Abstract
We describe a set of RNA molecules that work as transcriptional activators when tethered to DNA. These RNA activating regions were found amongst a randomized set of molecules bearing variants of a 10 nt loop attached to an RNA stem. The various RNA activating regions all bear an identical five- residue sequence with an interspersed sixth residue. The result shows that although all natural activating regions characterized thus far are peptidic, this function can be served by other kinds of moieties as well.
INTRODUCTION
The typical eukaryotic transcriptional activator bears two domains, one for DNA binding and another called an activating region (1,2). Many peptides work as activating regions when tethered to DNA. Members of the largest natural class of activating regions found in yeast, which also work in higher eukaryotes, contain an excess of acidic residues with crucial interspersed hydrophobic residues (1–7). New acidic activating regions have been found encoded in random bits of Escherichia coli DNA, and others have been created by peptide synthesis (8–10). A few non-acidic activating regions that work strongly in yeast (but not in higher eukaryotes) were isolated among randomly generated octameric peptides attached to a DNA-binding domain (11).
A variety of lines of evidence indicate that DNA-tethered activating regions contact, and thereby recruit, the transcriptional machinery to nearby promoters. Evidently, different activating regions interact with the transcriptional machinery in different ways (i.e. touch it in different places) to effect recruitment and hence activation (12). There is, in principle, no expectation that an activating region must comprise a peptide: any DNA-tethered compound that interacts with the machinery (directly or indirectly) with the appropriate affinity (and within certain limited stereo-specific constraints) could work as an activating region. Indeed, the finding that many RNA sequences (called aptamers) can be selected to bind specific proteins (13–17), including components of the transcriptional machinery, suggested to us that certain RNA sequences, tethered to DNA, might work as activating regions.
Here we describe a class of RNA molecules that, when tethered to DNA, activate transcription with modest strengths in yeast. Our experimental strategy was to isolate a minimal RNA sequence that can function as a transcriptional activator. We screened a variable loop of 10 nt that was presented on top of a well-characterized and often used stem-forming sequence. The strategy was based on the observation that many of the nucleic acid aptamers that have been characterized so far are overwhelmingly stem–loop-forming structures. In contrast to many of the in vitro aptamer selections, we chose a smaller library so that the screen could cover all the possible variants of the library. A 30–70 nt variable region would perhaps have yielded more potent activators. However, covering all the variants would have been impossible considering the combinatorial enormity of the library.
The recovered activators maintained the common stem and bore loop sequences closely related to one another. The activity of these activators (where tested) was changed only modestly by attaching them to heterologous stems. As this work was in progress, we learned of the isolation of RNA molecules (as a byproduct of a search for specific protein-binding RNA molecules) that activate transcription (18). We compare our activators with theirs (see Discussion).
MATERIALS AND METHODS
Construction of the 10 nt random library
An oligonucleotide, called ‘Oligo 2’, was synthesized to contain a 15 nt stem (U1) with a 10 nt loop. The loop part of the oligonucleotide was randomized. An additional 50 nt vector (pIII/MS2) (19) sequence was added to both sides of the stem so that the PCR-amplified Oligo2 can recombine (gap repair) (20) in yeast with linearized vector plasmid. Oligo2 had the sequence CTCTGGGAGCTGCGATTGGCAGAAT TCCGGCTAGAACTAGTGGATCCCCCGGGCGAGGCT TATCCNNNNNNNNNNGGATGTGCTGACCCCGGGC AGCTTGCATGCCTGCAGGTCGACTCTAGAAAACAT GAGGATCACCC. Two other oligonucleotides, ‘Oligo1’ and ‘Oligo3’ with the sequences CTCTGGGAGCTGCG ATTGGC and GGGTGATCCTCATGTTTTCT are used to PCR amplify the stem–loop-forming Oligo2. Oligo2 was amplified in 10 simultaneous PCRs utilizing 72 µl of water, 10 µl of 10× PCR buffer (Promega), 2 µl of nucleotide mix (10 nM each), 3 µl of Oligo1 (500 ng/µl), 3 µl of Oligo3 (500 ng/µl), 5 µl of Oligo 2 (50 ng/µl) and 1 µl of Taq DNA polymerase (5 U).
