Abstract
In addition to its well-established role in the activation of herpes simplex virus immediate-early gene transcription, VP16 interacts with and downregulates the function of the virion host shutoff protein (vhs), thereby attenuating vhs-mediated destruction of viral mRNAs and translational arrest at late times of infection. We have carried out two-hybrid analysis in vivo and protein-protein interaction assays in vitro to identify determinants in VP16 necessary for interaction with vhs. The minimal amino-terminal subfragment of VP16 capable of binding to vhs encompassed residues 1 to 345. Alteration of a single leucine at position 344 to alanine (L344A) in the context of the amino-terminal fragment of VP16 containing residues 1 to 404 was sufficient to abolish interaction with vhs in vitro and in vivo. Leu344 could be replaced with hydrophobic amino acids (Ile, Phe, Met, or Val) but not by Asn, Lys, or Pro, indicating that hydrophobicity is an important property of binding to vhs. VP16 harboring a loss-of-function mutation at L344 was not compromised in its ability to interact with host cell factor (HCF-1) or to activate transcription of viral immediate-early genes in transient-transfection assays. Virus complementation assays using the VP16-null virus 8MA and the VP16/vhs double-mutant virus 8MAΔSma showed that VP16(L344A) was able to complement the growth of 8MAΔSma but not 8MA. Thus, a single point mutation in VP16 uncouples binding to vhs from other functions of VP16 required for virus growth and indicates that direct physical association between VP16 and vhs is necessary to sustain a productive infection.
Herpes simplex virus type 1 (HSV-1) is a large, enveloped DNA virus whose genome encodes some 80 genes. These genes fall into three broad kinetic classes, depending on their order of appearance during a lytic infection: immediate early (IE or α), early (E or β), and late (L or γ). Members of each class are coordinately and temporally regulated in a cascade fashion, primarily at the transcriptional level, by interactive networks that involve both virus and host factors. HSV-1 is noteworthy in that several important viral regulatory proteins exist as preformed structural components of the virion. These factors are delivered into the host cell by the infecting virus particle and are thus poised to affect the earliest events of viral infection and initiation of replication (reviewed in reference 41). The most prominent of these is the viral transactivator VP16, an abundant 490-amino-acid phosphoprotein contained in the viral tegument, an amorphous protein layer present between the viral capsid and envelope (5). VP16 possesses a potent C-terminal transactivation domain and triggers the lytic cycle by initiating IE gene expression via conserved cis-acting TAATGARAT elements present in the promoter regions of all the IE genes (2, 33, 38, 42, 51). VP16 on its own has only weak DNA binding activity, and efficient recruitment to promoter target sites relies on the assembly of a multicomponent protein/DNA binding transcription regulatory complex (the VP16-induced complex, VIC) that involves the participation of two cellular factors; the ubiquitous transcription factor Oct-1, which binds independently to TAATGARAT elements, and host cell factor-1 (HCF-1) (also called C1 or VCAF), which binds directly to VP16 and delivers it to DNA-bound Oct-1 (21, 22, 23, 26, 34, 59; reviewed in references 17 and 18). Once IE gene transcription is engaged, the lytic cycle becomes self-sustaining, since the IE proteins are sufficient to maintain IE transcription and induce the subsequent expression of early and late genes (reviewed in reference 55). Interestingly, the transactivation function of VP16 is not absolutely required for viral growth since viral mutants compromised in the carboxyl-terminal transactivation domain or in VIC assembly can be propagated in tissue culture, albeit only when high multiplicities of infection (MOI) are used (2, 49). Aside from its role in viral gene transactivation, VP16 is crucial at late times of an infection, possessing an essential but undetermined role in virus maturation and assembly (32, 36, 54).
We have previously shown that VP16 plays another important regulatory role in viral growth, manifested through its ability to modulate the activity of the virion host shutoff protein (vhs) (28, 48). vhs, a 489-amino-acid phosphoprotein that, like VP16, exists as a preformed viral structural component, disables host protein synthesis and triggers mRNA degradation following infection (13, 24, 25, 35, 40, 43, 47, 52). vhs is not required for HSV-1 propagation in tissue culture, although in its absence, the expression of viral proteins is altered during an infection and there is a significant reduction in virus yield (39, 40). Recent in vitro and in vivo findings indicate that vhs targets both the 5′ and 3′ ends of mRNA for non-sequence-specific endoribonucleolytic cleavage (10, 20), although it is not clear if vhs is itself an RNase or is instead a component of a larger endonucleolytic complex (reviewed in reference 50). The latter is suggested by evidence that vhs nuclease activity in vitro requires the presence of cellular factors (12, 30).
Vhs is synthesized as a late protein and indiscriminately destabilizes both viral and cellular mRNAs (50). Consequently, activity of newly synthesized vhs must be suppressed at late times of infection in order to sustain progression of the lytic cycle. Our previous in vitro and in vivo studies strongly suggest that VP16 fulfills this role (28, 48). Thus, a VP16-null virus (8MA), which is defective in the production of infectious progeny because of the absence of newly synthesized VP16, displays unrestrained vhs activity, resulting in accelerated host and viral mRNA turnover and arrest of viral protein synthesis at intermediate and late times of an infection. In contrast, an 8MA derivative that also harbors an inactive vhs (8MAΔSma) does not display the severe inhibition of viral protein synthesis and mRNA decay that is observed with 8MA. Importantly, the lethal phenotype in 8MA can be partially rescued by wild type as well as by a transactivation-defective form of VP16. The foregoing suggests that in addition to its roles in transcription and virus assembly, newly synthesized VP16 is required to sustain viral protein synthesis at late times of infection by blocking vhs-mediated translational arrest and destruction of viral mRNAs. In agreement with this, stable cell lines that constitutively express VP16 are resistant to host shutoff by superinfection with HSV (28). VP16 is thus a multifunctional viral protein that, through a complex regulatory network of protein-protein interactions, is involved in at least two distinct facets of HSV-1 gene regulation.
We have previously shown that VP16 and vhs form a stable complex in vitro and in vivo in infected cells (48), suggesting that physical association is necessary for VP16 to attenuate vhs activity during an infection. However, this has been difficult to directly demonstrate given the fact that in the absence of newly synthesized VP16, unrestrained vhs activity is lethal and that VP16 plays multiple essential roles in the viral life cycle. The VP16-binding interface on vhs is well defined. We have shown that a 21-amino-acid region in vhs (between residues 310 and 330) is necessary and sufficient for interaction with VP16 in vitro and in vivo and that alteration of a single tryptophan residue (residue 321) within this domain inhibits binding (44). In contrast, interfaces within VP16 that are targeted by vhs have not been mapped in detail. Mutational analysis reported thus far indicates that vhs targets regions of VP16 that overlap with domains known to be important for HCF binding, VIC assembly, and/or DNA binding (48). In order to gain further insights into the functional consequences of interaction between VP16 and vhs, we sought to identify minimal determinants in VP16 targeted by vhs and determine whether VP16/vhs binding could be uncoupled from other roles of VP16 in transcriptional activation and/or virus assembly. We show here that alteration of a single leucine residue at position 344 of VP16 abolishes interaction with vhs in vitro and in vivo while having no effect on interaction with HCF-1 or on transcriptional activation of an IE promoter. Importantly, VP16 harboring an L344 mutation was able to substitute for wild-type VP16 in virus complementation assays with 8MAΔSma, but was unable to do so with 8MA. These findings indicate that a single point mutation in VP16 can uncouple binding to vhs from VP16-mediated transactivation and virus assembly and, moreover, suggest that a direct interaction between VP16 and vhs is required for VP16-mediated downregulation of vhs activity.
