Abstract
The LysR-type transcriptional regulators (LTTRs) comprise the largest family of prokaryotic transcription factors. These proteins are composed of an N-terminal DNA binding domain (DBD) and a C-terminal cofactor binding domain. To date, no structure of the DBD has been solved. According to the SUPERFAMILY and MODBASE databases, a reliable homology model of LTTR DBDs may be built using the structure of the Escherichia coli ModE transcription factor, containing a winged helix– turn–helix (HTH) motif, as a template. The remote, but statistically significant, sequence similarity between ModE and LTTR DBDs and an alignment generated using SUPERFAMILY and MODBASE methods was independently confirmed by alignment of sequence profiles representing ModE and LTTR family DBDs. Using the crystal structure of the E.coli OxyR C-terminal domain and the DBD alignments we constructed a structural model of the full-length dimer of this LTTR family member and used it to investigate the mode of protein–DNA interaction. We also applied the model to interpret, in a structural context, the results of numerous biochemical studies of mutated LTTRs. A comparison of the LTTR DBD model with the structures of other HTH proteins also provides insights into the interaction of LTTRs with the C-terminal domain of the RNA polymerase α subunit.
INTRODUCTION
The LysR-type transcriptional regulators (LTTRs), first described by Henikoff et al. (1) are present in diverse bacterial genera, archaea and algal chloroplasts (2). They are thought to constitute the largest family of prokaryotic DNA binding proteins. According to recent compilations (3), the reportoire of 314 DNA binding proteins encoded by the genome of Escherichia coli K12 contains 45 LTTRs (18 experimentally verified and 27 predicted). Over the last 10 years, the number of protein sequences recognized as LTTRs has expanded from ∼50 (2) to ∼800 according to the PFAM database (4) of protein families. Biochemical studies of various LTTRs show that they are similarly sized molecules (300–350 amino acids) that activate the transcription of operons and regulons involved in extremely diverse cellular functions including nitrogen fixation, oxidative stress response and bacterial virulence (2). Most LTTRs, while activating expression of target genes, repress their own expression, frequently by the use of divergent promoters. In common with other bacterial transcription factors (e.g. LacI, AraC), LTTRs act as homodimers or homotetramers. Mutational studies have mapped the DNA binding activity to the N-terminal part of the sequence (2,5). According to secondary structure prediction methods this part of the sequence is likely to form a helix–turn–helix (HTH) motif (1). The highest sequence similarity between LysR-type proteins exists within the 66 N-terminal amino acids containing the putative HTH motif. Most LTTRs require a small molecule ligand to act as a coinducer. The ligand binding site has been mapped by mutational studies to the C-terminal part of the sequence (2,6).
Structural studies of LTTRs have been limited to the crystal structures of the cofactor binding domains of Klebsiella aerogenes CysB (7) and E.coli OxyR (8) proteins. Both structures are composed of two α/β domains linked by two interdomain strands. They are structurally similar to the periplasmic ligand binding proteins (PBP) which, according to SCOP (9) classification, belong to the PBPII superfamily. In the case of CysB the two α/β domains enclose the ligand binding cavity (7). Similarly to PBPs, LTTRs bind many different classes of compound including sugars, amino acids and inorganic anions (2). The LTTR structures are also highly similar to those of the cofactor binding domains of the LacI family of transcriptional repressors, which are also composed of two α/β domains (7). For both CysB and OxyR the availability of structures of the C-terminal domain dimers allows comparison with the dimer of the Lac repressor. Surprisingly, the arrangement of monomers in the LTTR dimers is significantly different to that of the Lac repressor (7). The two N-terminal DNA binding domain (DBDs) are located at the same end of the dimer in the Lac repressor, whereas in LTTR dimers they are present at opposite ends.
At present, there is no experimentally determined structure of an N-terminal DBD for any LysR-type protein. DBDs of LTTRs, although highly conserved, are not closely related to any protein of known structure. Recent advances in sequence analysis techniques permit reliable fold assignment even in the cases of remote sequence similarity. In two genome-scale fold recognition efforts the LTTR DBDs were recognized as sharing the fold of the winged HTH DBD from the ModE transcriptional regulators. Both the SUPERFAMILY (10) and the MODBASE (11) services propose the same alignment between various LTTRs and the E.coli ModE transcription regulator, the only member of ModE family for which the structure is known (12). The remote sequence similarity between ModE and LTTR proteins is also noted in the PFAM (4) database, although no alignment is given. In this study we use a homology model of the LTTR DBD, built according to alignment with the ModE protein, to propose a structural model of the full-length LTTR dimer and its mode of protein–DNA interaction. Using this new structural model we propose a possible arrangement for the DBDs with respect to the cofactor binding domains.
