. In this study, we examined the envelopment of HCMV particles in human lung fibroblasts (HF) HL 411 and HL 19, human umbilical vein endothelial cells, human pulmonary arterial endothelial cells, and arterial smooth muscle cells at different time points after infection by electron microscopy (EM), immunohistochemistry, and confocal microscopy analysis. Double-immunofluorescence labeling experiments demonstrated colocalization of the HCMV glycoprotein B (gB) with the Golgi resident enzyme mannosidase II, the Golgi marker TGN (trans-Golgi network) 46, and the secretory vacuole marker Rab 3 in all cell types investigated. Final envelopment of tegumented capsids was observed at 5 days postinfection by EM, when tegumented capsids budded into subcellular compartments located in the cytoplasm, in close proximity to the Golgi apparatus. Immunogold labeling and EM analysis confirmed staining of the budding compartment with HCMV gB, Rab 3, and mannosidase II in HL 411 cells. However, the markers Rab 1, Rab 2, Rab 7, Lamp 1 (late endosomes and lysosomes), and Lamp 2 (lysosomes) neither showed specific staining of the budding compartment in the immunogold labeling experiments nor colocalized with gB in the immunofluorescent colocalization experiments in any cell type studied. Together, these results suggest that the final envelopment of HCMV particles takes place mainly into a Golgi-derived secretory vacuole destined for the plasma membrane, which may release new infectious virus particles by fusion with the plasma membrane.
Human cytomegalovirus (HCMV) is a member of the herpesvirus family, which includes members characterized by a restricted host range, the ability to establish latency, a long reproductive cycle, and a linear double-stranded DNA genome packaged within an icosahedral capsid. Packaging of viral DNA into capsids occurs within the nucleus, whereupon nucleocapsids are transported out of the nucleus to the cytoplasm by an unknown pathway. The nuclear egress of herpes simplex virus (HSV) has been suggested to involve a step of envelopment by budding through the inner leaflet of the nuclear membrane, followed by a de-envelopment when exiting through the perinuclear space (49). In the cytoplasm, CMV capsids start to sequentially become surrounded by a protein structure known as the tegument or matrix. An envelope membrane, containing several virus-specific glycoproteins, in turn covers tegumented HCMV nucleocapsids (42). This envelope consists of a lipid bilayer, which is similar in structure and composition to host cell membranes (15). Although a number of studies have examined the different steps in the HCMV assembly process, these studies have not conclusively identified the site of final envelope acquisition of the HCMV particles (17, 43-45, 47).
Most enveloped viruses, such as alpha-, orthomyxo-, paramyxo-, rhabdo-, and retroviruses, acquire their lipid envelope by budding at the plasma membrane (15). Upon this type of budding, mature virus particles are released directly into the extracellular space. In contrast, viruses that mature intracellularly bud into the lumen of intracellular compartments to acquire their final envelope (15, 56). Mature virus particles are then transported within the cell in membrane vacuoles, which upon fusion with the plasma membrane release virions extracellularly. For example, coronaviruses and poxviruses bud at the endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC) (28, 51), Bunyaviridae and rubella virus bud at the Golgi complex (36), and HSV has been suggested to bud at the inner nuclear membrane (8, 31, 55) or into cytoplasmic vacuoles (29). In the case of budding at the inner nuclear membrane, subsequent maturation of the herpesvirus particle is believed to take place during passage of virus particles through the ER (56) or the Golgi complex (24). In contrast, more recent studies on HSV morphogenesis suggest that nucleocapsids undergo a sequential envelopment and de-envelopment crossing the nuclear membrane (5, 49), and thereafter the capsid will receive its final envelope by budding into subcellular vacuoles in the cytoplasm (50). A number of studies have presented evidence of different budding sites for HCMV. For example, structural electron microscopy (EM) studies have suggested that capsids receive their final envelope by budding into early endosomes (60), or into trans-Golgi cisternae containing viral glycoproteins (27, 47). Such vacuoles containing mature virions have been suggested to release infectious virus particles by excocytosis (27). Furthermore, studies using the fungal metabolite brefeldin A have demonstrated a cytoplasmic accumulation of naked virus particles in HSV-infected (6) and HCMV-infected (10) cells. This finding supports the hypothesis that the final envelopment occurs in the cytoplasm and implies an important role of the Golgi apparatus in this process. However, the site of the final envelopment for HCMV has not been clearly defined by methods examining the origin of the subcellular compartment for budding.
Since assembly processes are believed to be initiated by specific interactions between the nucleocapsid and a cytoplasmic extension of one or more of the viral glycoproteins (36, 48), the site of virus maturation can be examined by determining the accumulation of the viral glycoproteins in the budding compartment. To study the site of budding of HCMV particles, we performed EM and immunohistochemical analysis of HCMV-infected human embryonic lung fibroblasts (HF) at different time points after infection. Structural EM analysis suggested that tegumented virus particles bud into cytoplasmic vacuoles to acquire their final envelope. Colocalization studies of HCMV glycoprotein B (gB) and different markers for specific subcellular compartments were performed with HF, human umbilical vein endothelial cells (HUVEC), human pulmonary arterial endothelial cells (HPAEC), and arterial smooth muscle cells (ASMC) and revealed colocalization of gB with the Golgi resident enzyme mannosidase II, TGN (trans-Golgi network) 46, and the Rab 3 GTPase, which is a specific marker for secretory vacuoles. Immunogold labeling and EM analysis confirmed staining of the budding compartment and the viral envelope with HCMV gB, mannosidase II, and Rab 3 in HF. In contrast, lysosomal and endosomal markers were not detected on the virus-containing vacuoles. These results suggest that HCMV buds into Golgi-derived vacuoles, which are destined for the plasma membrane.
MATERIALS AND METHODS
Cells and viruses.
HF HL 411 (kindly provided by V. Sundquist, Karolinska Institute, Stockholm, Sweden) and HL 19 were maintained in bicarbonate-free minimal essential medium with Hanks salts (GIBCO BRL, Grand Island, N.Y.) supplemented with 25 mM HEPES, 10% heat inactivated fetal calf serum, l-glutamine (2 mM), penicillin (100 U/ml), and streptomycin (100 μg/ml) (GIBCO BRL) at 37°C with 5% CO2. The HF cells were used in experiments until passage 20. The HF cells were infected with HCMV strain AD169 at a multiplicity of infection (MOI) of 1. The MOI was calculated as PFU × milliliters/number of cells. The virus supernatant was collected at 7 or 10 days postinfection (dpi), cleared from cell debris by low-speed centrifugation, and frozen at −70°C until used. Monolayers of HF grown in 96-well plates (Falcon) or on chamber slides (Nalge Nunc International) were infected with HCMV AD169 at an MOI of 1. Virus titers were determined as previously described (63).
HPAEC medium was obtained from Clonetics, San Diego, Calif. The cells were maintained in EBM-2 (Clonetics) with growth factors according to the manufacturer's instructions at 37°C with 5% CO2. At 1 h prior to infection, the medium was changed to endothelial basal medium with 5% fetal calf serum, and the cells were cultured in this medium during the infection process. The cells were cultured in 0.2% gelatin-coated 175-cm2 tissue culture flasks (Falcon) or eight-well chamber slides (Nalge Nunc International) and used until passage 8 in the experiments.
HUVEC were cultured in EGM medium (Clonetics). At 1 h prior to infection, the medium was changed to endothelial basal medium with 5% fetal calf serum, and the cells were cultured in this medium during the infection process. The cells were cultured in 175-cm2 tissue culture flasks (Falcon) and eight-well chamber slides (Nalge Nunc International) and used until passage 8. The cells were infected at an MOI of 1 with an endothelium-adapted strain, TB40, kindly provided by G. Jahn and C. Sinzger, Department of Medical Virology, University of Tübingen, Tübingen, Germany.