MATERIALS AND METHODS
Mammalian cell culture.
Vero, COS-1, and 16-8 cells were propagated and maintained in Dulbecco's modified Eagle medium containing 1% l-glutamine and 1% penicillin-streptomycin and supplemented with 10% calf or fetal bovine serum. 16-8 cells constitutively express VP16, and this cell line is used to propagate VP16-null virus (54). 16-8 cell stocks were maintained in the presence of G418 (450 μg/ml; Gibco BRL).
Plasmids.
Carboxyl-terminal deletions of VP16 were generated by PCR amplification, using a forward (F200) and reverse primers specific for each of the respective mutants (R340, R345, R350, R355; numbering refers to the carboxyl-terminal endpoint of VP16) as outlined in Table 1. PCR-derived fragments were digested with MscI/SacI and subcloned into the corresponding sites of pCADVP299, a Saccharomyces cerevisiae GAL4 activation domain (AD) plasmid that encodes amino acids 1 to 299 of VP16 linked to the GAL4 acidic AD (6, 48). pCDBvhs(Apa/Sma) contains the ApaI/SmaI region of vhs from HSV-1(KOS) (amino acids 179 to 344) fused to the DNA binding domain (DB) of GAL4 within the yeast expression plasmid pPC97 (6, 48). pCDBHCF380 contains amino acids 1 to 380 of human HCF-1 fused to the GAL4 DB and was generated by cloning a PCR-derived EcoRI/SalI fragment from pCGNHCF (57) into pPC97.
TABLE 1.
Oligonucleotides used for mutagenesisa
| Sequence | Oligonucleotide |
|---|---|
| 5′-GAACCGTGTTGGCCAACTTCTGCTC-3′ | FVP200 |
| 5′-CCCCGAGCTCAATCAACACCATAAAGTACCCAGAGG-3′ | RVP340 |
| 5′-CCCCGAGCTCGTCCAACTTCGCCCGATCAACAC-3′ | RVP345 |
| 5′-CCCCGAGCTCGAAGCTGGAATACGAGTCCAAC-3′ | RVP350 |
| 5′-CCCCGAGCTCGGAGGGCGAGGTCGTGAAGCT-3′ | RVP355 |
| 5′-GTACTTTATGGTGTTGATTGCGGCGAAGTTGGACTCG-3′ | VP(R341A) |
| 5′-GGTGTTGATTCGGGGGAAGTTGGACTCG-3′ | VP(A342G) |
| 5′-GTGTTGATTCGGGCGTTGGACTCGTATTC-3′ | VP(K343A) |
| 5′-GTTGATTCGGGCGAAGCGGGACTCGTATTCCAG-3′ | VP(L344A) |
| 5′-GGCGAAGTTGGCCTCGTATTCCAGC-3′ | VP(D345A) |
| 5′-CGGGAACACGCGGCCAGCCGCGCGCGTAC-3′ | VP(Y364A) |
| 5′-GTTGATTCGGGCGAAGCCGGACTCGTATTCCAGCTTC-3′ | VP(L344P) |
| 5′-GTTGATTCGGGCGAAGAAGGACTCGTATTCCAGCTTC-3′ | VP(L344K) |
| 5′-GTGTTGATTCGGGCGAAGAACGACTCGTATTCCAGCTTC-3′ | VP(L344N) |
| 5′-GTGTTGATTCGGGCGAAGATCGACTCGTATTCCAGCTTC-3′ | VP(L344I) |
| 5′-GTTGATTCGGGCGAAGGTGGACTCGTATTCCAGCTTC-3′ | VP(L344V) |
| 5′-GTGTTGATTCGGGCGAAGTTCGACTCGTATTCCAGCTTC-3′ | VP(L344F) |
| 5′-GTTGATTCGGGCGAAGATGGACTCGTATTCCAGCTTC-3′ | VP(L344M) |
Underlined nucleotides correspond to restriction endonuclease sites incorporated in forward (F) and reverse (R) primers used to construct the carboxyl-terminal truncation derivatives, while the numbering corresponds to the position of the carboxyl-terminal truncation in VP16. Oligonucleotides used for site-directed mutagenesis are shown with the altered nucleotides in boldface type, and the corresponding amino acid changes are indicated. Complementary oligonucleotides are not shown.
Site-specific mutations in VP16 were constructed using the QuikChange site-directed mutagenesis protocol using a commercially available kit (Stratagene) with the mutagenic oligonucleotide primers listed in Table 1. The template plasmid for each of the mutants was pGEM5Zf(-)-VP404, an in vitro transcription vector which contains the NcoI/SacII fragment corresponding to the amino-terminal residues 1 to 404 of VP16, cloned into the corresponding sites of pGEM5Zf(-) (Promega). Mammalian expression vectors for the various VP16 point mutants were generated using pEVRF65, an expression vector for full-length VP16 (45). pEVRF65 was first altered by site-directed mutagenesis to create a silent mutation that eliminated the SalI site at amino acid 411 of VP16 to generate pEVR65Δsal (5′-CGCAGACTGTCTACGGCCCCCCC-3′; and its complement oligonucleotide). The SalI/SacII fragment (encompassing the unique SalI site at amino acid residue 5 to the SacII at residue 404 of VP16) was excised from pEVRF65Δsal and replaced with the corresponding SalI/SacII fragments from the various VP16 point mutant derivatives generated in pGEM5Zf(-)-VP404. pα4Luc, a mammalian luciferase reporter gene was constructed by excising a 3.5kB BamHI fragment from pα4-CAT (56) containing the promoter/regulatory region of the immediate-early α4 gene of HSV-1 and subcloning into the BglII site in pGL3Basic-Luc (Promega). pSPAS is an in vitro transcription/translation vector which contains the ApaI/SmaI fragment of vhs cloned downstream of the SP6 promoter in the plasmid pSPUTK (48). pGEM7Zf(-)-HCF902 is an in vitro expression vector for HCF-1 (residues 1 to 902) under control of the T7 promoter and was constructed by inserting the PCR derived EcoRI/KpnI fragment of HCF-1 from pCGNHCF into the corresponding sites of pGEM7Zf(-) (Promega). All plasmid constructs were verified by DNA sequence analysis.
Yeast transformation and β-galactosidase assay.
S. cerevisiae strain PCY2 (MATα Δgal4 Δgal80 his3 trp1-901 ade2-101 ura3-52 leu2-3112 URA3:GAL-lacZ lys2:gal HIS3 Cyhrr) was transformed with GAL4 AD and GAL4 DB expression plasmids (6), or derivatives thereof (as described in the figure legends), using the modified lithium acetate protocol (9). Colonies were selected by plating the transformation mixture on synthetic complete plates lacking appropriate amino acids. Quantitative β-galactosidase liquid assays were carried out on cultures of transformed yeast grown in media lacking the appropriate amino acids, using the liquid nitrogen permeabilization method (3). Overlay β-galactosidase assays were performed by placing a molten mixture of 0.5% (wt/vol) agarose dissolved in sodium phosphate buffer (0.5 M NaPO4 pH 7, 0.1% sodium dodecyl sulfate [SDS], 2% dimethylformamide, 0.05% [wt/vol] X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, Sigma]) directly over transformed yeast colonies grown on culture plates (31). The plates were incubated at 37°C and monitored for blue color formation (∼30 min). Plates were left overnight to detect weak activity.
Solid-phase capture assays.