As the basis of our model we have used the example of E.coli OxyR protein, from which the C-terminal domain has been solved by Choi et al. (8). The OxyR protein, which activates the oxidative stress response, works as a redox switch, with its C-terminal domain undergoing a conformational change as the result of the formation of a disulfide bridge (8). This model system was chosen because of the available data, which proved useful for verification of the relative arrangement of the DBD and the cofactor binding domains.
Due to the high degree of sequence similarity among LTTR family members the model presented in this work may be used to gain structural insights not only into the function of the OxyR protein but also of other members of the LTTR family. We demonstrate that the model can be used to interpret and understand the results of numerous mutational and biochemical studies conducted on these proteins and to direct further experimental work.
MATERIALS AND METHODS
Database searching and sequence alignment
The SUPERFAMILY (10) and MODBASE (11) internet services have been used to find a template structure for homology modeling of the OxyR DBD. Both databases were searched for proteins with known structures aligned with the sequence of the OxyR N-terminal domain (residues 1–85). The alignment of sequence profiles representing OxyR and ModE DBDs was calculated in the following way. First, a BLAST (13) search of the SwissProt/trEMBL (14) database was performed to collect all the sequences significantly (E-value < 10–4) similar to the E.coli ModE and OxyR DBDs. To eliminate near identical sequences from the two sequence sets, the BLASTCLUST (13) program (E-value < 10–6) was applied. The two resulting sets of sequences, representing the ModE and LTTR DBDs were separately aligned using the CLUSTALX (15) program with default parameters. Subsequently, two sequence profiles, computed from the alignments of the ModE and LTTR DBDs, were aligned with the CLUSTALX program.
Model building
The model of a single OxyR DBD was built using Swiss-Pdb Viewer (16) software. The OxyR and ModE DBDs were aligned according to the results of the sequence analyses described above (see Fig. 1). The initial coordinates of the OxyR DBD, assigned by the Swiss-Pdb Viewer were submitted for refinement to the Swiss Model (16) service. The dimer of OxyR DBDs was built by superimposing the model structure onto the two monomers composing the dimer of the ModE DBD.
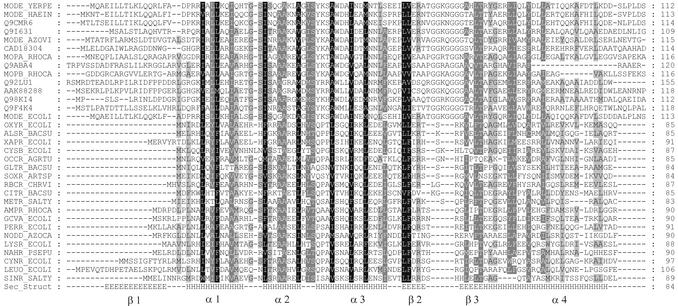
Figure 1.
Alignment of the LTTR and ModE transcription factor families. Sequence profiles of both families were aligned as described in Materials and Methods. The Figure shows 33 sequences selected from the profile–profile alignment. Sequences are designated by their SwissProt/trEMBL identifiers. An alignment of the E.coli OxyR and ModE proteins (OXYR_ECOLI, MODE_ECOLI) is identical to that computed by the SUPERFAMILY and MODBASE services. Shading was added to assist interpretation. Amino acids were divided into the following groups {FYVLIVMCA} (hydrophobic), {DE}, {KR}, {NQ} and {ST}. Residues belonging to the same group were considered similar and shading was added to indicate 100, 80 and 60% column similarity levels. The bottom line shows the secondary structure assignment according to the ModE structure (H, helix; E, strand). Secondary structure elements were labeled following the ModE structure.