Smooth muscle cells were isolated from aortic grafts obtained from patients undergoing surgery for aortic aneurysm with insertion of aortic grafts or from either aortic grafts or grafts of the iliaca vessels from transplantation donors. The cells were isolated by first removing the endothelial cells by cell scraping with a sharp instrument. The vessel was then divided into two different layers, the innermost layer (corresponding to the intima of the vessel) and the outermost layer (corresponding to the vessel media), by gentle separation of the two layers with a pair of tweezers. The aortic graft was cut into 1-mm pieces and cultured on cell culture dishes in SMGM 2 medium (Clonetics) with the addition of SMGM 2 SingleQuots (human epidermal growth factor [0.5 μg/ml], 0.5 ml; insulin [5 mg/ml], 0.5 ml; human fibroblast growth factor b [1 μg/ml], 1 ml; fetal bovine serum, 25 ml; GA-1000, 0.5 ml) for about 2 weeks, until smooth muscle cells had migrated from the explants. The cells were cultured in 175-cm2 cell culture flasks (Falcon). To determine the purity of the smooth muscle cell cultures, the cells were grown on chamber slides, fixed in 3% formaldehyde for 45 min at 4°C, and permeabilized with 0.3% Triton X-100 for 5 min. After fixation, the cells were stained for smooth muscle cell-specific fluorescein isothiocyanate (FITC)-conjugated antiactin (Sigma) and evaluated by fluorescence microscopy. The cells were fixed with methanol-acetone (1:1) at 4°C for 5 min, stained with FITC-conjugated anti-von Willebrand factor and endothelial cell-specific von Willebrand factor, and evaluated by fluorescence microscopy. Isotype controls for the antibodies were used in the experiments. The cultures were 100% positive for antiactin antibody and negative for von Willebrand factor.
Antibodies.
For immunohistochemical analysis, antibodies specific for the markers Rab 1 to 6 (rabbit polyclonal immunoglobulin G), Rab 7, Lamp 1, and Lamp 2 (goat polyclonal immunoglobulin G), purchased from Santa Cruz Biotechnology, Santa Cruz, Calif., were used as 1:100 dilutions (1:10 dilutions in the immunogold experiments) from stocks of 200 μg/ml. TGN 46 (sheep anti-human antibody; Serotec, Oxford, United Kingdom) was detected with FITC-labeled rabbit anti-sheep antibody (Dako A/S, Glostrup, Denmark). Polyclonal mouse anti-human antibody directed against mannosidase II was kindly provided by Kelley Moreman and Marylin Farquhar, The University of Georgia, Athens. These antibodies were used in colocalization experiments with mouse or human anti-HCMV gB and detected with FITC- or rhodamine-phalloidin-conjugated swine, goat, or mouse F(ab)′2 fragments against rabbit, goat, or mouse Ig, respectively; FITC-conjugated rabbit anti-mouse immunoglobulin; FITC-conjugated goat F(ab)′2 anti-mouse antibody (Dako A/S); or phycoerythrin-conjugated goat F(ab)′2 fragments against human immunoglobulin (Southern Biotechnology Associates Inc., Birmingham, Ala.). Secondary antibodies were diluted in DAKO antibody diluent (DAKO A/S) and used as 1:100 dilutions. The following mouse monoclonal antibodies (MAbs), produced by William Britt, were used for CMV protein localization studies with immunofluorescence detection at 1:100 dilutions: UL99 (pp28), UL83 (pp65), UL82 (pp71), UL32 (pp150), UL55 (gB), UL75 (gH), and UL115 (gL) (44, 45).
Immunofluorescence staining of HCMV-infected cells.
Monolayers of HF, HUVEC, HPAEC, or ASMC grown in 24- to 96-well tissue culture plates or on 8-well chamber slides (Nalge Nunc International) were infected with HCMV at an MOI of 1 and fixed at 1, 3, 5, 7, or 9 dpi. Uninfected and infected cells were washed in phosphate-buffered saline, fixed in either ice-cold methanol-acetone or 3% paraformaldehyde for 15 min at room temperature (RT), and permeabilized by 0.3% Triton X-100 treatment for 5 min. Samples were incubated with Serum-Free Protein Block (DAKO A/S) for 10 min at RT before being washed and stained with antibodies diluted in antibody diluent (DAKO A/S) or appropriate isotype controls. Both infected and uninfected cells were used in control experiments. Samples were mounted on glass slides by using fluorescent mounting medium containing 15 mM NaN3 (DAKO A/S). The secondary antibodies were made in the same species when two different antibodies were stained for colocalization experiments. For control purposes, the secondary antibody alone or cells treated with human, mouse, rabbit, or goat serum (DAKO A/S) (incubation in 1 to 3% serum for 15 min at RT) were stained for false fluorescence and autofluorescence (same control with both infected and noninfected cells). 4,6-Diamide-2-phenylindol (Sigma-Aldrich, St. Louis, Mo.) at 4 ng/ml was used to stain the nucleus blue.
Fluorescence microscopy and image analysis.
Fluorescence microscopy analysis was performed with a DMRXA microscope (Leica Microsystems Gmbh, Wetzlar, Germany) equipped with a cooled charge-coupled device camera (model S/N 370 KL 0565; Cooke Corporation, Auburn Hills, Mich.). Filter sets for 4,6-diamide-2-phenylindol-Hoechst, FITC, Cy3, and Cy5 were obtained from Chroma Technology (Brattleboro, Vt.). The images were acquired and analyzed with the image processing software SlideBook 2.1.5 (Intelligent Imaging Innovations Inc., Denver, Colo.). Images were postprocessed and mounted in Photoshop 5.0 (Adobe Systems Inc., San Jose, Calif.) and printed on a Tektronix Phaser 860DP color printer (Xerox, Stamford, Conn.) or on a DS 8650 printer (Kodak, Rochester, N.Y.).
EM analysis. (i) Tissue preparation for structural EM analysis.
To examine virus-infected cells (MOI of 1) by EM, uninfected and HCMV-infected cells were harvested at 1, 3, 5, 7, and 10 dpi and fixed in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer-0.1 M sucrose with 3 mM CaCl2 (pH 7.4) at 4°C for 20 h. Fixed cells were collected by scraping with a wooden peg and pelleted in Eppendorf tubes. The pellets were washed in 0.15 M sodium cacodylate containing 3 mM CaCl2 (pH 7.4). Specimens were postfixed in 2% osmium tetroxide in 0.1 M cacodylate buffer containing 3 mM CaCl2 for 1 h at 4°C, dehydrated in ethanol followed by acetone, and embedded in LX-112. Sections were cut, placed on Formvar-coated copper grids, and stained with uranyl acetate followed by lead citrate. Sections were examined in a calibrated Philips 420 electron microscope at 80 kV.
(ii) Immunohistochemical gold labeling of HCMV-infected cells.
Monolayers of uninfected and HCMV-infected HF were washed with phosphate-buffered saline and fixed in 3% paraformaldehyde-0.1% glutaraldehyde in 0.1 M phosphate buffer (PB) (pH 7.2) for 30 min at RT. Following fixation, cells were scraped with a wooden peg and pelleted in Eppendorf tubes. The pellet was infiltrated with 4% gelatin in 0.1 M phosphate buffer (pH 7.4) for 1 h, followed by fixation in the same fixative for 1 h at RT. The specimens were thereafter infiltrated with 2.3 M sucrose and frozen in liquid nitrogen. Sectioning was performed in an ultracryotome (Leica UCT) at −90°C by the method previously described by Tokuyasu (59). The sections were washed in 0.1 M PB, blocked with 10% normal goat serum for 10 min, and incubated with the MAbs at same dilutions as described above overnight at 4°C. Sections were rinsed in 0.1 M PB plus 0.1% normal goat serum. Bound antibodies were detected by secondary goat anti-mouse antibodies conjugated with 10-nm-diameter gold particles. After an additional washing step with 0.1 M PB, the specimen was fixed in 2% glutaraldehyde for 30 min and washed with distilled water four times for 1 min each, followed by final contrasting in 0.1 M uranyl acetate for 45 min.
(iii) Quantification of immunogold labeling.