Glutathione-S-transferase (GST) fusion protein expression plasmids containing wild-type or the various mutant derivatives of VP16 were constructed in pGEX2T (Pharmacia) and transformed into BL21(DE3) cells (Stratagene). Cultures were grown to an optical density at 600 nm of 0.6 and induced for 2 h at 37°C with 0.1 mM isopropyl-β-d-thiogalactopyranoside (Biosynth AG) and GST fusion proteins were purified using GST beads (Pharmacia) as instructed by the manufacturer. Purified GST fusion protein beads were resuspended in 250 μl of IPAB (5 mM MgCl2, 20 mM HEPES [pH 7.9], 150 mM KCl, 0.1% NP-40, 0.1% Triton X-100) plus BSA (20 μg/ml). Equivalent amounts of purified fusion proteins were incubated with [35S]methionine-labeled proteins synthesized in vitro by coupled transcription-translation using a commercially available rabbit reticulocyte lysate kit (Promega) according to the manufacturer's instructions. Beads were extensively washed and bound material was eluted and subjected to SDS-polyacrylamide gel electrophoresis and detected by autoradiography as described previously (31).
Transfections.
Transfections were carried out in COS-1 cells using Lipofectamine (Invitrogen) essentially as outlined by the manufacturer. Briefly, COS-1 cells were grown on six-well plates to 70% confluency and transfected with a mixture of 4 μl of Lipofectamine reagent and 10 ng of pEVRF65 (or the various VP16 mutant derivatives thereof) together with 100 ng of the pα4Luc reporter plasmid. Promoter and plasmid dosage was kept constant with the addition of appropriate amounts of the corresponding empty vector pEVRF0. Cells were incubated at 37°C for 4 h followed by replacement with fresh media. Luciferase activity was measured 48 h posttransfection as described (31).
Electrophoretic mobility shift analysis (EMSA).
Gel retardation assays were carried out essentially as described previously using HeLa cell nuclear extracts and VP16 proteins transcribed and translated in vitro (45, 56, 59). Briefly, 2 μl of unprogrammed reticulocyte lysate or lysate programmed with the various VP16 derivatives was incubated with 10 μg of HeLa cell nuclear extract and 32P-labeled DNA probe corresponding to the promoter-proximal TAATGARAT element of the HSV-1 ICP0 gene in a buffer containing 20 mM HEPES (pH 7.9), 50 mM KCl, 0.5 mM EDTA, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 0.05% NP-40, 10 μg of bovine serum albumin, and 4 μg of a 1:2 mixture of poly(dI-dC)-denatured salmon sperm DNA. Protein/DNA complexes were resolved by electrophoresis on a 4% polyacrylamide gel.
Western blot analysis.
Western blot analysis was carried out using the ECL detection system (Amersham) according to the manufacturer's instructions. Briefly, Vero cells were transfected with pEVRF65, pEVRF65(L344A) or pEVRF0 (as detailed in the figure legends) and cell extracts were prepared 48 h posttransfection (37). Equivalent amounts of protein (50 μg) were subjected to SDS-polyacrylamide gel electrophoresis and probed using rabbit anti-VP16 polyclonal antibody (diluted to 0.1 μg/ml; Clontech) as the primary antibody and anti-rabbit horse radish peroxidase conjugated secondary antibody (used at a 104-fold dilution; Santa Cruz).
Virus complementation assays.
Wild-type HSV-1 (KOS) stocks were prepared in Vero cells while 8MA and 8MAΔSma virus stocks were prepared and titrated on VP16-complementing 16-8 cells. VP16 complementation assays were performed as follows: Vero cells, grown on six-well plates, were transfected with 2 μg of pEVRF65, pEVRF65(L344A), or pEVRF0 as described above and incubated at 37°C for 24 h. Medium was removed and cells were infected at an MOI of 5 with either 8MA or 8MAΔSma for 1 h with occasional rocking. The media was replaced with fresh growth media and cells were incubated until the appearance of cytopathic effects (48 to 72 h postinfection). Cells and supernatants were collected and extracts were prepared by three freeze-thaw cycles. Infectious virus yield was measured by plaque assay on 16-8 cells.
RESULTS
Requirements in VP16 for interaction with vhs.
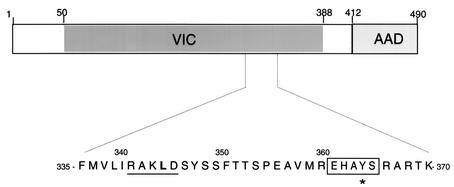
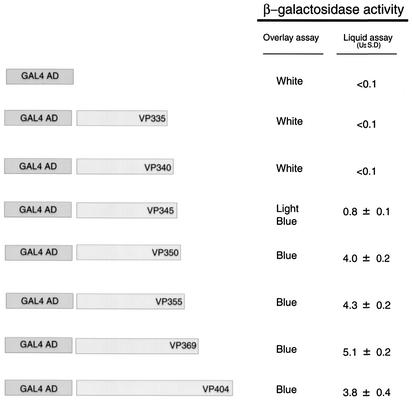
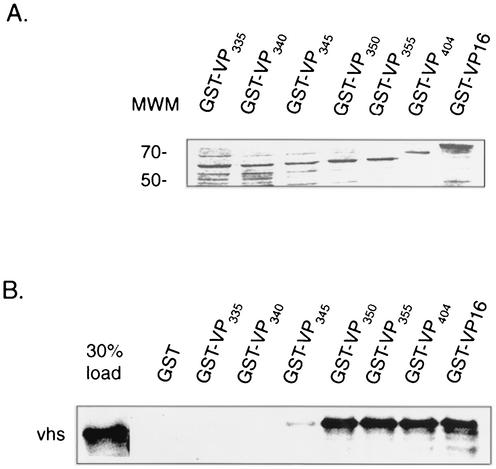
Our previous deletion analysis demonstrated that the VP16 region spanning amino acids 335 to 369 was critical for interaction with vhs (48). This subregion also contains determinants required for interaction with HCF-1 and Oct-1, and for VIC assembly (Fig. 1). To map the carboxyl-terminal endpoint for interaction with vhs more precisely and to identify residues within this region that may be uniquely required for vhs association, we generated additional carboxyl-terminal deletions in VP16 and tested these for interaction with vhs protein using yeast two-hybrid and GST pull down assays. As previously described, VP16404 and VP16369, fused to the GAL4 AD interacted with vhs fused to the GAL4 DB in the two-hybrid system (48) (Fig. 2). Similarly, VP16 derivatives truncated at residue 355 and 350 interacted with vhs and did so at levels comparable to VP16404, as determined by quantitative β-galactosidase assays. In contrast, a derivative truncated at 345 showed only weak binding, while derivatives truncated at amino acid 340 or 335 were unable to bind to vhs. The results from the two-hybrid analysis was corroborated by GST pull down assays carried out in vitro using equivalent amounts of the various GST-VP16 derivatives (Fig. 3A) and in vitro-labeled vhs (residues 179 to 344). As shown in Fig. 3B, labeled vhs bound equally well to full-length GST-VP16 and GST-VP16 derivatives truncated at residue 404, 355, and 350, respectively (approximately 20% of the input vhs was retained in each case). In contrast, binding of vhs to GST-VP16345 was greatly diminished (less than 1% of input radioactivity), whereas vhs binding to derivatives truncated at 340 or 335 was not detected.
FIG. 1.