RNA activator screening
L40-coat expressing yeast cells (19) were grown in rich YPDA media to make competent cells according to Sherman et al. (21). A 40 µg aliquot of linearized (SmaI-cleaved) pIII/MS2 plasmid (19) and 20 µg of PCR-amplified DNA library were introduced into yeast cells as gap-repaired (20) replicating plasmids. Cells were plated on solid selective medium [His–, Ura3–, 5–10 mM 3-aminotriazole (3-AT)] and grown at 30°C until colonies appeared. Approximately 200 colonies were picked up from the primary screen and tested for β-galactosidase gene expression by looking at the colony color on X-gal-containing plates. Yeast colonies that turned blue were used to isolate (gap-repaired pIII/MS2) plasmids for further tests. Fifteen plasmids were tested again with the L40-coat strain, and only seven were linked to the activations of the reporter genes. A control experiment was done without the 3-AT to determine the transformation efficiency. Around 10 million yeast colonies were screened to yield eight RNA activators.
β-Galactosidase activity
β-Galactosidase activity was assayed as described in Himmelfarb et al. (22), and each assay was performed in triplicate. The sequences of RNA activators were determined by sequencing the stem–loop part of the plasmids. Oligo 1 and Oligo 3 were used to sequence both strands of the plasmids. A Sequenase kit (version 2.0) was purchased from USB. Activating regions of activators 2 and 8 were identical, and only activator 2 was used in the work.
Yeast strains/plasmids
L40-coat and TAT-7 yeast strains were generous gift from Dr Marvin Wickens and Dr Stenglanz. Plasmid pIII/MS-2 was also obtained from Dr Marvin Wickens. LexA-Gal4 and LexA-Gal11 are described in Himmelfarb et al. (22). LexA-Taf23 was obtained from the Strubin laboratory. LexA-yTBP is described in Chatterjee and Struhl (23). Plasmids used in Figure 5 were constructed by annealing oligos of appropriate sequences (shown in the figure) and ligating with XmaI-linearized plasmid pIII/MS-2. All plasmids were sequenced to determine their sequence and orientation.
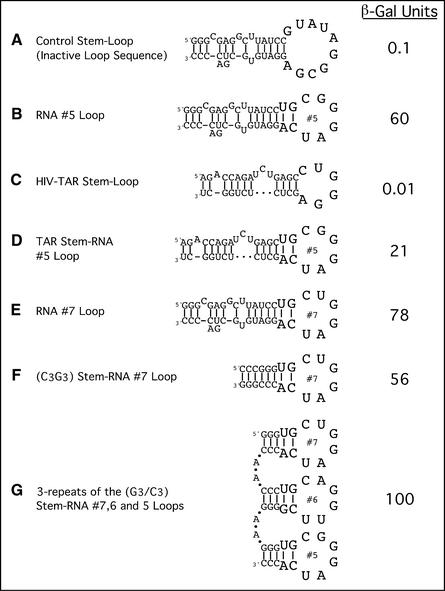
Figure 5.
The activator loop is transferable. Different nucleotide stems were used to carry the activator loops from either RNA activator nos 7, 6 or 5. Multiple loops on multiple stems are also tested for transcriptional activity. Stem–loop structures are described in the figure as (A–G).
Primer extension
Primer extension analysis of the His3 promoter was performed according to Ma and Ptashne (1). The primer that was used to analyze the His3 transcripts is identical in sequence to that used in Chen and Struhl (24). An internal control (Pgk1) (25) was also used in all reactions to probe that the reactions contained equal amounts of RNA and that its general transcriptional levels were not affected by LexA-fused activators. Total RNA was extracted from yeasts containing appropriate LexA-fused activators and control vector using a standard extraction protocol. Equal amounts (20 µg) of RNA were used for the analysis.
RESULTS
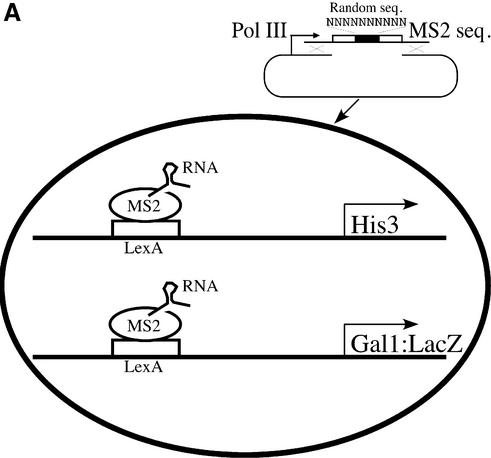
Figure 1A shows the method used to isolate RNA activating regions. The yeast strain contains two reporters, one expressing His3 (from the His3 promoter) and the other β-galactosidase (from the Gal1 promoter). Each reporter bears LexA-binding sites upstream of the respective transcriptional start site. The strain also expresses a hybrid protein comprising LexA fused to the coat protein of the RNA bacteriophage MS2. This fusion protein, LexA-MS2, binds to the LexA sites on DNA as indicated. The MS2 coat protein, in turn, binds a specific and well-characterized RNA sequence (26,27), also as indicated.