The relative position of the DBD dimer (residues 1–85) and the C-terminal domain dimer (residues 86–298) was found using the following protocol implemented with the XPLOR (17) software. Both dimers were randomly positioned in space with respect to each other. The peptide bonds joining residues 85 and 86 in both polypeptide chains of the OxyR dimer were defined. As is a common practice in the NMR structure determination of homodimers, non-crystallographic symmetry restraints (18) were added to the force field. A molecular dynamics (MD) simulation was subsequently started in which the DBD dimer was treated as a rigid body, the loops (residues 86–90) were flexible and residues 91–298 of C-terminal domains remained in fixed positions. The calculations were stopped when the RMS deviation of DBD positions converged to the values <0.1 Å. The MD simulations were repeated 100 times, starting from a different random configurations, and the main chain bond lengths were examined in the loop regions of resulting alternative models. The models containing the bonds with the lengths deviating by >1.0 Å from the equilibrium values were rejected. These models represented relative configurations of the DBD and C-terminal domain dimers in which the distances between the corresponding ends of the polypeptide chains were too large to allow joining of the domains by the flexible loops. The average structure of the remaining models was computed and refined by a standard simulated annealing protocol (19,20).
The final structures were evaluated by the PROCHECK (21), PROSAII (22) and MATCHMAKER (23) programs. To test if the Z-score values computed by PROSAII software are within the range characteristic for the properly folded polypeptide chains of the given length the pG values (24) were computed at http://guitar.rockefeller.edu/pg/. Swiss-Pdb Viewer software was used to create structural alignments of the OxyR DBD model and experimental structures of other transcription factors by superimposition of the Cα atoms of the residues belonging to HTH motif. Solvent accessible surface areas were computed using the XPLOR program with default parameters. The electrostatic potential was calculated and mapped to the surface using numerical integration of the Poisson–Boltzmann equation as implemented in the Swiss-Pdb Viewer.
The structures used in this work
The following experimentally determined structures were used in this work: C-terminal domain of E.coli OxyR (PDB codes: 1I69, 1I6A), E.coli ModE (1B9M), Corynebacterium diphtheriae DtxR (1F5T), E.coli CAP (1J59), Human RFX1-DBD (1DP7), E.coli ROB (1D5Y), E.coli Fis (1F36), E.coli αCTD (1COO), E.coli OmpR (10DD).
RESULTS AND DISCUSSION
Sequence similarity among DNA binding domains of LTTR and ModE family transcriptional regulators
The DBDs of LTTRs do not share high sequence similarity with any protein of known structure. A variety of contemporary sequence analysis methods, based on sequence profiles (13) and hidden Markov models (HMM) (25) allow reliable detection of remote sequence similarity, which reflects a common fold in the proteins analyzed. The SUPERFAMILY (10) and MODBASE (11) databases contain the results of the application of these methods to all protein sequences from fully sequenced genomes and all sequences from the SwissProt/trEMBL database, respectively. We searched both databases for the proteins of known structure sharing remote sequence similarity with the N-terminal domain (residues 1–85) of the E.coli OxyR transcription factor. Both services indicated similarity between the OxyR N-terminal domain and the winged HTH DBD of the ModE transcription factor from E.coli. The alignments of the OxyR sequence (1–85) and the ModE DBD were the same in both databases. The alignment generated by MODBASE also erroneously included part of the OxyR C-terminal domain aligned with the ModE cofactor binding domain. Due to this error the homology model, automatically generated by MODELLER (26) software, has very low overall quality.
The sequence similarity between the ModE and OxyR DBDs, although remote (12% identity), is statistically significant. The E-value of an alignment of the OxyR DBD with a HMM representing the ModE DBD is 4.6 × 10–25, according to SUPERFAMILY. In MODBASE, the sequence similarities were detected by PSI-BLAST searches with an E-value < 10–4.
Correctness of the alignment is the most important factor determining the quality of the model in homology modeling. To further confirm the alignment of the LTTR and ModE DBDs we aligned sequence profiles representing both families. The profile–profile alignment procedure applied here used information about the sequence variability within the two families under comparison, whereas in the database searches described above the single protein sequences were aligned with profiles or HMMs representing the protein families. An alignment including members of the LTTR family other than OxyR was also necessary for the analysis of results of mutational studies. The sequences of the ModE family of transcriptional regulators and those representing the LTTR family members were collected from the SwissProt/trEMBL (14) database by standard BLAST (13) sequence similarity searches (see Materials and Methods for details). The sequence sets representing LTTR and ModE families were separately aligned by the CLUSTALX (15) program. The same program was subsequently used to align profiles representing the alignments of both families. The final alignment is shown in Figure 1. It is clear that the hydrophobic residues are conserved in both families. As will be described below, these residues form the hydrophobic core of both the ModE and LTTR DBDs. Another conserved position is represented by serine 44 of ModE, a residue located in the turn of the HTH motif and probably involved in the interaction with the phosphate backbone of DNA.