Gold beads are the number of gold particles (designated Au) counted on a selected micrograph or on a certain structure seen in the micrograph, such as the envelope membrane of the virion, dense bodies, or vacuoles. The area of the micrograph where gold staining was counted was measured in square micrometers. Background staining is given as Au per square micrometer. The quantification is based on the comparison between the parameters Au per square micrometer on virus particles, Au per square micrometer on dense bodies, and Au per square micrometer on virus-containing vacuoles in relation to the background staining. Several antibodies (e.g., Lamp 1, Lamp 2, Rab 4, and Rab 7) were not suitable or optimal for use in gold labeling analysis due to instability of the antibodies.
RESULTS
Structural EM analysis of HCMV-infected HF.
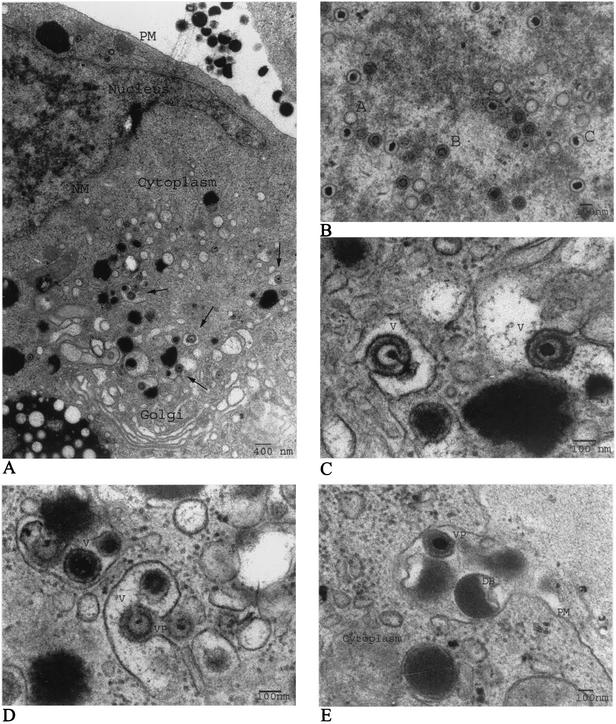
The mechanism and subcellular site of final envelopment of HCMV have previously not been well defined. In this study, we performed EM and immunohistochemical analysis with HCMV-infected HF cells, at 1, 3, 5, 7, and 10 dpi as our model system, and in addition, we examined HCMV-infected HUVEC, HPAEC, and ASMC harvested at 5, 7, and 9 dpi. As has previously been reported, we observed the formation of three different capsid particles in the nuclei of infected HF starting at 3 dpi, i.e., A capsids (empty capsids without DNA), B capsids (capsids with a translucent core), and C capsids (nucleocapsids containing DNA), all of which were approximately 100 nm in diameter (Fig. 1A and B and data not shown). DNA appeared to be encapsulated into empty A capsids in the nucleus, in accordance with the previous suggestion that A, B, and C capsids might reflect different maturity stages of the capsid (Fig. 1B). The number of nucleocapsids observed in the nucleus increased significantly from 5 dpi. Whether this accumulation of nucleocapsids was due to high production or a limited release of the nucleocapsids from the nucleus remains to be investigated. Nontegumented enveloped virus particles (NTEV) undergoing de-envelopment were observed at the rim of the nuclear membrane (Fig. 2I), which further supports the same mechanism of action for the nuclear exit of HCMV nucleocapsids as for HSV nucleocapsids, by envelopment and de-envelopment when traversing the nuclear membrane. All three capsid forms were observed to exit the nucleus by the same mechanism (Fig. 2I). NTEV were observed in close association with the nuclear membrane in HL 411, HL 19, and HUVEC (data not shown). In the cytoplasm, an amorphous material, the tegument, surrounded nonenveloped A, B, and C capsids, giving the tegumented capsids a diameter of approximately 130 nm (Fig. 2A and J and data not shown). The amorphous structure, or the tegument, was never observed around capsids present in the nucleus. Furthermore, the majority of the tegumented capsids present in the cytoplasm were observed in cytoplasmic clusters, surrounding vacuole-like structures (Fig. 2A and J). Although tegumented A, B, and C capsids can be seen in the cytoplasm, the vast majority of capsids surrounding the cytoplasmic clusters (Fig. 2A and J) were of the C type. The acquisition of tegument proteins around capsids seems to be a rapid process following nuclear exit, since only fully tegumented capsids were observed in the cytoplasm, with the exception of NTEV that appeared to be in the process of loosing a possibly temporal nuclear membrane envelope.
FIG. 1.
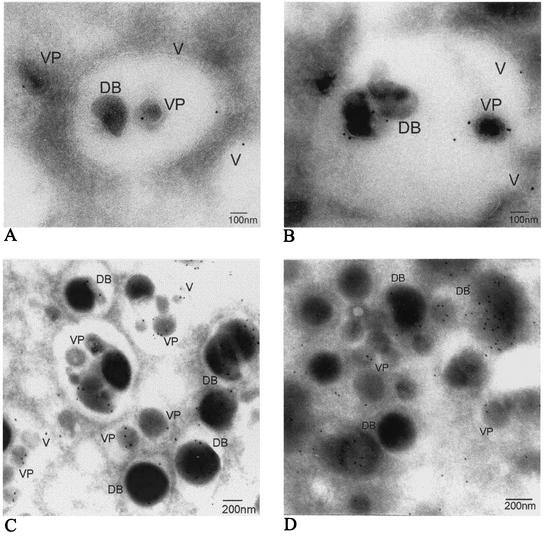
HCMV buds into cytoplasmic vacuoles. Structural EM was performed on HCMV-infected human lung fibroblasts at 5 dpi. An overview of the HCMV replication cycle at 5 dpi in an HCMV-infected HF is shown. (A) Virus particles undergoing envelopment in cytoplasmic vacuoles are indicated by arrows. PM, plasma membrane; NM, nuclear membrane. (B) Three morphologically different capsids, A, B, and C, are formed in the nucleus. (C and D) Tegumented capsids bud into cytoplasmic vacuoles (V) to acquire their envelope. (E) Mature virus particles (VP) and dense bodies (DB) exit the cell by fusion of the transporting vacuole and the plasma membrane to release its content to the extracellular space.
FIG. 2.
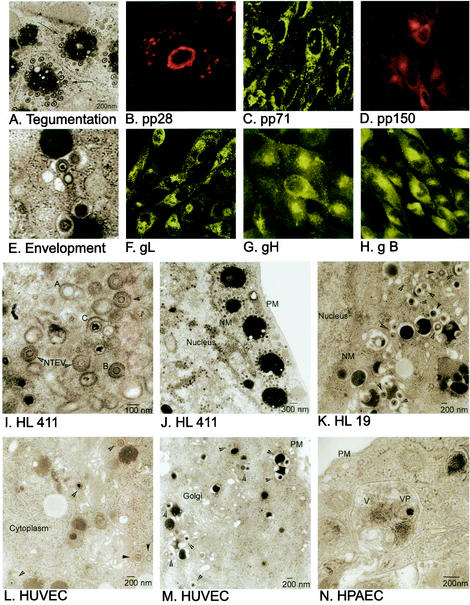
Immunofluorescent staining localizing HCMV protein expression in HCMV-infected HF at 5 dpi. The tegument proteins pp28 (B), pp71 (C), and pp150 (D) are found in the cytoplasm and in high concentrations around the nuclear membrane (pp28 in panel B) and in the Golgi region (pp150 in panel D). A possible cytoplasmic site of tegument protein acquisition is showed in the EM micrographs (A and J), where tegumented capsids accumulate around structures of dense stained material. The viral glycoproteins gL (F), gH (G), and gB (H) are expressed in the Golgi apparatus and in punctuate structures in the cytoplasm. Notable is the dot-like staining underneath the plasma membrane and in the cytoplasm close to the Golgi apparatus. Naked A, B, and C capsids that have traversed the nuclear membrane lose the acquired membrane when entering the cytoplasm (I). The different cell types HL 411 (E), HL 19 (K), HUVEC (L), and HPAEC (N) showed the same mechanism of envelopment by budding into cytoplasmic vacuoles. PM, plasma membrane; NM, nuclear membrane.
HCMV acquires its envelope by budding into cytoplasmic vacuoles in close proximity to the Golgi apparatus.