Figure 1.
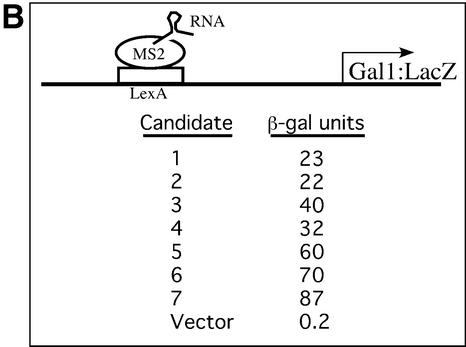
Schematic of the RNA activator screen. (A) A yeast strain with an integrated LexA-MS2 and His3 and LacZ reporters carrying upstream LexA sites was used to introduce gap-repaired plasmids to express MS-2 hybrid RNAs. The DNA library containing the well-characterized stem (open bars) and 10 nt long randomized loop sequence (black bar) was PCR amplified with an additional 50 nt sequence (black lines) on both the 5′ and 3′ sides of the library DNA so that the resulting fragments could recombine with the linearized plasmid vector. Recombined plasmids were expected to express hybrid RNAs that could bind to LexA operator sequences through the LexA-MS2 protein and have RNA sequences that should possess activator motifs. Transformed yeast colonies that grew on 5 and 10 mM 3-AT-containing media and turned blue on X-gal plates were selected as potential candidates. The screen was modeled on the three-hybrid system developed by SenGupta et al. (19). We simply omitted the ‘third hybrid’ (a protein bearing an attached transcriptional activating region), and thereby screened for RNA molecules that activate ‘directly’. (B) Plasmid linkage test and strength of the RNA activating regions. Plasmids from the potential candidates were extracted and tested for their ability to activate β-galactosidase gene-bearing LexA operators.
A library of RNA molecules, each 140 nt long, was expressed in yeast as indicated in Figure 1A. The RNA molecules, encoded in genes transcribed by RNA polymerase III, are neither capped nor polyadenylated and thus are thought not to be translated. Each of these RNA molecules bears two copies of a sequence that binds the MS2 coat protein and, at its other end, a sequence known to form a stem–loop structure with 10 residues in the loop. The stem sequence resembles the second stem of U1 RNA and forms a stable structure in vivo as well as in vitro (28,29). We chose a 10 nt loop as this is among the longest sequences found in naturally occurring RNA loops (including that in U1), and it offers approximately one million different sequence combinations. DNA encoding the 10 unpaired residues in the loop was randomized, and RNA activating regions were sought by picking colonies that grew on 5–10 mM 3-AT and that also expressed β-galactosidase as judged by colony color. (Growth on 3-AT requires expression of the His3 gene, and the appropriate colony color is conferred by expression of the LacZ gene.)
From 107 colonies screened, seven were found to contain RNA molecules that upon re-testing proved to be activating regions (Fig. 1B). The strengths of these varied, the strongest (no. 7 in the figure) eliciting a level of expression of β-galactosidase some 400-fold above the background. In no case was activation observed in the absence of LexA-MS2 or when the corresponding ‘antisense’ RNA was expressed (not shown). Activation was also not dependent on the identity of the DNA-binding domain used to tether the RNA activators. The various RNAs in Figure 1B were re-tested in a strain that expressed a hybrid protein bearing the Gal4 DNA-binding domain fused to the MS2 coat protein, and that had an integrated LacZ reporter gene regulated by five Gal4-binding sites. The relative activator strength of each of the seven RNA molecules in this strain was similar to that shown in Figure 1B for the LexA-MS2-containing strain (data not shown).