Schematic representation of VP16 indicating the region involved in assembly of the VP16-induced complex (VIC) with Oct-1 and HCF-1 (residues 49 to 388) and the position of the carboxyl-terminal acidic AD (AAD). The underlined amino acids correspond to residues that have been altered in this study, with the critical leucine at position 344 shown in boldface type. The boxed region corresponds to the HCF-binding motif. Mutation of Y364, indicated by (*) has been shown to inhibit interaction with HCF-1 (26, 27).
FIG. 2.
Interaction of VP16 carboxyl terminal deletions with vhs using the yeast two-hybrid system. S. cerevisiae PCY2 harboring the GAL4-DB-vhs expression plasmid was transformed with each of the indicated GAL4 AD-VP16 derivatives and assayed for β-galactosidase activity. Values for specific activity are averages ± standard deviations from at least 3 independent transformants assayed in duplicate and normalized to cell density.
FIG. 3.
Mapping the vhs binding domain within VP16 in vitro. In vitro-translated, [35S]methionine-labeled vhs (amino acids 179 to 344) was incubated with equivalent amounts of purified GST-bound GST-VP16 or various carboxyl-terminal deletions (as determined by SDS-polyacrylamide gel electrophoresis and Coomassie blue staining [A]) or with GST beads alone, as indicated (numbering refers to carboxyl-terminal end points). Bound material was eluted from the beads and analyzed by SDS-polyacrylamide gel electrophoresis (B). The left lane of panel B represents 30% of the total input radioactivity used in each of the binding assays. The positions of molecular weight markers (MWM) (in thousands) are shown.
Alteration of leucine 344 in VP16 abolishes binding to vhs while having no effect on interaction with HCF-1.
The above findings indicate that residues 340 to 345 of VP16 play a critical role in interaction with vhs. Two positively charged amino acids within this region, R341 and K343, have previously been shown to be important for mediating DNA binding and intramolecular contacts within VP16, respectively (27, 29). To identify individual amino acids important for vhs binding, we altered each residue to alanine (except A342 which was changed to G) by site-directed mutagenesis in the context of VP16404 to generate R341A, A342G, K343A, L344A, and D345A. Mutations to alanine were chosen since these would not be expected to significantly affect the structure of the polypeptide backbone (7). The point mutant derivatives were subcloned into the GAL4 AD vectors and GST expression vectors for analysis by two-hybrid and in vitro pull down assays, respectively.
The results of the two-hybrid analysis are shown in Table 2. All of the point mutant derivatives, with the notable exception of L344A, interacted with vhs in yeast as determined by the β-galactosidase overlay assay. The same mutant derivatives were also tested for interaction with HCF-1 using GAL4 DB-HCF380, a GAL4 DB vector expressing amino acids 1 to 380 of HCF-1. This amino-terminal region of HCF-1, known as HCFVIC, is necessary and sufficient for interaction with VP16 in vitro and in vivo (19). As shown in Table 2, all of the mutant derivatives interacted with HCF-1. This is not surprising since the critical determinants in VP16 that are targeted by HCF-1 lie between amino acids 360 and 370 (Fig. 1) (27, 45, 46, 58). As a control to ensure specificity of the two-hybrid assay, we tested VP16(Y364A), a point mutant derivative previously shown to be defective in interaction with HCF-1 (16, 26). As shown in Table 2, Y364A interacted with vhs but failed to interact with HCF-1. The foregoing indicates that alteration of L344 to alanine is sufficient to selectively abolish binding to vhs while having no apparent effect on HCF-1 binding.
TABLE 2.
Two-hybrid analysis of site-specific point mutants within VP16a
| GAL4-AD-VP16 construct | β-Galactosidase activity
|
||
|---|---|---|---|
| DB | DB-vhs | DB-HCF | |
| AD | − | − | − |
| AD-VP404 | − | + | + |
| AD-VP404 (R341A) | − | + | + |
| AD-VP404 (A342G) | − | + | + |
| AD-VP404 (K343A) | − | + | + |
| AD-VP404 (L344A) | − | − | + |
| AD-VP404 (D345A) | − | + | + |
| AD-VP404 (Y364A) | − | + | − |
| AD-VP404 (L344P) | − | − | + |
| AD-VP404 (L344K) | − | + | + |
| AD-VP404 (L344N) | − | ± | + |
| AD-VP404 (L344I) | − | + | + |
| AD-VP404 (L344V) | − | + | + |
| AD-VP404 (L344F) | − | + | + |
| AD-VP404 (L344M) | − | + | + |
S. cerevisiae PCY2 was cotransformed with expression plasmids for the indicated GAL4-AD-VP16 fusion proteins and GAL4-DB-vhs or HCF-1, as indicated and assayed for β-galactosidase activity by the qualitative overlay assay. Where indicated, blue color formation was observed in <30 min (+), observed after overnight incubation (±), or not observed (−). The data are the summary of at least three independent transformations.
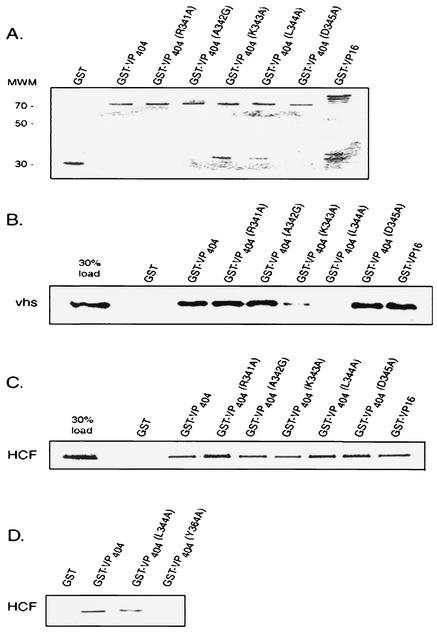
To confirm the yeast in vivo results, GST-VP16 fusion proteins harboring the respective point mutations within the context of the amino-terminal 404 residues of VP16 (Fig. 4A) were used in GST pull down assays with radiolabeled vhs or HCF-1 (residues 1 to 902). Consistent with the yeast findings, L344A failed to form a complex with vhs in vitro, while retaining its ability to interact with HCF-1 (Fig. 4B and C). As expected, Y364A did not interact with HCF-1 under the same experimental conditions that wild-type and L344A bound (Fig. 4D). In contrast to the yeast findings, vhs binding to K343A was also diminished, albeit not to the extent observed with L344A. K343A was also compromised in its interaction with HCF-1 in vitro. Previous structural studies of VP16 point to a role for K343 in maintaining critical intramolecular contacts within the VP16 core structure and as such, substitution of this lysine residue may cause perturbations in overall tertiary and/or secondary structure that may have generalized effects on interaction with both vhs and HCF-1 in vitro (29).
FIG. 4.
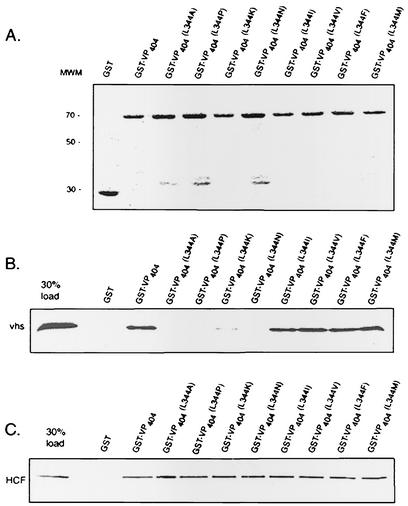
A mutation at leucine 344 of VP16 abolishes binding to vhs. Equivalent amounts of full-length GST-VP16, GST-VP16 (residues 1 to 404), or derivatives thereof harboring point mutations at the indicated amino acids (or control GST beads) were incubated with in vitro-translated, [35S]methionine-labeled vhs (B) or HCF-1 (amino-terminal residues 1 to 902) (C), and bound material was analyzed by SDS-polyacrylamide gel electrophoresis. The left lane in each case represents 30% of the total input radioactivity used in the respective binding assays. (D) Control experiment that compares binding of VP16(L344A) and VP16(Y364A), a derivative that is known to be defective in interaction with HCF-1 (26). (A) Coomassie blue-stained gel of GST or the various GST-VP16 fusion proteins used in the binding assays.