Alignments of the OxyR N-terminal domain and the N-terminal domain of ModE family members calculated by the SUPERFAMILY and MODBASE services were exactly the same as the one produced by profile–profile alignment of LTTR and ModE families. The statistical significance of the sequence similarity, conservation of the hydrophobic core and agreement between the alignments calculated by three independent and well established methods indicated that the alignment of LTTR and ModE families, shown in Figure 1, may be used for reliable homology modeling of the LTTR DBD.
The structure of the full-length OxyR transcriptional regulator
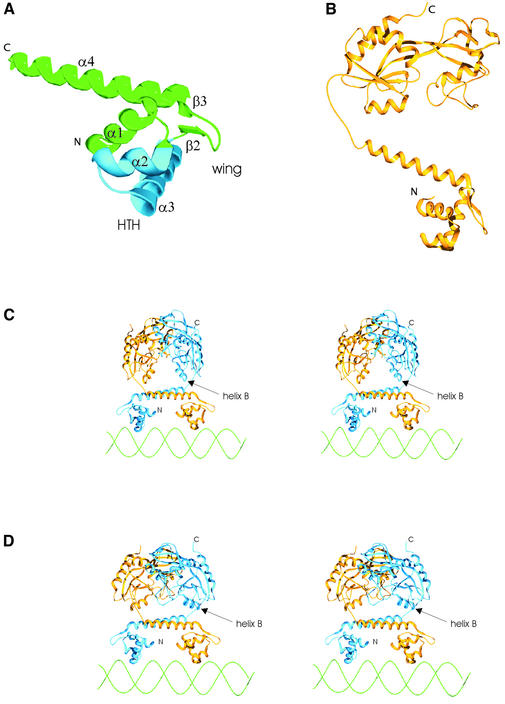
To build a homology model of the DBD of the E.coli OxyR transcription factor we used as a template the structure of the ModE protein from E.coli (12), the only member of ModE family for which the structure has been solved. Figure 2A shows the model of the OxyR DBD monomer built according to the alignment shown in Figure 1. A model of the OxyR DBD dimer was built by superimposition of two monomer structures on the monomers composing the dimer of ModE DBDs. According to our model, the DBD is composed of a globular part plus a long α-helix forming the backbone (α4), which is responsible for most of the interactions between the DBD monomers. The globular part, containing the winged HTH structural motif is formed by three alpha helices (α1, α2 and α3). Helices α2 and α3 form the HTH structure and the wing is formed by the loop joining the strands β2 and β3. In the dimer, the two long backbone helices (helices α4) form the surface, which in the ModE protein is turned towards the cofactor binding domains. We will refer to this surface as the flat face of the DBD domain.
Figure 2.
Schematic ribbon diagrams of the structure of the full-length OxyR transcription factor dimer. (A) Homology model of the DBD. The HTH is marked in blue. Secondary structure elements are named according to Figure 1. (B) The full-length monomer of the OxyR reduced form. (C and D) The models of the oxidized (C) and reduced (D) forms of OxyR dimer bound to DNA (stereo images). Helix B in the regulatory domain is named according to Choi et al. (8). Helices B are located on the face of the cofactor binding domain dimer that is turned towards the dimer of the DBDs.