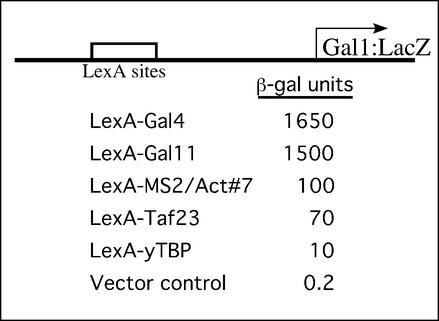
The β-galactosidase expression levels shown in Figure 2 indicate that the RNA activating region designated no. 7, tethered to DNA by interaction with Lex-MS2, worked (i.e. elicited transcription) 10-fold more efficiently than LexA-TBP did, somewhat better than LexA-Taf23, and ∼10- to 15-fold less well than LexA-Gal11 or LexA-Gal4. The latter two are among the strongest activators in yeast. Each of these hybrids, except for LexA-Gal4, comprises a DNA-binding domain fused to a component of the transcriptional machinery, and is sometimes called a ‘non-classical’ activator (12). Parallel experiments showed that activator no. 7 also activated transcription of reporters bearing the His4, Cyc1 and Pho5 core promoters with LexA-binding sites located upstream (unpublished data).
Figure 2.
Comparison of the strength of an RNA activator with LexA fusions of classical (Gal4) and non-classical (components of the transcriptional machinery) activators.
Figure 3 shows an mRNA primer extension analysis of the transcripts elicited by LexA-Gal11 and RNA no. 7 (bound to LexA-MS2 coat protein). This experiment was done to investigate whether RNA activators would activate transcription from the previously defined His3 start sites. These major start sites map at +1 and +12, and a minor site maps at +22 (24). As seen in Figure 3, the major start site for transcription stimulated by RNA no.7 is at +1, whereas the stronger activator LexA-Gal11 stimulated transcription from all three sites. The relative level of transcript in the two cases is consistent with the relative levels of growth on 3-AT-containing media. Cells containing RNA no. 7 grew on 50 mM 3-AT media, whereas LexA-Gal11 grew on media containing >100 mM 3-AT (data not shown). The mRNA levels of Pgk1 (25), a housekeeping gene without any LexA-binding sites in its promoter, were unchanged in cells expressing LexA-Gal11 or RNA no. 7.
Figure 3.
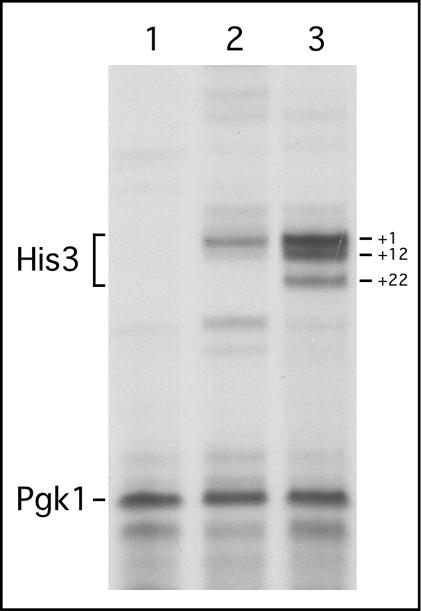
Primer extension analysis of the His3 promoter-bearing LexA operators. Primers from His3 and Pgk1 (internal control) were used to probe the transcriptional activities from His3 and Pgk1 promoters, respectively. Primer extension products are labeled. Major and minor transcriptional initiation sites (+1, +12 and +22) of the His3 gene are also labeled. Lanes 1, 2 and 3 show primer extension products from a vector (stem with an inactive loop), LexA-MS2-bound RNA activator no. 7, and LexA-Gal11, respectively.
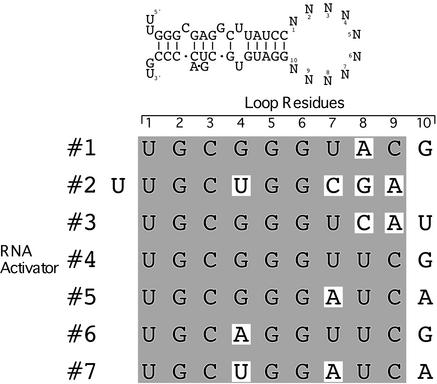
As shown in Figure 4, the seven activating regions have a strong six base consensus sequence; all were identical in this stretch except for three (nos 2, 6 and 7), which bear variants at position 4. The consensus begins in all cases except one at position 1 of the loop; the exception is no. 2, in which the consensus begins at position 2 of the loop. If one takes into account the formation of G:U base pairs in RNA, five of these activating regions (nos 1, 4, 5, 6 and 7) have the potential to form stems extended by 2 bp, whereas the no. 2 stem can be extended by 5 bp, forming a hairpin lacking a loop.