Abundant large intracellular vacuoles containing one or several virus particles were observed in the cytoplasm of infected HL 411 (Fig. 1C and D), HL 19 (Fig. 2K), HUVEC (Fig. 2L), and HPAEC (Fig. 2N) after 5 dpi by EM analysis. The final envelopment of tegumented capsids appeared to take place through budding into vacuoles of different sizes, which were formed in close proximity to the Golgi apparatus (Fig. 1A and Fig. 2M). The cytoplasmic vacuoles ranged in diameter from 300 to 600 nm, with an average diameter of approximately 400 nm, and were not observed in uninfected cells (data not shown). The vacuoles appeared to fuse with the plasma membrane and released virions to the extracellular space (HF in Fig. 1E and HPAEC in Fig. 2N). Virus particles were observed neither within the Golgi apparatus itself nor in association with the nuclear membrane. Tegumented capsids were never observed in the nucleus. Furthermore, dense bodies seemed to be formed in a similar way in HCMV-infected HF, where an amorphous electron-dense material became enveloped to form dense bodies of different sizes (200 to 500 nm) through budding into the same type of vacuoles (Fig. 1D and E). These observations suggest that the membranes of dense bodies and enveloped virus might have the same composition.
HCMV tegument proteins and glycoproteins localize to cytoplasmic compartments in close proximity to the Golgi apparatus.
To localize the site of viral capsid tegument acquisition in the infected cells, MAbs reactive with the tegument proteins pp28, pp65, pp71, and pp150 were used to stain HCMV-infected HF, HUVEC, HPAEC, and ASMC. In these experiments, the viral tegument protein pp28 was present mainly in the nuclear membrane and in large vacuole compartments in the cytoplasm, which may correspond to the large sites of accumulation of electron-dense material (500 to 2,000 nm) that were observed in the cytoplasm by EM analysis (Fig. 2A). These round structures of large electron-dense material were generally surrounded by numerous tegumented capsids (Fig. 2A). While the pp71 protein was strongly localized to the nuclear membrane (Fig. 2C), the pp150 protein was present mainly in the cytoplasm, with a high concentration in the Golgi region as well as in punctuate structures in close association with the cell nucleus and the Golgi apparatus (Fig. 2D). While pp28, pp71, and pp150 were detected exclusively in the cytoplasm, pp65 was observed in the nucleus at 1 and 3 dpi and in both the nucleus and the cytoplasm at 5 and 7 dpi (Fig. 2 and data not shown). In contrast to the previously reported presence of the HCMV tegument proteins pp28, pp71, and pp150 in the nucleus (22), we did not observe either pp28, pp71, or pp150 in the nucleus at any time point examined between 1 and 9 dpi. Further confocal microscopy analyses of the cellular localization of the HCMV envelope glycoproteins gB, gH, and gL demonstrated a staining pattern similar to that of the tegument proteins pp71 and pp150 in the cytoplasm, with a high-intensity staining pattern around, and in, the Golgi region. In addition, the envelope glycoproteins gB, gH, and gL were present in small vacuoles around the Golgi apparatus and underneath the plasma membrane, which may correspond to the small (300- to 600-nm) virus-containing vacuoles observed by EM analysis (Fig. 1C, D, and E and 2E). Furthermore, while antibodies specific for gH and gL stained the nuclear membrane at early times in infection (Fig. 2F and G), gB was only rarely detected at the nuclear membrane (Fig. 2H). Thus, these results suggest that the tegumentation and envelopment appear to be separate processes.
Confocal microscopy analyses demonstrate colocalization of gB with Rab 3, TGN 46, and mannosidase II.
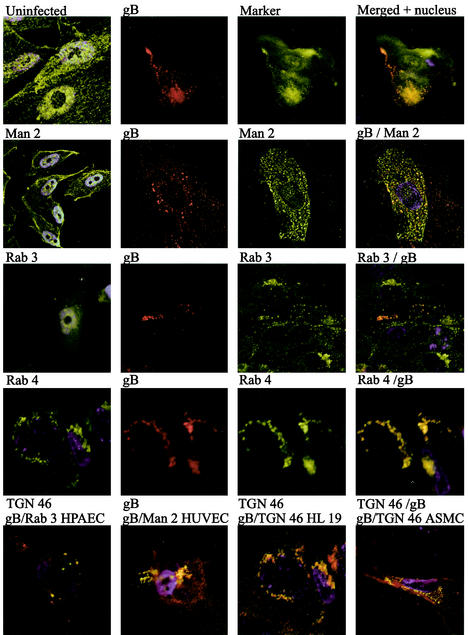
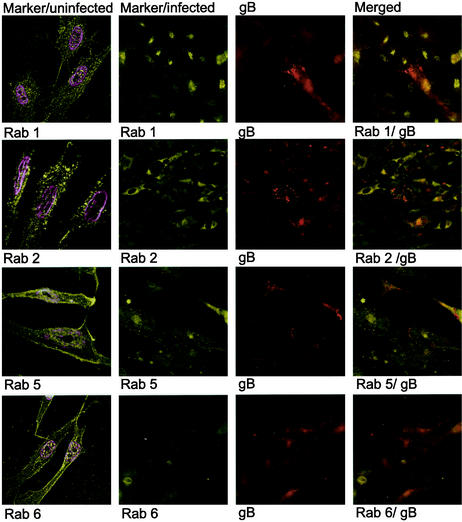
To identify the site of intracellular accumulation of the major HCMV envelope glycoprotein gB by using specific intracellular markers, we performed colocalization experiments with antibodies directed against HCMV gB as well as markers for the following specific subcellular compartments: Rab 1 (ER and Golgi complex), Rab 2 (transitional ER and cis-Golgi network), Rab 3 (secretory vacuoles), Rab 4 (early endosomes) Rab 5 (early endosomes), Rab 6 (medial- and trans-Golgi cisternae), TGN 46 (trans-Golgi network), and mannosidase II (medial Golgi) (Table 1). The Golgi resident enzyme mannosidase II colocalized with HCMV gB in cytoplasmic vacuoles, in close association with the plasma membrane (HUVEC and HL 411 in Fig. 3) Furthermore, Rab 3 also demonstrated a strong colocalization with gB in cytoplasmic vacuoles (HL 411 and HPAEC in Fig. 3). In contrast, Rab 1, 2, 5, and 6 rarely colocalized with gB in the infected cells (HL 411 in Fig. 4). Similar results were obtained with HUVEC (data not shown). To further examine whether the site of accumulation of viral proteins coincided with a site where virus particles could be degraded, we performed colocalization experiments with antibodies directed against gB and the following markers: Rab 7 (late endosomes), Lamp I (late endosomes and lysosomes), and Lamp II (lysosomes). The HCMV gB-positive vacuole compartments were negative for all of these markers, which suggests that the cellular compartments containing enveloped virus particles do not enter a degradation pathway (data not shown). As expected, all markers tested showed a weak colocalization signal with gB in the Golgi region, most likely due to their common pathway of production. However, colocalization of gB and either Rab 1, Rab 2, Rab 4, Rab 5, Rab 6, Lamp I, or Lamp II was never observed in any distinct cytoplasmic compartment at any time point during infection (Fig. 4 and data not shown). Antibodies directed against Lamp 1 and Lamp 2 showed background staining in the immunofluorescence detection experiments.
TABLE 1.
Antibody markers and their intracellular targetsa
| Marker | Intracellular compartment (reference) |
|---|---|
| Rab 1 | ER-Golgi (46) |
| Rab 2 | Trans-ER (46) |
| Rab 3 | Secretory vacuoles (46) |
| Rab 4 | Early endosomes (46) |
| Rab 5 | Early endosomes, plasma membrane (46) |
| Rab 6 | Medial- and trans-Golgi cisternae (46) |
| Rab 7 | Late endosomes (46) |
| Mannosidase II | Medial Golgi (11, 62) |
| TGN 46 | Golgi (61) |
| Lamp 1 | Late endosomes (44) |
| Lamp 2 | Lysosomes (44) |
The intracellular markers were used in colocalization studies with gB. Rab proteins (46) are considered directors of vacuole trafficking. Mannosidase 2 is a Golgi resident enzyme, and TGN 46 is a marker for the trans-Golgi compartment.