The arrangement of the DBDs in functional LTTR molecules has remained a puzzle since the structure of the CysB cofactor binding domain dimer was solved (7). This structure shows that the DBDs are located at the opposite ends of the dimer, which is contrary to the well known example of the LacI family transcription factors in which the N-terminal ends of the cofactor binding domains point in the same direction. To study a possible arrangement of the DBD and C-terminal domain dimers we have applied a conformational sampling protocol described in Materials and Methods. During the simulation we were looking for the conformations in which the C-terminal ends of the DBD domains (residues 1–85) could be joined with the N-terminal ends of the regulatory domains (residues 90–298) by a flexible loop without introducing steric clashes and distorted conformations of a polypeptide chain. The conformational sampling, starting from 100 random initial configurations, was performed for both the oxidized and reduced forms of the OxyR. The simulations yielded 23 and 30 alternative conformations for the reduced and oxidized forms, respectively. In both cases, alternative models represented very similar structures. The RMS deviation of non-hydrogen atom positions from the averaged structure of the reduced form was 1.31 Å. A similar value of 1.33 Å was obtained for the oxidized form of the OxyR molecule. Therefore, the averaged structures were subjected to the simulated annealing refinement and considered as final models of the OxyR transcription factor dimers. The stereochemistry of the refined models was evaluated using the PROCHECK (21) software. The quality of the models was assessed by statistical potentials implemented in PROSAII (22) and MATCHMAKER (23) programs. The Z-score values calculated by PROSAII were in the ranges observed for the correctly folded protein chains of the same length. The pG values (see Materials and Methods) were equal to 1 for full-length models of the reduced and oxidized forms, and the pG value for the DBD dimer alone was 0.98. The statistical potential, as calculated by MATCHMAKER, had negative values for both the DBD dimer model (–0.12 kT) and the models of full-length reduced and oxidized OxyR molecules (–0.26 and –0.29 kT, respectively). Both the pG values close to 1 and a negative statistical potential indicate that the models are valid. PDB files containing the coordinates of the full-length OxyR dimers are available on request.
The models of the reduced and oxidized forms of OxyR dimers are shown in Figure 2. In both cases, the flat face of the DBD domain is turned towards the face of the C-terminal domain formed by helices B, C and strands 7, 8 and 11. The stretch of residues linking the DBD and C-terminal domains is shorter than is the case in the ModE family. In E.coli ModE protein, the backbone helix of the DBD is linked with the cofactor binding domain through a loop followed by a short helix (12). The stretch of residues linking the DBD and regulatory domains in the OxyR and other LTTR proteins is too short to accommodate a helix. Our models show that these domains are joined by flexible loops. The sensitivity of the linker sequence joining the DBD and the C-terminal domains of the OxyR to proteolytic digestion (8) supports the presence of a flexible solvent exposed loop in this region.
The activation of OxyR occurs following the formation of a disulfide bridge between Cys199 and Cys208 (27). In the oxidized form of the OxyR dimer the regulatory domains are rotated by 30°, with respect to the reduced form, in such a way that the N-terminal ends are moved towards the DNA and towards the interface with the DBD (8). Hydroxyl radical footprinting and interference assays have been used by Toledano et al. (28) to compare the DNA binding sites of the reduced and oxidized forms of the OxyR. The results obtained with the oxyR/oxyS divergent promoter demonstrated that the oxidized form of OxyR tetramer binds four adjacent major grooves (grooves 1, 2, 3, 4), whereas the reduced form binds two pairs of adjacent major grooves separated by one helical turn (grooves 1, 2, 4, 5). Moreover, it has been also shown that groves 1 and 2 are exactly the same parts of the DNA molecule in both cases. This observation lead authors to propose that each of the two OxyR dimers, occupying the DNA binding site, binds two adjacent major grooves. According to this model, reduction of the OxyR causes conformational change that influences interaction between dimers in such a way that one of them remains bound at grooves 1 and 2, while the other is shifted to the neighboring binding site (grooves 4, 5). The model presented in our work further supports the mechanism described above as it shows that similarly to the ModE transcription factor the OxyR dimer indeed binds the two adjacent major grooves of the DNA molecule. As both forms of the OxyR were shown to bind exactly the same site on the DNA (grooves 1, 2), it is also very unlikely that conformational change of the regulatory domain dimer causes significant conformational change within the DBD dimer. Such a conformational change would alter the distance between recognition helices and prevent the binding to the site present at grooves 1 and 2. Therefore, the conformational change within the regulatory domains most probably alters the DNA binding process by modulating the interaction among dimers. We would also like to point out that the above considerations justify the rigid body assumption of the conformational sampling protocol used to assemble OxyR models. If the DBDs present in both forms of the OxyR are capable of binding exactly the same site on the DNA, it is safe to assume that their conformations are not significantly different and use the same rigid structure of the DBD dimer to build the full-length models of both the reduced and oxidized OxyR dimers.