Figure 4.
Nucleotide sequence alignment of loops from RNA activators. DNA sequences from the sense strand of the loops were determined and, when aligned, they revealed an activator consensus sequence: UGC(G>U>A)GG(U>A>C)(U>ACG)(C>A)(G>A>U). The first residue in each alignment is the N1 residue of the loop, whereas the last residue is N10. The RNA activating region of no. 2 is aligned with the rest of the activating regions, with a one residue shift to the left with respect to other activating regions. The shaded sequence represents the activator consensus sequence.
Figure 5 shows that the activating function can be conferred on a heterologous RNA stem by fusing to that stem the appropriate loop sequence. Thus the stem–loop structure comprising TAR RNA (the site on HIV mRNA that binds the protein TAT) activated when its loop was replaced with that from activator no. 5 (compare Fig. 5C and D). The activity of this hybrid RNA is some 3-fold lower than that of the intact activator no. 5 (compare Fig. 5D and B).
Figure 5E versus F shows that the sequence of the stem of activator RNA no. 7 can be replaced with a string of G and C residues, with only modest loss of activity. Figure 5E and G shows that three contiguous iterations of a stem–loop structure bearing loops from RNA activators nos 5, 6 and 7 modestly increase activity compared with the case of the single stem–loop structure.
DISCUSSION
Our results show that certain RNA molecules, tethered to DNA, can work as activating regions in yeast. All of the functional sequences we found bore a 6 nt consensus, with five of the bases being invariant. These sequences were found within a 10 nt sequence that was attached to a double-stranded stem and tethered to DNA. The limited set of sequences we recovered suggests that the RNA activators see a limited set of target proteins, perhaps just one. What that target might be is not known; the finding that these molecules bind TBP (S. Saha, unpublished) is consistent with the fact that many acidic peptidic activating regions have been found to interact with TBP, but the finding alone does not establish that the interaction is functional in vivo. Our strongest RNA activator (RNA no. 7), when tethered to DNA, works ∼7–10% as efficiently as does the strongest known yeast activator, Gal4. Our primer extension study shows that RNA activator no. 7 works at the level of transcription and is able to stimulate transcription initiating from a previously characterized start site.
The RNA activators described by SenGupta et al. (18) were detected in a screen essentially identical to the one we use here, except that their RNA library was unconstrained for sequence or size. Members of our library, as we have described, were designed to all have a common stem and loop structure. Their RNA molecules do not show a consensus, are of varying size, and the most powerful of those (called TA1) works some 5-fold less efficiently than does the best activator (RNA no. 7) we recovered (S. Saha, unpublished). It is possible that these activators may interact with different target proteins or with different surfaces of the same protein.
It might seem surprising that we isolated such a restricted set of RNA sequences that work as activating regions in view of the finding that there are so few constraints on peptide sequences with that function. Perhaps the stem constrains the rather short (10 nt) loop such that a very limited set of sequences can find a physiologically relevant target. The stem–loop sequence we examined was used previously by Keene and co-workers to find RNA epitopes that interact with a specific antibody (30). In that case, 45 clones were isolated, each of which bore an identical 6 nt sequence. That sequence, when tested by us, did not work as an activating region (data not shown).
We searched the yeast genome database to identify yeast transcripts that naturally contain the RNA activator consensus sequence. Twenty-seven transcribed regions that encode a 10 nt region identical to one or another of our RNA activators were found. Twelve of these naturally occurring yeast sequences are identical to our strongest activator (RNA no. 7). In contrast, the sequences matching the other six RNA activators were found only two or three times. We do not know whether any of those RNA molecules are involved in initiation or elongation of the transcript.
A role for RNA molecules in gene regulation is, of course, not without precedent. The TAR sequence in a nascent transcript of HIV recruits the HIV protein TAT, which in turn recruits cyclin-dependent kinases that stimulate transcript elongation (31). RNA molecules that serve as co-activators in transcriptional initiation have been found for steroid receptors as well (32). These are thought to function as structural components of a co-activator complex of which the acetyl transferase SRC-1 is a member.