According to the crystal structure of the conserved core of VP16, L344 is partially solvent exposed and exists as the terminal residue of an α-helix structure (29). The carbonyl group of the corresponding peptide bond, however, remains available to participate in hydrogen-bond formation, and the hydrophobic side chain is able to participate in “hydrophobic interactions” (8). To yield insight into the structural and functional importance of L344 in mediating binding to vhs, we substituted various amino acids at this position. The intent was to delineate between the strict requirements of the leucine side chain and/or the importance of the alpha helix structure and peptide bond at this position. Additional substitutions at L344 of VP16 included L344P, L344K, L344N, L344I, L344V, L344F, and L344 M.
None of the substitutions at L344, including proline, compromised interaction with HCF-1 in vivo or in vitro as determined by two-hybrid analysis (Table 2) or GST pull down assays (Fig. 5C), respectively. Analysis of vhs interaction in yeast showed that L344I, L344V, L344F, and L344 M bound to vhs whereas L344N displayed a relatively weak interaction. Interaction with L344P was not observed. Similarly, L344I, L344V, L344F, and L344 M bound with vhs in vitro to levels comparable to wild-type VP16, whereas L344P, L344N, and L344K were compromised in their ability to bind to vhs (Fig. 5B). Thus, the in vitro and in vivo results are generally concordant, however, as shown above, L344N and L344K still retained some binding activity in the yeast system. The reasons for this is not clear but may reflect differences in binding efficiencies between two-hybrid and in vitro binding assays, as noted previously by others (11). It is possible that transient vhs interaction with L344N and L344K is sufficient to induce reporter gene activity in yeast, whereas these complexes are less stable under the conditions used for the pull down assays.
FIG. 5.
Hydrophobic amino acids substitutions at leucine 344 of VP16 are compatible with binding to vhs. Equivalent amounts of full-length GST-VP16, GST-VP16 (residues 1 to 404), or derivatives harboring the indicated amino acids substitutions at position 344 were incubated with in vitro-translated, [35S]methionine-labeled vhs (B) or HCF-1 (amino-terminal residues 1 to 902) (C) as described in the legend to Fig. 4. (A) Coomassie blue-stained gel of GST or the various GST-VP16 derivatives used in the binding assays is shown.
The above findings indicate that L344 can be substituted with a variety of hydrophobic amino acids (methionine, isoleucine, valine, and phenylalanine), but not with charged residues (asparagine and lysine) without loss in vhs binding. Moreover, proline, an amino acid that is expected to introduce structural perturbations or affect hydrogen bonding in the polypeptide backbone, inhibits VP16/vhs binding in vitro and in vivo.
Transcriptional activation by VP16 mutant proteins.
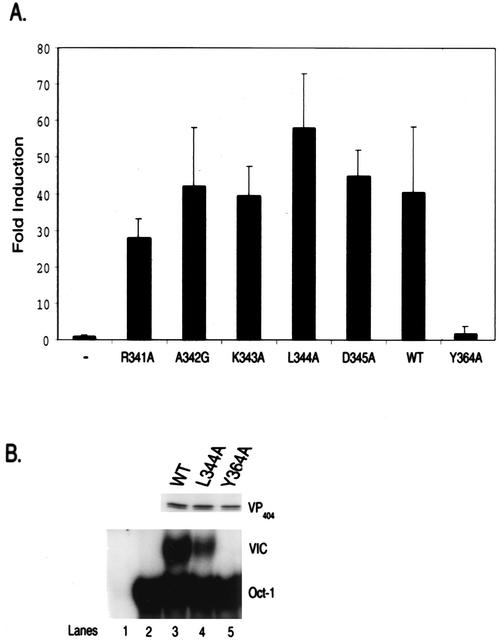
While none of the point mutants generated above affected interaction with HCF-1, the possibility remained that the functional integrity of VP16 was compromised by the changes. In order to address this, the various point mutations between residues 340 and 345 were introduced into full-length VP16 in a mammalian expression vector and tested for their ability to activate transcription of a luciferase reporter gene linked to the immediate-early HSV-1 α4 promoter. As shown in Fig. 6A, all of the VP16 point mutant derivatives, including L344A and K343A, activated expression of the reporter gene and did so to levels comparable to that observed with wild-type VP16. (Each mutant derivative was tested at various plasmid concentrations to ensure that reporter gene activation was not saturating; data not presented). As expected, Y364A was unable to activate reporter gene expression under the same experimental conditions.
FIG. 6.
VP16(L344) is able to direct VIC formation and activate transcription of IE promoters. (A) Transcriptional activation by VP16 mutant proteins. COS-1 cells was transfected with mammalian expression vectors for VP16 harboring the indicated point mutations along with the pα4-luc reporter gene and luciferase activity was measured. Wild-type (WT) VP16 and VP16(Y364A), a derivative which is unable to activate transcription, served as positive and negative controls, respectively. Values shown are averages (± standard deviations [error bars]) from three independent experiments done in duplicate and normalized to the value obtained from control transfections carried out with the reporter gene and empty expression vector (-) which was taken as 1. (B) Wild-type VP16, VP16(L344A), and VP16(Y364A) were synthesized in vitro (shown in upper panel) and tested for VIC formation by EMSA with HeLa cell nuclear extracts and a labeled TAATGARAT probe. The position of the Oct-1 protein/DNA complex and VIC is indicated. Lane 1 represents probe incubated with in vitro-synthesized WT VP16 in the absence of HeLa cell nuclear extract, while lane 2 represents probe incubated with nuclear extract and unprogrammed reticulocyte lysate.
To further confirm the functional integrity of L344A, we examined the ability of this derivative to direct VIC formation by EMSA analysis using HeLa cell nuclear extracts (HeLa cells provide both Oct-1 and HCF-1) and in vitro translated VP16. As shown in Fig. 6B both wild-type VP16 and L344A generated the VIC complex when incubated with labeled TAATGARAT DNA probe. In contrast, Y364A failed to form VIC. Thus, alteration of L344 does not compromise the ability of VP16 to direct complex formation and activate target gene expression.
VP16(L344A) is able to complement the growth of HSV-1 8MAΔSma but not 8MA.
VP16 is essential for growth of HSV-1 since deletion of the VP16 open reading frame is lethal, as demonstrated with 8MA, a VP16-null virus which can only be propagated in VP16 complementing cells (54). Recent studies indicate that the lethal 8MA phenotype is due to both uncontrolled vhs activity as well as a structural requirement for VP16 at later stages of virion formation (28, 32). We hypothesized that if VP16-dependent downregulation of vhs activity requires direct physical association between VP16 and vhs, then VP16(L344A) should be unable to complement 8MA. In contrast, VP16(L344A) should be able to complement the VP16/vhs double mutant 8MAΔSma since this mutant virus lacks functional vhs. Alternatively, VP16(L344A) would not be expected to complement the growth of 8MAΔSma if this VP16 mutant is also defective in some later step required for virus assembly.