In their work describing crystal structure of the OxyR C-terminal domain, Choi et al. (8) also proposed a model of the interactions in the tetramer, based on the crystal contacts. They have aligned the tetramers of reduced and oxidized form, built according to crystal contacts, with the approximate model of a DNA region protected from the DNase I digestion by the OxyR binding. The model shows that in both the reduced and oxidized forms, the face of the C-terminal domain, formed by helices B, C and strands 7, 8 and 11, is turned towards the DNA molecule. This is in agreement with our model that shows the same face turned towards the DBD and DNA. However, the exact location of the contacts among dimers is not in agreement with our model. For example, the interface implicated by the crystal contact in the structure of the oxidized OxyR regulatory domain (residues 202–207 and 168–173) cannot be explained by our model if the two dimers occupy four consecutive major grooves on an approximately linear DNA molecule, as was indicated by the biochemical data. As is clear from Figure 2C, an interface between two oxidized dimers may involve the contacts between the wing regions of the HTH. Also, the C-terminal helix is exposed for the interaction with the neighboring dimer. This could explain results of the experimental studies of the OxyR (5) and E.coli CysB (6) that show involvement of the C-terminal end of the molecule in the tetramer formation.
The hydrophobic core of the DBD domain and intersubunit contacts
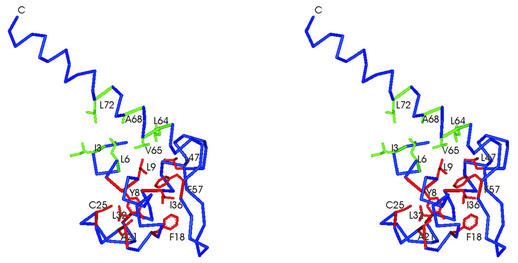
According to our model the DBD of the OxyR protein consists of a globular part, containing the winged HTH structure, and the long C-terminal, backbone helix (α4). The core of the globular domain is formed by a cluster of hydrophobic residues: Y8, L9, L12, F18, A21, A22, C25, L32, I36, L47, L48, F57 (Fig. 3). The side chain of Y8 is buried close to the center of the globular part of the DBD domain. It is surrounded by side chains of L9, L12, F18, A21, L32, I36 and L48, positioned within 6 Å distance. The side chain of L47 is located close to L9. Residues L48 and F57 are positioned at the base of the wing in strands β2 and β3. They form the hydrophobic contact that may play a role in stabilization of the wing structure. All the residues mentioned above are not exposed to the solvent (accessible surface area <30 Å2). According to the sequence alignment (Fig. 1), the hydrophobic character of these positions is conserved among both the LTTRs and ModE proteins. Moreover, in both the LTTR and ModE families, residues aligned to positions L12, V27, L32, I36 and L47 of the OxyR molecule are aliphatic. The alanine residues in positions 21 and 22 are highly conserved in both protein families. The fact that the residues forming the hydrophobic core of the DBD are highly conserved between the ModE and LTTR families further supports the validity of the alignment shown in Figure 1 and the homology modeling studies presented in this work.
Figure 3.
Hydrophobic core of the OxyR DBD (stereo view). Residues belonging to the hydrophobic core of the globular part of the DBD are colored red. The cluster of hydrophobic residues formed by helices α1 and α4 is colored green.
The findings of several mutational studies of LTTRs can be attributed to disruption of the hydrophobic core in the globular part of the DBD. The L32F mutation in OxyR causes weak DNA binding and low transcriptional activity in vivo (5). Similarly, the V32I and V32A mutations in GcvA (29) (position equivalent to V27 of OxyR) and A27T in NahR (30) (A21 in OxyR) result in the loss of DNA binding. The V32I mutation in GcvA shows that even subtle changes within the hydrophobic core of the DBD may lead to impaired DNA binding activity. It is surprising, therefore, that a hydrophobic character of the positions corresponding to V27 in the OxyR is not strictly conserved among LTTRs (Fig. 1). In a few exceptional cases (CysB E.coli, MetR Salmonella typhimurium, LysR E.coli) the presence of the hydrophilic residue in this position may be compensated by the ‘context’, i.e. mutations in other parts of the DBD and subtle rearrangements of the hydrophobic core. Other examples of the mutations that are located in the hydrophobic core of the DBD and influence DNA binding are I48T in E.coli CysB (6) (L48 in OxyR) and I33N (L32 in OxyR) in E.coli CysB (31). In the case of the E.coli CysB I48T mutation, it has been suggested on the basis of genetic analysis that this residue is involved in oligomerization. According to our model, residue I48 of CysB does not take part in the interaction between monomers forming the dimer. Its direct involvement in the interaction of dimers is also unlikely since the residue is buried within the core of the DBD. However, disruption of the hydrophobic core of the DBD could lead to alterations in the surface of the DBD that could interfere with tetramer formation. The examples described above show that even the relatively small changes to the residues conserved in the hydrophobic core of the DBD may change its structure enough to impair efficient DNA binding.