Nucleic acid aptamers have been isolated that bind various components of the gene expression machinery, but to our knowledge have only been used to inhibit one or another step in expression (16,17). We do not know whether RNA aptamers that bind pre-selected components of the transcriptional machinery [e.g. those of Sentenac and co-workers (17)] would work as transcriptional regulators when tethered to DNA.
The first three conserved residues UGC of our RNA activators are identical to the sequences found in two loops of U1 RNA and to a loop in U2 RNA. Each of these loops binds specific proteins that contain so-called RNA recognition motifs (RRMs) (33,34). In each case, a β-hairpin penetrates the RNA loop, and makes common interactions with the conserved triplet. Interactions with bases 3′ to the conserved triplet then further determine specificity. Whether our RNA activators interact with an RRM in a target(s) in a similar fashion remains to be determined. Nonetheless, we show that non-peptidic molecules can serve as activating regions.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Marvin Wickens and Dr Stenglanz for yeast strains (L40-coat and TAT-7) and plasmids (pIII/MS-2, LexA-MS-2 and Gal4-MS2), Drs Michael Green, Kevin Struhl, Grace Gill and Zaf Zaman for plasmids and reagents, Dennis Underwood for helping us with RRM motifs in yTBP, William Donahue for the initial gene bank search for activator sequences, Angie You for sequencing help, and Dr Terrell Gibbs for helping with the figures. This work was supported by grants from the National Institutes of Health to M.P. and K.J.
REFERENCES
- 1.Ma J. and Ptashne,M. (1987) Deletion analysis of Gal4 defines two transcriptional activating segments. Cell, 48, 847–853. [DOI] [PubMed] [Google Scholar]
- 2.Hope I. and Struhl,K. (1986) Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell, 46, 885–894. [DOI] [PubMed] [Google Scholar]
- 3.Ma J., Przibilla,E., Hu,J., Bogorad,L. and Ptashne,M. (1988) Yeast activators stimulate plant gene expression. Nature, 334, 631–633. [DOI] [PubMed] [Google Scholar]
- 4.Kakidani H. and Ptashne,M. (1988) Gal4 activates gene expression in mammalian cells. Cell, 52, 161–167. [DOI] [PubMed] [Google Scholar]
- 5.Cress W.D. and Triezenberg,S.J. (1991) Critical structural elements of the VP16 transcriptional activation domain. Science, 251, 87–90. [DOI] [PubMed] [Google Scholar]
- 6.Drysdale C.M., Duenas,E., Jackson,B.M., Reusser,U., Braus,G.H. and Hinnebusch,A.G. (1995) The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol. Cell. Biol., 15, 1220–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan S.M., Horn,P.J., Olson,V.A., Koop,A.H., Niu,W., Ebright,R.H. and Triezenberg,S.J. (1998) Mutational analysis of a transcriptional activation region of the VP16 protein of herpes simplex virus. Nucleic Acids Res., 26, 4487–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma J. and Ptashne,M. (1987) A new class of yeast transcriptional activators. Cell, 51, 113–119. [DOI] [PubMed] [Google Scholar]
- 9.Giniger E. and Ptashne,M. (1987) Transcription in yeast activated by a putative amphipathic alpha helix linked to a DNA binding unit. Nature, 330, 670–672. [DOI] [PubMed] [Google Scholar]
- 10.Gerber H.P., Seipel,K., Georgiev,O., Hofferer,M., Hug,M., Russconi,S. and Schaffner,W. (1994) Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science, 263, 808–811. [DOI] [PubMed] [Google Scholar]
- 11.Lu X., Ansari,A.Z. and Ptashne,M. (2000) An artificial transcriptional activating region with unusual properties. Proc. Natl Acad. Sci. USA, 97, 1988–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ptashne M. and Gann,A. (1997) Transcriptional activation by recruitment. Nature, 386, 569–577. [DOI] [PubMed] [Google Scholar]
- 13.Patel D.J. and Suri,A.K. (2000) Structure, recognition and discrimination in RNA aptamer complexes with cofactors, amino acids, drugs and aminoglycoside antibiotics. J. Biotechnol., 74, 39–60. [DOI] [PubMed] [Google Scholar]