FIG. 3.
Colocalization of gB (red) with the subcellular markers (green) mannosidase II (Man 2), Rab 3, Rab 4, and TGN 46 in HL 411. Colocalization was performed to identify the intracellular site of gB accumulation. Mannosidase II, Rab 3, and TGN 46 showed clear colocalizationwith gB in distinct vacuole compartments in the cytoplasm and in the Golgi region. The cytoplasmic, punctuate colocalization staining pattern of Rab 3, TGN 46, and mannosidase II is enlarged to show the distribution of cytoplasmic vacuoles (yellow dots). Rab 4 colocalized with gB to a lesser extent, but only in compartments next to the nuclear membrane or the Golgi stack. Uninfected cells stained with the same markers are shown in the left columns. The colocalization of Rab 3, mannosidase II, and TGN 46 with gB was confirmed in HPAEC, HUVEC, HL 19, and ASMC (bottom panels).
FIG. 4.
Colocalization of gB and the markers Rab 1, Rab 2, Rab 5, and Rab 6 in HCMV-infected HF cells at 5 dpi. Colocalization was performed to identify the intracellular site of gB accumulation. Rab 1, Rab 2, Rab 5, and Rab 6 did not show colocalization with gB. The four markers demonstrated a weak colocalization with gB in the Golgi apparatus but not in any intracellular compartment.
Immunogold EM analyses confirm the presence of gB, Rab 3, and mannosidase II on HCMV-containing vacuoles.
Although the confocal microscopy experiments show colocalization of viral and cellular proteins, these experiments cannot identify which vacuoles contain virus particles. The fact that gB colocalizes with Rab3, TGN 46, and mannosidase II implies that HCMV utilizes the secretory vacuoles for its exit. To identify the origin of the virus-containing cytoplasmic vacuoles and to confirm the presence of virus and dense bodies in the same compartments, we performed immunogold labeling experiments with HCMV-infected HF and the specific subcellular markers described in Table 1. MAbs specific for gB and the proteins mannosidase II, gB, Rab 1, Rab 2, Rab 3, Rab 4, Rab 5, Rab 6, Rab 7, Lamp I, and Lamp II were used to stain ultrathin cryosections of LX-112-embedded HCMV-infected HF. Immunohistochemical gold labeling experiments confirmed the presence of gB (Fig. 5A), mannosidase II (Fig. 5B), and Rab 3 (Fig. 5C) on the virus-containing vacuoles, as well as on virus particles and dense bodies. While the Rab 5 marker was present on the membrane of the virus-containing vacuole and on dense bodies (Fig. 5D), Rab 6 often stained the vacuole compartment but only occasionally stained the virus envelope membrane. In contrast, Rab 1, Rab 2, Rab 4, Rab 7, Lamp I, and Lamp II did not specifically stain the HCMV envelope or the vacuole compartment at any time point during the virus life cycle. The quantification of gold particles on the different membranes is summarized in Table 2. Due to the low statistical probability of capturing several gold grains on the same two-dimensional surface on the electron micrograph when staining a three-dimensional 3D structure (vacuole compartment or virus envelope membrane), the value for Au per square micrometer is highly significant when it is higher than 10. This value is the number of gold grains (Au) per square micrometer captured on a two-dimensional level. Although Rab 3 is recycled between the plasma membrane and intracellular membranes, it was found to be highly specific in its binding to the virus membrane, the vacuole compartments containing enveloped virus particles, and dense bodies (Fig. 5C).
FIG. 5.
Immunogold localization of subcellular markers in HCMV-infected HF. Immunogold labeling was performed to identify the vacuole compartment detected by confocal microscopy. Secondary antibodies marked with colloidal gold were used to visualize the staining of gB (A), mannosidase II (B) Rab 3 (C), and Rab 5 (D). The HCMV gB, Rab 3, and mannosidase II antibodies stained the envelope membrane of virus particles (VP), dense bodies (DB), and the vacuole membrane (V).
TABLE 2.
Statistical analysis of the immunogold labeling experimentsa
| Subcellular marker | Colocalization with gB | Au/μm2 (mean ± SD)
|
|||
|---|---|---|---|---|---|
| Background | Virus | Dense bodies | Vacuoles | ||
| Rab 2 | − | 17 ± 2 | 25 | 5 ± 1 | 0 |
| Rab 3 | +++ (cytoplasm) | 5 ± 2 | 48 ± 4 | 22 ± 2 | 5 ± 2 |
| Rab 5 | + (Golgi) | 6 ± 2 | 15 ± 2 | 43 ± 6 | 22 ± 2 |
| Rab 6 | + (Golgi) | 13 ± 2 | 5 ± 3 | 5 ± 1 | 67 ± 2 |
| gB | 5 ± 2 | 75 ± 7 | 9 ± 2 | 10 ± 2 | |
Specific binding of gold beads compared to background staining (see text).
DISCUSSION
The details of the cytoplasmic events during HCMV morphogenesis have been a matter of controversy. In this study, we present descriptive new evidence which suggest that the major site of HCMV envelopment in fibroblasts, endothelial cells, and smooth muscle cells is in virally induced cytoplasmic vacuoles rather than at the nuclear membrane. We utilized a combination of EM, immunohistochemistry, and immunofluorescent protein localization techniques to determine the subcellular origin of these virus-containing cytoplasmic vacuoles in these cell types and obtained similar results regarding protein localization and possible assembly pathway for the virus. The structural EM analysis confirmed the previous proposed hypothesis that the envelopment of tegumented HCMV capsids occurs into cytoplasmic vacuoles (44, 50, 60). Using protein localization techniques, we showed that while the tegument proteins pp150 and pp71 preferably localized to the ERGIC, the glycoproteins gB, gH, and gL were present both in the ERGIC and in small cytoplasmic vacuoles in close proximity to the plasma membrane. Further confocal microscopy analyzes showed that HCMV gB colocalized with mannosidase II and Rab 3, and immunogold labeling experiments confirmed that intracellular gB was detected on vacuoles containing both virus particles and dense bodies. These vacuoles stained positive for Rab 3 and mannosidase II, which suggests that the vacuoles are derived from the Golgi compartment and are entering Rab 3 secretory pathways. Furthermore, since the vacuoles were negative for the endosome and lysosome markers Rab 7, Lamp 1, and Lamp 2, these observations eliminate the possibility that virus-containing vacuoles are entering default pathways by fusing with endosomes or lysosomes for degradation. These results suggest that HCMV acquires its final envelope by budding into virally induced cytoplasmic vacuoles, which are derived from the Golgi apparatus and destined for the plasma membrane apparently without entering endosomal and lysosomal degradation pathways.
A number of investigations on herpesvirus assembly and egress have suggested two hypotheses for herpesvirus assembly (for reviews, see references 29 and 57). The earliest studies suggested that the final envelopment of the virion takes place at the inner nuclear membrane (7, 8). This hypothesis was expanded further to include modifications of the enveloped particle as it traversed and matured inside the ER-Golgi network compartments (9, 25). Thereafter, studies of the assembly process suggested that the envelopment of capsids occurred by passage though the inner leaflet of the nuclear membrane followed by a de-envelopment of capsids at the outer nuclear membrane. Finally, envelopment takes place by budding into cytoplasmic compartments (5, 41, 49). More recent models suggest that the final envelopment of the tegumented capsid occurs in the TGN by budding into vacuoles containing membranes with a high concentration of processed viral envelope glycoproteins (16, 18, 19, 32, 40, 65, 66). Our findings fit well with the latter hypothesis and extend the model to include a method for the virus to exit different infected cell types.