The second hydrophobic cluster present in the DBD of OxyR involves residues I3 and L6 positioned in the helix α1 and residues L64, V65, A68 and L72 located in the backbone helix (α4). The interaction between helices α1 and α4 may be important for maintaining the relative orientation of the globular part of the DBD and the backbone helix. This in turn is necessary for maintaining the distance and relative orientation of the two HTH motifs which are important for DNA binding (Fig. 3).
The long backbone helices (α4) participate in the interactions between two DBDs and the interactions of the DBD and C-terminal domain dimers. In the OxyR, the interface between two helices buries 1338 Å2 of the accessible surface area. The aliphatic residues L63, L64, V65, V75 and L78 form the hydrophobic core of this interface. The contact between the DBD and C-terminal domains is formed mainly by the N-terminal part of backbone helices α4 and the helices B belonging to the C-terminal domain. The contact is formed between helices α4 and B belonging to different chains of the dimer rather than between the parts of the same protein molecule (Fig. 2C). The surface areas buried in this interface are 778.616 and 778.068 Å2 in the reduced and oxidized forms, respectively; they change very little during conformational change between the two forms. Our models suggest that the interfaces between the DBD and C-terminal domain dimers lack an extensive network of hydrogen bonds and salt bridges. In the reduced form, hydrogen bonds are formed by residues Q59, S138 and D66, H130. Both these interactions are not present in the oxidized form. It seems, therefore, that the interactions between the DBD and regulatory domains are not very strong, which facilitates conformational change between non-active and active forms.
Protein–DNA interactions
Hall et al. (12) proposed that interaction of the ModE protein with DNA resembles that of DtxR, another transcriptional regulator containing a winged HTH motif. It was assumed that like DtxR, ModE does not bend the DNA molecule. Moreover, Choi et al. (8) in the discussion of the possible arrangement of the regulatory domain dimers in the tetramer structure argued that DNA bending by the reduced form of OxyR is caused by arrangement of the dimers in the tetramer structure rather than by the dimer itself. Therefore, we have followed the example of the ModE structure and built a model of the OxyR–DNA complex in which two recognition helices of DBD dimer are positioned in two adjacent major grooves of an unbent B DNA molecule. This model of the OxyR dimer–DNA complex is shown in Figure 2C and D.
We are aware of the fact that resolution of the theoretical model does not allow the study of the fine details of the protein–DNA interactions exclusively on the basis of the interatomic distances measured within the model. It is possible, however, to explore the structural similarity between the OxyR DBD and experimental structures of the DNA-bound HTH proteins to find conserved residues that most probably play an important role in the protein–DNA interactions. These data can be used to interpret, in a structural context, results of numerous mutational studies performed on the LTTR family members. We have, therefore, superimposed the HTH structure of the OxyR DBD model onto the HTH structures of several transcription factors for which experimental structures with the DNA are available. We have found that several residues, which are conserved in the LTTR family, are aligned with identical residues of the bacterial transcription factors DtxR (32), Rob (33), CAP (34) and Fis (35). Fis has been included in the analysis in spite of the lack of experimental structures of the protein–DNA complex due to the fact that available models of Fis–DNA interactions were confirmed by experimental data. We have also found conserved residues among HTH regions of OxyR and eukaryotic transcription factor RFX1 (36) containing the winged HTH motif. The analysis of the model structure, presented below, is supported by the conservation of the residues found in the structures compared.
The known structures of HTH proteins bound to oligonucleotides show that the recognition helix may assume various orientations with respect to the major groove of the DNA molecule. Similarly, the wing structure in the winged HTH motif may play different roles in protein–DNA interactions (37). It may interact either with the phosphate backbone or the minor groove of DNA. According to our model, the recognition helix in the LTTR family is longer by one helical turn than in other HTH proteins. It contacts the major groove of DNA only via its N-terminal part. In OxyR the C-terminal part contains the negatively charged residues D41 and E42 and does not form protein–DNA contacts.