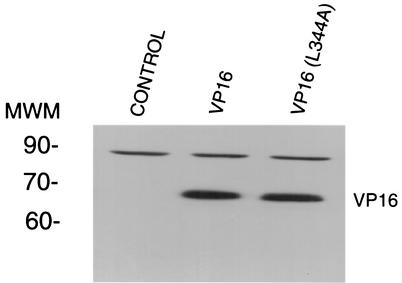
To examine the biological consequences of the L344A VP16 mutation, we undertook complementation assays with 8MA and 8MAΔSma. Both 8MA and 8MAΔSma produced from 16-8 cells can infect nonpermissive cells, since the virus contains normal amounts of functional VP16 acquired from the complementing cell line. However, mutant virus fails to produce infectious progeny in nonpermissive Vero cells because of the absence of newly synthesized VP16. Infectious progeny can be generated if VP16 is supplied in trans. Vero cells were transiently transfected with expression vectors for wild-type VP16 or VP16(L344). Cell cultures were infected 24 h posttransfection with HSV-1 8MA or 8MAΔSma at a MOI of 5 and incubated until cytopathic effects were observed. Infected cell extracts and supernatants were harvested and used to measure infectious virus production by plaque assay on 16-8 cells. As summarized in Table 3, extracts from Vero cells transfected with wild-type VP16 and infected with 8MA produced an average of 2.8 × 106 PFU/ml, compared to 5 × 102 PFU/ml obtained from cells transfected with VP16(L344A). Thus, wild-type VP16 is 3 to 4 orders of magnitude more efficient in complementing the lethal defect in 8MA compared to VP16(L344A). This difference cannot be attributed to variation in expression levels of wild-type VP16 versus VP16(L344A) in transfected cells since expression levels of the two proteins differed by less than 50% as determined by Western blot analysis of transfected cells (Fig. 7).
TABLE 3.
Virus complementation assays of 8MA and 8MAΔSmaa
| Expression plasmid | Virus yields (PFU/ml) from plaque assays on 16-8 cells after superinfection with:
|
|
|---|---|---|
| 8MA | 8MAΔSma | |
| Control | 0-4 | 20 |
| VP16 | 2.5 × 106-3 × 106 | 1 × 103-2 × 103 |
| VP16(L344A) | 0.8 × 102-9 × 102 | 3.5 × 102-5 × 102 |
Virus yields from Vero cells transfected with empty vector or expression plasmids for wild-type VP16 or VP16(L344A) followed by superinfection with 8MA or 8MAΔSma was quantitated by plaque assay on 16-8 cells. The data represent the range of PFUs obtained from two independent experiments.
FIG. 7.
Western blot analysis of wild-type VP16 and VP16(L344A) expressed in Vero cells. Vero cells were transfected with expression vectors for wild-type VP16 or VP16(L344A) (1 and 1.5 μg, respectively) or 1.5 μg of the corresponding empty vector control and examined by Western blot analysis with polyclonal anti-VP16 antibody. The position of the molecular weight markers (MWM) (in thousands) is indicated.
As expected, complementation of 8MAΔSma by wild type was less efficient in comparison to 8MA, since the former virus grows more poorly (Table 3) (32). However, in contrast to the results obtained with 8MA, there was only a threefold difference in the titer of infectious virus recovered from cells that were transfected with wild-type VP16 or VP16(L344A) and infected with 8MAΔSma (1.5 × 103 and 4 × 102 PFU/ml, respectively). Thus, VP16(L344A) is able to complement 8MAΔSma but not 8MA, suggesting that this point mutation does not compromise the essential role of VP16 in virus assembly.
The foregoing indicates that the only function of VP16 that appears to be compromised by alteration of L344 to alanine is its ability to associate with vhs. Moreover, the findings demonstrate that direct physical interaction between VP16 and vhs is critical for sustaining the lytic cycle and generating infectious virus.
DISCUSSION
VP16 is a multifunctional protein involved in distinct facets of the HSV viral life cycle, manifesting its functions through an intricate network of protein-protein and protein-DNA interactions with both viral and host factors. Previous studies have established that a key role of VP16 is to attenuate the activity of newly synthesized vhs, thus sustaining the lytic cycle by preventing vhs-mediated arrest of viral protein synthesis at late times of infection (28). The results reported herein extend this finding to show that a single point mutation at leucine 344 in VP16 can selectively inhibit VP16/vhs binding, while having no effect on the other functions of VP16 in viral gene expression or in viral assembly and maturation. Our findings confirm the importance of VP16 in restraining vhs function in the infected cell, underscoring the requirement for direct physical interaction between VP16 and vhs in this process.
Previous studies undertaken to identify regions in VP16 that are required for interaction with vhs showed that an amino-terminal region encompassing residues to 250, and a region between residues 335 and 369 to be of importance (48). These regions have previously been shown to contain determinants variously important for protein-protein and protein-DNA interactions, VIC assembly, transactivation, and in virion assembly (1, 14, 15, 16, 53, 56). Residues 360 to 370, which are embedded in a major surface-exposed site, are of particular significance as extensive mutational studies indicate that amino acids in this region are differentially required for interaction with HCF-1, contact with Oct-1, and/or direct DNA binding (27, 45, 46, 58). Structural determination of the core VP16 protein (residues 49 to 412) indicates that a central domain forms part of a seat-like structure that may be important in DNA binding by VP16 (4, 29). Furthermore, this region is postulated to be critical in the maintenance of VP16 secondary structure and in virion assembly (29), consistent with the enhanced sensitivity of this region to loss-of-function mutations (1, 53, 56).
Residues 340 to 345 are shown here to be critical for interaction with vhs. This segment is conserved in VP16 homologs present in other herpes viruses, containing invariant R341 and K343, as well as a conservative change at L344 (29). Specific amino acids in this region have previously been implicated in DNA binding (R341) and intramolecular stabilization of VP16 structure (K343) (27, 29). Based on the core VP16 crystal structure, residues 333 to 344 form a α-helix, while the adjacent residues up to amino acid 350 form a coordinated loop structure (29). The leucine side chain is partially solvent exposed and may thus utilize vhs to mask an unfavorable environment through a “hydrophobic interaction” (8). Consistent with this, alanine, proline, lysine and asparagine substitutions at L344 inhibited binding to vhs, while hydrophobic residues including methionine, isoleucine, phenylalanine, or valine could functionally replace leucine at this position. The incompatibility of proline at position 344 suggests that a α-helix is structurally required and/or that available hydrogen bonding partners within the polypeptide backbone at this position may play a role in stabilizing VP16/vhs interaction. The fact that these mutant VP16 proteins, including the proline substitution, are able to bind HCF and activate transcription, suggests that the overall structural integrity of the protein derivatives are preserved.
Our previous studies have demonstrated that VP16 targets a small, modular region of vhs encompassing residues 310 to 330 (PAAGGTEMRVSQWTEILTQQIA) (44). Tryptophan 321 is critical as alteration of this single residue abolished interaction with VP16 both in vitro and in vivo. Tryptophan 321 could potentially contribute both a hydrogen bond acceptor for L344 and a hydrophobic side chain at the VP16-binding interface, although this interpretation awaits more detailed structural investigation.