The tegument contains several proteins which form the characteristic proteinaceous material that surrounds the icosahedral herpesvirus capsids. Although the number of proteins and the size of the tegument seem to differ between the herpesviruses HSV (29), varicella-zoster virus (VZV) (54), and HCMV (17), the tegument proteins are expressed predominantly in the cytoplasm, where most of tegument acquisition likely takes place. Furthermore, EM analyses of the tegument acquisition for human herpesvirus 6 (HHV-6) suggest that this process takes place in close connection to the envelopment and de-envelopment processes when capsids are passing through the nuclear membrane in compartments termed tegusomes (41). Thereafter, the tegumented capsids have been implied to bud into cytoplasmic vacuoles to achieve their final envelope (41). In a number of studies examining HCMV tegument acquisition (43-45), the evidence suggests that virion tegument and envelope proteins accumulate in stable cytoplasmic compartments. These compartments express markers for the ERGIC, which is a dynamic compartment of the secretory pathway in close connection to both the ER and the Golgi apparatus. Here, we also localized the tegument proteins pp28, pp65, pp71, and pp150 to the ER and Golgi apparatus, in accordance with previous observations in HF, HUVEC, HPAEC, and ASMC. Since the tegument is located underneath the envelope in mature virus particles, the process of tegument acquisition has to take place earlier than the envelopment. We here observed an important difference between the accumulated expression of cytoplasmic tegument proteins and the cytoplasmic gathering of the viral glycoproteins in vacuoles around the Golgi apparatus and adjacent to the plasma membrane. Viral tegument proteins did not show punctuate staining closer to the plasma membrane but were limited to regions around the Golgi apparatus.
Our EM analyses showed that tegumented nucleocapsids were inserted into vacuole cavities by curving of the vacuole membrane, which then seems to wrap around the capsid in the budding process. The original concave face of the vacuole forms the viral envelope, and mature virions accumulate inside the vacuole compartment. These vacuoles were positive for gB, the Golgi marker mannosidase II, and Rab 3 and contained virus particles and dense bodies. Cytoplasmic vacuole compartments have previously been described for HHV-6 (41), VZV (26), and HCMV (50) but have not been characterized by staining for intracellular markers (39). Here, we observed an apparent increase in the number of vacuoles in HCMV-infected cells compared to uninfected cells by comparing the expression of intracellular markers in HCMV-infected compared to uninfected cells (data not shown). We further observed that the highly abundant large budding compartments for HCMV were not present in uninfected cells. Similar data that suggest that these vacuoles are virus induced have been reported for VZV (26). Interestingly, one study which examined the role of the PrV protein UL20 suggested an important role of this protein in the egress of vacuoles containing virus particles. Infection of Vero cells with a deletion mutant of PrV UL20 resulted in retention of enveloped virus particles in cytoplasmic vacuoles (13). Furthermore, infection of cells with a deletion mutant of the HSV type 1 UL20 gene results in an accumulation of virions in the perinuclear space, suggesting a role of this protein in nuclear egress (2). The HSV type 1 UL20 membrane protein has also been demonstrated to interfere with the exocytosis of virions and viral membrane glycoproteins at late times of infection (1). In addition, deletion of the PrV UL3.5 gene blocks the tegumentation and envelopment of cytoplasmic PrV capsids (14). However, it is presently unknown which proteins are responsible for the budding and egress of HCMV.
In contrast to our observations, a number of investigators have suggested that nuclear capsids bud through the inner nuclear membrane to acquire their final envelope. However, a final envelopment at the nuclear membrane is impossible in this case, since the capsid tegument acquisition must take place in the cytoplasm where most tegument proteins are accumulating. A consensus is now being developed that herpesviruses appears to exit the host cell by the pathway of envelopment, de-envelopment, and reenvelopment, based on studies mainly examining HSV and pseudorabies virus (29). Our data further substantiate this hypothesis, since we provide similar descriptive evidence for HCMV assembly. We suggest that the final viral envelope originates from the Golgi apparatus, as demonstrated by the detection of mannosidase II and TGN 46 on the budding compartment as well as on virus particles. The presumed involvement of the Golgi complex in HCMV morphogenesis is also supported by previous experiments using the fungal metabolite brefeldin A (10). Treatment of cells with this drug results in disintegration of the Golgi complex, and virus-infected cells treated with brefeldin A accumulate nonenveloped capsids in the cytoplasm. Our study presents the first evidence of the presence of Golgi-specific markers on intracellular virus-containing vacuoles. Similar to the case for HCMV, other members of the herpesvirus family, such as HHV-6 and VZV, as well as other viruses, including bunyaviruses and coronaviruses, have also been suggested to utilize Golgi-derived compartments for the final envelopment (reviewed in reference 26).
The prerequisite for budding to take place is the accumulation of glycoproteins in the budding compartment. A number of studies of intracellular trafficking of herpesvirus envelope glycoproteins have demonstrated the retrieval or endocytosis of viral glycoproteins from the plasma membrane into the budding compartment (4). HCMV gB is the major glycoprotein present on HCMV particles, and the protein is essential for production of new viral progeny. A previous study has found that the majority of infectious intracellular virus is contained within gB-coated vacuoles (11). Studies of the intracellular transport of gB initially proposed that endocytosis brings viral gB from the plasma membrane to a trans-Golgi vesicular compartment where they are used for envelopment (38). However, later studies examining the retrieval of HCMV gB in astrocytoma cells (12, 23), a mutant PrV gB (33), or PrV gE (58) have shown that endocytosis is not required for virion incorporation of these glycoproteins into the final envelope. Interestingly, not only viral proteins are present in the virus envelope. We and others have found that host cell-derived proteins such as CD13 (52), β2-microglobulin (20), annexin II (64), a 45-kDa actin-related protein (3), and CD55 and CD59 (53) are present in the viral envelope. The reason why the virus incorporates host cell-derived proteins in the envelope is unknown, but evidence suggest a role of some of these proteins in the ability of the virus to avoid immune recognition (21).
Viral glycoproteins are processed through the Golgi network, where they are segregated into specific transport vacuoles and dispatched to their final destinations, i.e., the plasma membrane, secretory vacuoles, endosomes, or lysosomes (61). The secretory pathway of eukaryotic cells consists of a series of compartments that are interconnected by vesicular transport steps. At present, three groups of intracellular vacuole coats have been identified: membranes containing clathrin and plasma membrane-specific adaptation molecules that mediate endocytosis, membranes with TGN-specific adaptins that mediate transport from the TGN to late endosomes, and coatamer protein complex-containing coats found on Golgi-derived transport vacuoles (35). In this study, we identified the Golgi resident enzyme mannosidase II on the virus-containing vacuoles. Mannosidase II is present in the Golgi stack and processes mannoses on oligosaccharides to form a complex of oligosaccharides that are resistant to attacks by endoglycosidase H. TGN 46 is a trans-Golgi marker that has been indicated to play a pivotal role in directing HCMV glycoproteins in the secretory pathway (61). We further identified Rab 3 on the virus-containing vacuole, which suggest that the vacuole may enter a secretory pathway (37, 46). The Rab GTPases, which belong to a family of Ras-like enzymes, play key roles in the secretory pathway and membrane trafficking and have been used to identify specific vacuole membranes (34). Rab 3 is a marker for secretory vacuoles that are shuttled between the plasma membrane and the trans-Golgi compartment and would therefore be optimal for the virus to utilize for transport out of the infected cell. In support of this hypothesis, Rab 3D cycles between TGN 46-positive membranes and the plasma membrane (61). A study on HSV type 1 in rat dorsal root ganglion neurons identified the Golgi markers giantin, mannosidase II, and TGN 38 on cytoplasmic vacuoles, intracytoplasmic virions, and extracellular virions (30), further strengthening the hypothesis of a trans-Golgi origin of the cytoplasmic vacuoles utilized for HCMV envelopment.