- 14.Hermann T. and Patel,D.J. (2000) Adaptive recognition by nucleic acid aptamers. Science, 287, 820–825. [DOI] [PubMed] [Google Scholar]
- 15.Zhai G., Iskandar,M., Barilla,K. and Romaniuk,P.J. (2001) Characterization of RNA aptamer binding by the Wilms’ tumor suppressor protein WT1. Biochemistry, 40, 2032–2040. [DOI] [PubMed] [Google Scholar]
- 16.Shi H., Hoffman,B.E. and Lis,J.T. (1999) RNA aptamers as effective protein antagonists in a multicellular organism. Proc. Natl Acad. Sci. USA, 96, 10033–10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas M., Chedin,S., Carles,C., Riva,M., Famulok,M. and Sentenac,A. (1997) Selective targeting and inhibition of yeast RNA polymerase II by RNA aptamers. J. Biol. Chem., 272, 27980–27986. [DOI] [PubMed] [Google Scholar]
- 18.SenGupta D.J., Wickens,M. and Fields,S. (1999) Identification of RNAs that bind to a specific protein using the three-hybrid system. RNA, 5, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.SenGupta D.J., Zhang,B., Kraemer,B., Pochart,P., Fields,S. and Wickens,M. (1996) A three-hybrid system to detect RNA–protein interactions in vivo. Proc. Natl Acad. Sci. USA, 93, 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rothstein R. (1991) Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol., 194, 281–301. [DOI] [PubMed] [Google Scholar]
- 21.Sherman F., Fink,G.R. and Hicks,J.B. (1986) Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Himmelfarb H.J., Pearlberg,J., Last,D.H. and Ptashne,M. (1990) GAL11P: a yeast mutation that potentiates the effect of weak GAL4-derived activators. Cell, 63, 1299–1309. [DOI] [PubMed] [Google Scholar]
- 23.Chatterjee S. and Struhl,K. (1995) Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature, 374, 820–822. [DOI] [PubMed] [Google Scholar]
- 24.Chen W. and Struhl,K. (1985) Yeast mRNA initiation sites are determined primarily by specific sequences, not by the distance from the TATA element. EMBO J., 4, 3273–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Decker C.J. and Parker,R. (1993) A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev., 7, 1632–1643. [DOI] [PubMed] [Google Scholar]
- 26.Witherell G.W., Gott,J.M. and Uhlenbeck,O.C. (1991) Specific interaction between RNA phage coat proteins and RNA. Prog. Nucleic Acid Res. Mol. Biol., 40, 185–220. [DOI] [PubMed] [Google Scholar]
- 27.Stockley P.G., Stonehouse,N.J., Murray,J.B., Goodman,S.T., Talbot,S.J., Adams,C.J.L.L. and Valegard,K. (1995) Probing sequence-specific RNA recognition by the bacteriophage MS2 coat protein. Nucleic Acids Res., 23, 2512–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Venrooij W.J., Hoet,R., Castrop,J., Hageman,B., Mattaj,I.W. and van de Putte,L.B. (1990) Anti- (U1) small nuclear RNA antibodies in anti-small nuclear ribonucleoprotein sera from patients with connective tissue diseases. J. Clin. Invest., 86, 2154–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein Gunnewiek J.M., van de Putte,L.B. and van Venrooij,W.J. (1997) The U1 snRNP complex: an autoantigen in connective tissue diseases. An update. Clin. Exp. Rheumatol., 15, 549–560. [PubMed] [Google Scholar]
- 30.Tsai D.E., Kenan,D.J. and Keene,J.D. (1992) In vitro selection of an RNA epitope immunologically cross-reactive with a peptide. Proc. Natl Acad. Sci. USA, 89, 8864–8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones K.A. (1997) Taking a new TAK on tat transactivation. Genes Dev., 11, 2593–2599. [DOI] [PubMed] [Google Scholar]
- 32.Lanz R.B., McKenna,N.J., Onate,S.A., Albrecht,U., Wong,J., Tsai,S.Y., Tsai,M.J. and O’Malley,B.W. (1999) A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell, 97, 17–27. [DOI] [PubMed] [Google Scholar]
- 33.Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- 34.Birney E.S., Kumar,S. and Krainer,A.R. (1993) Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res., 21, 5803–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]