VP16(L344) was able to activate transcription of VP16-target genes in transient transfection assays and to complement the growth of 8MAΔSma as efficiently as wild-type VP16, demonstrating that the distinct functions of VP16 in transcriptional activation and in virus assembly/maturation were not compromised by this alteration. In contrast, VP16(L344A) was severely defective in its ability to complement 8MA, a VP16 null virus which harbors wild-type vhs. Assuming that the L344A mutant is solely defective for interaction with vhs, the findings strongly suggest that rescue of the lethal 8MA phenotype, and thus virus production, is critically dependent upon direct association between VP16 and vhs. Interestingly, although recovery of 8MA infectious virus by complementation with VP16(L344A) was greater than 3 orders of magnitude less than with wild-type VP16, it was still above that observed with the empty vector control. The reason(s) for this is unclear, but may reflect a residual ability of VP16(L344A) to associate with vhs in the infected cell.
In conclusion, the results presented here demonstrate that VP16 serves to block vhs activity through a direct interaction, an association that is ultimately required for virus production. The finding that the binding interfaces on VP16 for interaction with vhs, HCF-1, and Oct-1 are in close proximity suggests that vhs may also serve to modulate VP16 function. Consistent with this, we have previously shown that vhs can interfere with VIC assembly (48). However, this was only observed if vhs was first preincubated with VP16, as vhs did not interfere with VIC formation in the presence of Oct-1 and HCF-1. Moreover, overexpression of vhs does not interfere with transactivation by VP16 in vivo in transient assays (unpublished observations). The foregoing, coupled with the fact that VP16 is in vast excess over vhs in the virus and in the infected cell and that vhs is a nonessential viral protein, argues that vhs does not modulate VP16 activity. The mechanism(s) by which VP16 attenuates vhs activity remains to be addressed. It is possible that direct interaction of VP16 with vhs simply serves to direct vhs into the viral assembly pathway, consistent with findings that vhs derivatives that lack determinants necessary for association with VP16 are not packaged into virus (40). This hypothesis is further supported by our observation that VP16 colocalizes vhs to the nuclear periphery, perhaps downregulating vhs through changes in subcellular localization and/or promoting packaging of vhs within the viral tegument (J. Inglis, J. Knez, and J. P. Capone, unpublished observations). Alternatively, VP16 may directly affect the catalytic function of vhs and/or its associated cellular proteins (12, 30). The recent availability of systems that recapitulates vhs-mediated endoribonucleolytic activity in vitro may provide a means to directly test this possibility (50). However, an effect of VP16 on vhs activity in vitro has not been reported.
Finally, the results presented here, in conjunction with our earlier findings that VP16 targets a small, defined region in vhs, may provide an intriguing avenue for the identification of small molecule inhibitors of VP16/vhs complex formation that could be of potential use as antiviral therapeutics.
Acknowledgments
We thank W. Herr for plasmid pCGNHCF; J. Smiley for HSV-1 viral stocks of 8MA, 8MAΔSma, and 16-8 cells; P. Chevray for plasmids pPC97, pPC86, and yeast strain PCY2; and D. Piluso for constructing pα4luc, pCDBHCF380, and pGEM7Z-HCF902.
This work was supported in part by a grant from the National Cancer Institute of Canada.
REFERENCES
- 1.Ace, C. I., M. A. Dalrymple, F. H. Ramsey, V. O. Preston, and C. M. Preston. 1988. Mutational analysis of the herpes simplex virus type I trans-inducing factor Vmw65. J. Gen. Virol. 69:2595-2605. [DOI] [PubMed] [Google Scholar]
- 2.Ace, C. I., T. A. McKee, J. M. Ryan, J. Cameron, and C. M. Preston. 1989. Construction and characterization of a herpes simplex virus type I mutant unable to transinduce immediate-early gene expression. J. Virol. 63:2260-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1990. Current protocols in molecular biology. Wiley, New York, N.Y.
- 4.Babb, R., C. C. Huang, D. J. Aufiero, and W. Herr. 2001. DNA recognition by the herpes simplex virus transactivator VP16: a novel DNA binding structure. Mol. Cell. Biol. 21:4700-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, M. E. M., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate-early gene transcription. J. Mol. Biol. 180:1-19. [DOI] [PubMed] [Google Scholar]
- 6.Chevray, P. M., and D. Nathans. 1992. Protein interaction cloning in yeast: Identification of mammalian proteins that react with the leucine zipper of Jun. Proc. Natl. Acad. Sci. USA 89:5789-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou, P. Y., and G. D. Fasman. 1978. Empirical predictions of protein conformation. Annu. Rev. Biochem. 547:251-276. [DOI] [PubMed] [Google Scholar]
- 8.Creighton, T. E. 1983. Proteins: structures and molecular principles. W. H. Freeman and Co., New York, N.Y.
- 9.Elbe, R. 1992. A simple and efficient procedure for transformation of yeasts. BioTechniques 13:18-20. [PubMed] [Google Scholar]
- 10.Elgadi, M. M., C. E. Hayes, and J. R. Smiley. 1999. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J. Virol. 73:7153-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Estojak, J., R. Brent, and E. A. Golemis. 1995. Correlation of two-hybrid affinity data with in vitro measurements. Mol. Cell. Biol. 15:5820-5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng, P., D. N. Everly, Jr., and G. S. Read. 2001. mRNA decay during herpesvirus infections: interaction between a putative viral nuclease and a cellular translation factor. J. Virol. 75:10272-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenwick, M. L., and S. A. Owen. 1988. On the control of immediate early (alpha) mRNA survival in cells infected with herpes simplex virus. J. Gen. Virol. 69:2869-2877. [DOI] [PubMed] [Google Scholar]
- 14.Greaves, R., and P. O'Hare. 1989. Separation of the requirements for protein-DNA complex assembly from those for functional activity in the herpes simplex virus regulatory protein Vmw65. J. Virol. 63:1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haigh, A., R. Greaves, and P. O'Hare. 1990. Interference with the assembly of a virus-host transcription complex by peptide competition. Nature (London) 344:257-259. [DOI] [PubMed] [Google Scholar]
- 16.Hayes, S., and P. O'Hare. 1993. Mapping a major surface-exposed site in herpes simplex virus protein Vmw65 to a region of direct interaction in a transcription complex assembly. J. Virol. 67:852-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herr, W. 1998. The herpes simplex virus VP16-induced complex: mechanisms of combinatorial transcriptional regulation. Cold Spring Harbor Symp. Quant. Biol. 63:599-607. [DOI] [PubMed] [Google Scholar]
- 18.Herr, W., and M. A. Cleary. 1995. The POU domain: versatility in transcriptional regulation by a flexible two-in-one DNA binding domain. Genes Dev. 9:1679-1693. [DOI] [PubMed] [Google Scholar]
- 19.Hughes, T. A., S. LaBoissiere, and P. O'Hare. 1999. Analysis of functional domains of host cell factor involved in VP16 complex formation. J. Biol. Chem. 274:16437-16443. [DOI] [PubMed] [Google Scholar]
- 20.Karr, B. M., and G. S. Read. 1999. The virion host shutoff function of herpes simplex virus degrades the 5′ end of a target mRNA before the 3′ end. Virology 264:195-204. [DOI] [PubMed] [Google Scholar]
- 21.Katan, M., A. Haigh, C. P. Verrijzer, P. C. van der Vliet, and P. O'Hare. 1990. Characterization of a cellular factor which interacts functionally with Oct-1 in the assembly of a multicomponent transcription complex. Nucleic Acids Res. 18:6871-6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kristie, T. M., and P. A. Sharp. 1993. Purification of the cellular C1 factor required for the stable recognition of the Oct-1 homeodomain by the herpes simplex virus alpha-transinduction factor (VP16). J. Biol. Chem. 268:6525-6534. [PubMed] [Google Scholar]