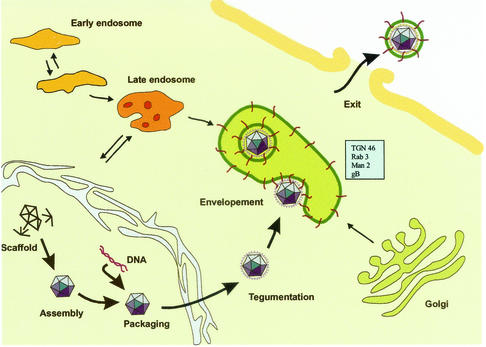
In summary, our observations suggest the following model for HCMV assembly in fibroblasts, endothelial cells, and smooth muscle cells (Fig. 6). Naked nucleocapsids, formed in the nucleus, traverse the nuclear membrane and become tegumented through the exit pathway by an unknown sequential process. Viral glycoproteins accumulate in Rab 3-, TGN 46-, and mannosidase II-positive Golgi-derived vacuoles. The tegumented nucleocapsids become enveloped by budding into the gB-, Rab3-, TGN 46-, and mannosidase II-positive vacuoles to acquire their final envelope. The vacuoles are then transported to the plasma membrane in Rab 3-positive secretory vacuoles, and virus particles are subsequently released from the cells by fusion of the vacuoles with the plasma membrane. Further molecular studies have to be performed to reveal the molecular events that govern the budding process of tegumented HCMV capsids and identify the specific protein-protein interactions involved in the envelopment process.
FIG. 6.
Model of the HCMV envelopment and exit pathway. Naked nucleocapsids, formed in the nucleus, traverse the nuclear membrane and become tegumented through the exit pathway by an unknown process. Viral glycoproteins are released from the Golgi apparatus within small transport vacuoles. Fusion of these vacuoles results in the formation of large gB-, mannosidase II-, TGN 46-, and Rab 3-positive cytoplasmic vacuoles. The tegumented HCMV capsids become enveloped when budding into the vacuoles, acquiring the vacuole membrane as their envelope. The vacuoles are then transported to the plasma membrane, and virus particles are subsequently released upon fusion of the vacuoles with the cellular plasma membrane.
Acknowledgments
We thank Ingrid Jusinski for skillful assistance in EM sample preparation and sectioning, Birger Kristensson for kindly sharing his confocal microscopy equipment, and Lars Branden for assistance with the confocal microscopy analysis. We thank Erna Möller and Jay Nelson for helpful discussions and Erna Möller for carefully reading the manuscript. We thank Kelley Moreman and Marylin Farquhar, The University of Georgia, Athens, for providing the antibody against mannosidase II; G. Jahn and C Sinzger, Department of Medical Virology, University of Tubingen, Tubingen, Germany, for providing the endothelium-adapted strain TB40; and Sara Gredmark, Department of Medicine, Karolinska Institutet, Stockholm, Sweden, for providing the ASMC.
This work was supported by grants from the Swedish Medical Research Council (12615-01A, 00793-36B, and K2001-16X-12615-04A), the Tobias Foundation (1313/98 and 33/02), the Swedish Children's Cancer Foundation (1998/065 and 01/046), the Swedish Society of Medicine (1999-02-0347), the Heart and Lung Foundation (1999-41-305), and the Emil and Wera Cornells Foundation. C.S.-N. is a fellow of the Wenner-Gren Foundation, Sweden.
REFERENCES
- 1.Avitabile, E., P. L. Ward, C. Di Lazzaro, M. R. Torrisi, B. Roizman, and G. Campadelli-Fiume. 1994. The herpes simplex virus UL20 protein compensates for the differential disruption of exocytosis of virions and viral membrane glycoproteins associated with fragmentation of the Golgi apparatus. J. Virol. 68:7397-7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines, J. D., P. L. Ward, G. Campadelli-Fiume, and B. Roizman. 1991. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J. Virol. 65:6414-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldick, C. J., Jr., and T. Shenk. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brideau, A. D., L. W. Enquist, and R. S. Tirabassi. 2000. The role of virion membrane protein endocytosis in the herpesvirus life cycle. J. Clin. Virol. 17:69-82. [DOI] [PubMed] [Google Scholar]
- 5.Browne, H., S. Bell, T. Minson, and D. W. Wilson. 1996. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J. Virol. 70:4311-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, P., B. W. Banfield, and F. Tufaro. 1991. Brefeldin A arrests the maturation and egress of herpes simplex virus particles during infection. J. Virol. 65:1893-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darlington, R. W., and L. H. Moss III. 1969. The envelope of Herpesvirus. Prog. Med. Virol. 11:16-45. [PubMed] [Google Scholar]
- 8.Darlington, R. W., and L. H. Moss III. 1968. Herpesvirus envelopment. J. Virol. 2:48-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Lazzaro, C., G. Campadelli-Fiume, and M. R. Torrisi. 1995. Intermediate forms of glycoconjugates are present in the envelope of herpes simplex virions during their transport along the exocytic pathway. Virology 214:619-623. [DOI] [PubMed] [Google Scholar]
- 10.Eggers, M., E. Bogner, B. Agricola, H. F. Kern, and K. Radsak. 1992. Inhibition of human cytomegalovirus maturation by brefeldin A. J. Gen. Virol. 73:2679-2692. [DOI] [PubMed] [Google Scholar]
- 11.Fish, K. N., W. Britt, and J. A. Nelson. 1996. A novel mechanism for persistence of human cytomegalovirus in macrophages. J. Virol. 70:1855-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fish, K. N., C. Soderberg-Naucler, and J. A. Nelson. 1998. Steady-state plasma membrane expression of human cytomegalovirus gB is determined by the phosphorylation state of Ser900. J. Virol. 72:6657-6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 1997. The UL20 gene product of pseudorabies virus functions in virus egress. J. Virol. 71:5639-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs, W., B. G. Klupp, H. Granzow, H. J. Rziha, and T. C. Mettenleiter. 1996. Identification and characterization of the pseudorabies virus UL3.5 protein, which is involved in virus egress. J. Virol. 70:3517-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garoff, H., R. Hewson, and D. J. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gershon, A. A., D. L. Sherman, Z. Zhu, C. A. Gabel, R. T. Ambron, and M. D. Gershon. 1994. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J. Virol. 68:6372-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson, W. 1996. Structure and assembly of the virion. Intervirology 39:389-400. [DOI] [PubMed] [Google Scholar]
- 18.Gong, M., and E. Kieff. 1990. Intracellular trafficking of two major Epstein-Barr virus glycoproteins, gp350/220 and gp110. J. Virol. 64:1507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granzow, H., F. Weiland, A. Jons, B. G. Klupp, A. Karger, and T. C. Mettenleiter. 1997. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J. Virol. 71:2072-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grundy, J. E., J. A. McKeating, and P. D. Griffiths. 1987. Cytomegalovirus strain AD169 binds beta 2 microglobulin in vitro after release from cells. J. Gen. Virol. 68:777-784. [DOI] [PubMed] [Google Scholar]
- 21.Hengel, H., W. Brune, and U. H. Koszinowski. 1998. Immune evasion by cytomegalovirus—survival strategies of a highly adapted opportunist. Trends Microbiol. 6:190-197. [DOI] [PubMed] [Google Scholar]
- 22.Hensel, G., H. Meyer, S. Gartner, G. Brand, and H. F. Kern. 1995. Nuclear localization of the human cytomegalovirus tegument protein pp150 (ppUL32). J. Gen. Virol. 76:1591-1601. [DOI] [PubMed] [Google Scholar]
- 23.Jarvis, M. A., K. N. Fish, C. Soderberg-Naucler, D. N. Streblow, H. L. Meyers, G. Thomas, and J. A. Nelson. 2002. Retrieval of human cytomegalovirus glycoprotein B from cell surface is not required for virus envelopment in astrocytoma cells. J. Virol. 76:5147-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, D. C., and P. G. Spear. 1982. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J. Virol. 43:1102-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, D. C., and P. G. Spear. 1983. O-linked oligosaccharides are acquired by herpes simplex virus glycoproteins in the Golgi apparatus. Cell 32:987-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones, F., and C. Grose. 1988. Role of cytoplasmic vacuoles in varicella-zoster virus glycoprotein trafficking and virion envelopment. J. Virol. 62:2701-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landini, M. P., B. Severi, L. Badiali, E. Gonczol, and G. Mirolo. 1987. Structural components of human cytomegalovirus: in situ localization of the major glycoprotein. Intervirology 27:154-160. [DOI] [PubMed] [Google Scholar]