- 23.Kristie, T. M., and P. A. Sharp. 1990. Interactions of the Oct-1 POU subdomains with specific DNA sequences and with the HSV alpha-trans-activator protein. Genes Dev. 4:2383-2396. [DOI] [PubMed] [Google Scholar]
- 24.Kwong, A. D., and N. Frenkel. 1987. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc. Natl. Acad. Sci. USA 84:1926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwong, A. D., J. A. Kruper, and N. Frenkel. 1988. Herpes simplex virus virion host shutoff function. J. Virol. 62:912-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaBoissiere, S., T. Hughes, and P. O'Hare. 1999. HCF-dependent nuclear import of VP16. EMBO J. 18:480-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai, J.-S., and W. Herr. 1997. Interdigitated residues within a small region of VP16 interact with Oct-1, HCF and DNA. Mol. Cell. Biol. 17:3937-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lam, Q., C. A. Smibert, K. E. Koop, C. Lavery, J. P. Capone, S. P. Weinheimer, and J. R. Smiley. 1996. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 15:2575-2581. [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, Y., W. Gong, C. C. Huang, W. Herr, and X. Cheng. 1999. Crystal structure of the conserved core of the herpes simplex virus transcriptional regulatory protein VP16. Genes Dev. 13:1692-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, P., F. E. Jones, H. A. Saffran, and J. R. Smiley. 2001. Herpes simplex virus virion shutoff protein requires a mammalian factor for efficient in vitro endoribonuclease activity. J. Virol. 75:1172-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyata, K. S., S. E. McCaw, H. V. Patel, R. A. Rachubinski, and J. P. Capone. 1996. The orphan nuclear hormone receptor LXRα interacts with the peroxisome proliferator-activated receptor and inhibits peroxisome proliferator signaling. J. Biol. Chem. 271:9189-9192. [DOI] [PubMed] [Google Scholar]
- 32.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2001. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Hare, P., and C. R. Goding. 1988. Herpes simplex virus regulatory elements and the immunoglobulin octamer domain bind a common factor and are both targets for virion transactivation. Cell 52:435-445. [DOI] [PubMed] [Google Scholar]
- 34.O'Hare, P., C. R. Goding, and A. Haigh. 1988. Direct combinatorial interaction between a herpes simplex virus regulatory protein and a cellular octamer-binding factor mediates specific induction of virus immediate-early gene expression. EMBO J. 7:4231-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oroskar, A. A., and G. S. Read. 1989. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J. Virol. 63:1897-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poon, A. P., and B. Roizman. 1995. The phenotype in vitro and in infected cells of herpes simplex virus 1 alpha trans-inducing factor (VP16) carrying temperature sensitive mutations introduced by substitution of cysteines. J. Virol. 69:7658-7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Popova, B., P. Bilan, P. Xiao, M. Faught, and J. P. Capone. 1995. Transcriptional activation by DNA-binding derivatives of HSV1 VP16 that lack their carboxyl-terminal acidic activation domain. Virology 109:19-28. [DOI] [PubMed] [Google Scholar]
- 38.Preston, C. M., M. C. Frame, and M. E. M. Campbell. 1988. A complex formed between cell components and an HSV structural polypeptide binds to a viral immediate early gene regulatory DNA sequence. Cell 52:425-434. [DOI] [PubMed] [Google Scholar]
- 39.Read, G. S., and N. Frenkel. 1983. Herpes simplex virus mutants defective in the virion associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of alpha (immediate early) viral polypeptides. J. Virol. 46:498-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Read, G. S., B. M. Karr, and K. Knight. 1993. Isolation of a herpes simplex virus type I mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J. Virol. 67:7149-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 42.Sadowski, I., J. Ma, S. Triezenberg, and M. Ptashne. 1988. GAL4-VP16 is an unusually potent transcriptional activator. Nature (London) 334:563-564. [DOI] [PubMed] [Google Scholar]
- 43.Schek, N., and S. L. Bachenheimer. 1985. Degradation of the cellular mRNAs induced by a virion-associated factor during herpes simplex virus infection of Vero cells. J. Virol. 55:601-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmelter, J., J. Knez, J. R. Smiley, and J. P. Capone. 1996. Identification and characterization of a small modular domain in the herpes simplex virus host shutoff protein sufficient for interaction with VP16. J. Virol. 70:2124-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaw, P., J. Knez, and J. P. Capone. 1995. Amino acid substitutions in the herpes simplex virus transactivator VP16 uncouple direct protein-protein interaction and DNA binding from complex assembly and transactivation. J. Biol. Chem. 270:29030-29037. [DOI] [PubMed] [Google Scholar]
- 46.Simmen, K. A., A. Newell, M. Robinson, J. S. Mills, G. Canning, R. Handa, K. Parkes, N. Borkakoti, and R. Jupp. 1997. Protein interactions in the herpes simplex virus type 1 VP16-induced complex: VP16 peptide inhibition and mutational analysis of host cell factor requirements. J. Virol. 71:3886-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smibert, C. A., D. C. Johnson, and J. R. Smiley. 1992. Identification and characterization of the virion-induced shutoff product of herpes simplex virus gene UL41. J. Gen. Virol. 73:467-470. [DOI] [PubMed] [Google Scholar]
- 48.Smibert, C. A., B. Popova, P. Xiao, J. P. Capone, and J. R. Smiley. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68:2339-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smiley, J. R., and J. Duncan. 1997. Truncation of the C-terminal acidic transcriptional activation domain of herpes simplex virus VP16 produces a phenotype similar to that of the in1814 linker insertion mutation. J. Virol. 71:6191-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smiley, J. R., M. M. Elgadi, and H. A. Saffran. 2001. Herpes simplex virus vhs protein. Methods Enzymol. 342:440-451. [DOI] [PubMed] [Google Scholar]
- 51.Spector, D., F. Purves, and B. Roizman. 1991. Role of α-transinducing factor (VP16) in the induction of alpha genes within the context of viral genomes. J. Virol. 65:3504-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strom, T., and N. Frenkel. 1987. Effects of herpes simplex virus on mRNA stability. J. Virol. 61:2198-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Triezenberg, S. J., R. C. Kingsbury, and S. L. McKnight. 1988. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 2:718-729. [DOI] [PubMed] [Google Scholar]
- 54.Weinheimer, S. P., B. A. Boyd, S. K. Durham, J. L. Resnick, and D. R. O'Boyle II. 1992. Detection of the VP16 open reading frame of herpes simplex virus type I. J. Virol. 66:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weir, J. P. 2001. Gene expression in herpes simplex virus. Gene 271:117-130. [DOI] [PubMed] [Google Scholar]
- 56.Werstuck, G., and J. P. Capone. 1989. Mutational analysis of the herpes simplex virus immediate-early trans-inducing factor Vmw65. Gene 75:213-224. [DOI] [PubMed] [Google Scholar]
- 57.Wilson, A. C., K. LaMarco, M. G. Peterson, and W. Herr. 1993. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell 74:115-125. [DOI] [PubMed] [Google Scholar]
- 58.Wu, T.-J., G. Monokian, D. F. Mark, and C. R. Wobbe. 1994. Transcriptional activation by herpes simplex virus type I VP16 in vitro and its inhibition by oligopeptides. Mol. Cell. Biol. 14:3484-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiao, P., and J. P. Capone. 1990. A cellular factor binds to the herpes simplex virus type I transactivator Vmw65 and is required for Vm65-dependent protein-DNA complex assembly with Oct-1. Mol. Cell. Biol. 10:4974-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]