- 28.Massalski, A., M. Coulter-Mackie, R. L. Knobler, M. J. Buchmeier, and S. Dales. 1982. In vivo and in vitro models of demyelinating diseases. V. Comparison of the assembly of mouse hepatitis virus, strain JHM, in two murine cell lines. Intervirology 18:135-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miranda-Saksena, M., R. A. Boadle, P. Armati, and A. L. Cunningham. 2002. In rat dorsal root ganglion neurons, herpes simplex virus type 1 tegument forms in the cytoplasm of the cell body. J. Virol. 76:9934-9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan, C., H. M. Rose, M. Holden, and E. P. Jones. 1954. Electron microscopic observations on the development of the herpes simplex virus. J. Exp. Med. 110:643-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nii, S., M. Yoshida, F. Uno, T. Kurata, K. Ikuta, and K. Yamanishi. 1990. Replication of human herpesvirus 6 (HHV-6): morphological aspects. Adv. Exp. Med. Biol. 278:19-28. [DOI] [PubMed] [Google Scholar]
- 33.Nixdorf, R., B. G. Klupp, A. Karger, and T. C. Mettenleiter. 2000. Effects of truncation of the carboxy terminus of pseudorabies virus glycoprotein B on infectivity. J. Virol. 74:7137-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novick, P., and P. Brennwald. 1993. Friends and family: the role of the Rab GTPases in vesicular traffic. Cell 75:597-601. [DOI] [PubMed] [Google Scholar]
- 35.Pelham, H. R., and S. Munro. 1993. Sorting of membrane proteins in the secretory pathway. Cell 75:603-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettersson, R. F. 1991. Protein localization and virus assembly at intracellular membranes. Curr. Top. Microbiol. Immunol. 170:67-106. [DOI] [PubMed] [Google Scholar]
- 37.Pfeffer, S. R., A. B. Dirac-Svejstrup, and T. Soldati. 1995. Rab GDP dissociation inhibitor: putting rab GTPases in the right place. J. Biol. Chem. 270:17057-17059. [DOI] [PubMed] [Google Scholar]
- 38.Radsak, K., M. Eickmann, T. Mockenhaupt, E. Bogner, H. Kern, A. Eis-Hubinger, and M. Reschke. 1996. Retrieval of human cytomegalovirus glycoprotein B from the infected cell surface for virus envelopment. Arch. Virol. 141:557-572. [DOI] [PubMed] [Google Scholar]
- 39.Risco, C., and J. L. Carrascosa. 1999. Visualization of viral assembly in the infected cell. Histol. Histopathol. 14:905-926. [DOI] [PubMed] [Google Scholar]
- 40.Rixon, F. J., C. Addison, and J. McLauchlan. 1992. Assembly of enveloped tegument structures (L particles) can occur independently of virion maturation in herpes simplex virus type 1-infected cells. J. Gen. Virol. 73:277-284. [DOI] [PubMed] [Google Scholar]
- 41.Roffman, E., J. P. Albert, J. P. Goff, and N. Frenkel. 1990. Putative site for the acquisition of human herpesvirus 6 virion tegument. J. Virol. 64:6308-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roizman, B. 1996. Herpesviridae, p. 2221-2230. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P Monath, B. Roizman, and S. E. Straus (ed.), Virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 43.Sanchez, V., P. C. Angeletti, J. A. Engler, and W. J. Britt. 1998. Localization of human cytomegalovirus structural proteins to the nuclear matrix of infected human fibroblasts. J. Virol. 72:3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez, V., E. Sztul, and W. J. Britt. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J. Virol. 74:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schimmoller, F., I. Simon, and S. R. Pfeffer. 1998. Rab GTPases, directors of vesicle docking. J. Biol. Chem. 273:22161-22164. [DOI] [PubMed] [Google Scholar]
- 47.Severi, B., M. P. Landini, and E. Govoni. 1988. Human cytomegalovirus morphogenesis: an ultrastructural study of the late cytoplasmic phases. Arch. Virol. 98:51-64. [DOI] [PubMed] [Google Scholar]
- 48.Simons, K., and H. Garoff. 1980. The budding mechanisms of enveloped animal viruses. J. Gen. Virol. 50:1-21. [DOI] [PubMed] [Google Scholar]
- 49.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment→deenvelopment→reenvelopment pathway. J. Virol. 75:5697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, J. D., and E. De Harven. 1973. Herpes simplex virus and human cytomegalovirus replication in WI-38 cells. I. Sequence of viral replication. J. Virol. 12:919-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sodeik, B., R. W. Doms, M. Ericsson, G. Hiller, C. E. Machamer, W. van 't Hof, G. van Meer, B. Moss, and G. Griffiths. 1993. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J. Cell Biol. 121:521-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soderberg, C., T. D. Giugni, J. A. Zaia, S. Larsson, J. M. Wahlberg, and E. Moller. 1993. CD13 (human aminopeptidase N) mediates human cytomegalovirus infection. J. Virol. 67:6576-6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spear, G. T., N. S. Lurain, C. J. Parker, M. Ghassemi, G. H. Payne, and M. Saifuddin. 1995. Host cell-derived complement control proteins CD55 and CD59 are incorporated into the virions of two unrelated enveloped viruses, human T cell leukemia/lymphoma virus type I (HTLV-I) and human cytomegalovirus (HCMV). J. Immunol. 155:4376-4381. [PubMed] [Google Scholar]
- 54.Spengler, M., N. Niesen, C. Grose, W. T. Ruyechan, and J. Hay. 2001. Interactions among structural proteins of Varicella-zoster virus. Arch. Virol. 17(Suppl.):71-79. [DOI] [PubMed] [Google Scholar]
- 55.Stackpole, C. W. 1969. Herpes-type virus of the frog renal adenocarcinoma. I. Virus development in tumor transplants maintained at low temperature. J. Virol. 4:75-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stephens, E. B., and R. W. Compans. 1988. Assembly of animal viruses at cellular membranes. Annu. Rev. Microbiol. 42:489-516. [DOI] [PubMed] [Google Scholar]
- 57.Steven, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. M. Burnett, and R. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 58.Tirabassi, R. S., and L. W. Enquist. 1998. Role of envelope protein gE endocytosis in the pseudorabies virus life cycle. J. Virol. 72:4571-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tokuyasu, K. T. 1973. A technique for ultracryotomy of cell suspensions and tissues. J. Cell Biol. 57:551-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tooze, J., M. Hollinshead, B. Reis, K. Radsak, and H. Kern. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163-178. [PubMed] [Google Scholar]
- 61.Traub, L. M., and S. Kornfeld. 1997. The trans-Golgi network: a late secretory sorting station. Curr. Opin. Cell Biol. 9:527-533. [DOI] [PubMed] [Google Scholar]
- 62.Velasco, A., L. Hendricks, K. W. Moremen, D. R. Tulsiani, O. Touster, and M. G. Farquhar. 1993. Cell type-dependent variations in the subcellular distribution of alpha-mannosidase I and II. J. Cell Biol. 122:39-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wentworth, B. B., and L. French. 1970. Plaque assay of cytomegalovirus strains of human origin. Proc. Soc. Exp. Biol. Med. 135:253-258. [DOI] [PubMed] [Google Scholar]
- 64.Wright, J. F., A. Kurosky, E. L. Pryzdial, and S. Wasi. 1995. Host cellular annexin II is associated with cytomegalovirus particles isolated from cultured human fibroblasts. J. Virol. 69:4784-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu, Z., M. D. Gershon, Y. Hao, R. T. Ambron, C. A. Gabel, and A. A. Gershon. 1995. Envelopment of varicella-zoster virus: targeting of viral glycoproteins to the trans-Golgi network. J. Virol. 69:7951-7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu, Z., Y. Hao, M. D. Gershon, R. T. Ambron, and A. A. Gershon. 1996. Targeting of glycoprotein I (gE) of varicella-zoster virus to the trans-Golgi network by an AYRV sequence and an acidic amino acid-rich patch in the cytosolic domain of the molecule. J. Virol. 70:6563-6575. [DOI] [PMC free article] [PubMed] [Google Scholar]