Residues corresponding to OxyR positions S33, R37 and K38 are well conserved among LTTRs. They are also present at structurally equivalent positions in the recognition helices of several other HTH transcription factors. Serine and threonine residues forming water-mediated hydrogen bonds with the phosphate backbone of DNA are present in RFX1 (36), Fis (35), CAP (34) and DtxR (32) in positions equivalent to T31 of OxyR. It is likely, therefore, that the side chains of the residues T31 and S33 form water-mediated hydrogen bonds with the phosphate backbone. In DtxR the serine residue is located at a position corresponding to OxyR S33. The lysine/arginine residues interacting with the phosphate backbone are present in RFX1, CAP, Fis and ROB (33) in positions structurally aligned with R37 of OxyR. A second positively charged residue, which is involved in the interaction with the phosphate backbone and is structurally aligned with K38 of OxyR is also present in Fis and DtxR. The location of R37 and K38 residues within the model of OxyR DBD and structurally equivalent positions of other HTH proteins suggests the interaction of these residues with the phosphate backbone of DNA.
Biochemical studies on several mutated LTTRs confirm the importance of residues equivalent to T31, S33 and K38 in protein–DNA interactions. The T31M and S33N mutations abolishes DNA binding in the E.coli OxyR protein (5). The loss of DNA binding was also observed in the S34R mutant of S.typhimurium CysB (31) and the S38P mutant of E.coli GcvA (29). Residues S34 of CysB and the S38 of GcvA correspond to position S33 in OxyR, according to the alignment shown in Figure 1. The mutation R43H in NahR of Pseudomonas putida, which corresponds to position K38 of OxyR, resulted in the complete loss of DNA binding activity (30).
The P30 residue of OxyR, located in the part of the recognition helix buried within the major DNA groove, is well conserved among LTTRs. The P35S mutant in NahR of P.putida (position aligned with OxyR P30) caused a 70% reduction in DNA binding (30). It is difficult to determine the role of this proline residue in protein–DNA interaction. However, it is also present in DtxR at position 39 (structurally equivalent to P30 in OxyR), and it was shown that this residue is involved in the unusual van der Waals contact with the methyl groups of thymine bases and the serine residue at a neighboring position (32). Although the resolution of our model does not allow us to study such subtle effects in detail, one cannot exclude the possibility that a similar mechanism may also occur in LTTRs.
There are two residues of OxyR that are likely to interact with DNA, and these are positioned in parts of the HTH motif other than the recognition helix. Residue S28, located in the turn preceding the recognition helix is likely to interact with the phosphate backbone, similarly to serine residues present in structurally equivalent positions in CAP and DtxR. This residue is well conserved within the LTTR family. An interaction with the phosphate backbone may also be formed by R19. This residue is located at the N-terminal end of the first helix of the HTH motif (α2) and extends toward the DNA molecule. It is likely that this interaction is unique to OxyR because lysine/arginine is not conserved in positions aligned with OxyR R19 in other members of the LTTR family.
According to our model the wing structure in LTTRs is five amino acids shorter than the wing structure of ModE. LTTRs lack the glycine-rich flexible region characteristic of ModE regulators. The wing structure in the LTTR family is therefore too short to interact with bases in the minor groove of DNA and its role is probably limited to interaction with the phosphate backbone. This is supported by the fact that LTTRs contain two conserved positively charged residues corresponding to positions R50 and R53 of OxyR. These residues may interact with the phosphate backbone but their side chains do not reach the minor groove of the DNA. The mutation R50W in OxyR abolished DNA binding (5). In OxyR, residue K54 in addition to R50 and R53 may also interact with the phosphate backbone.
The model of the OxyR DBD domain shows that residues located outside both the HTH and wing structural motifs may be implicated in protein–DNA interaction. These are N2, R4 and H17 located in the helix α1 and the first turn of the DBD. The mutation R4C in E.coli OxyR significantly decreases the rate of transcription initiation from the oxyS promoter (5).
We are aware that the protein–DNA interactions described above concern interactions with the phosphate backbone of DNA rather than with the bases in the major groove. A predominant role for interactions with the phosphate backbone is not unusual. For example, this was observed in the experimentally determined structure of the DtxR–DNA (32) complex. The study of the fine detail of sequence-specific interactions with the DNA bases requires a more detailed model and must await the solution of the OxyR DBD–DNA complex structure by X-ray crystallography or NMR methods.
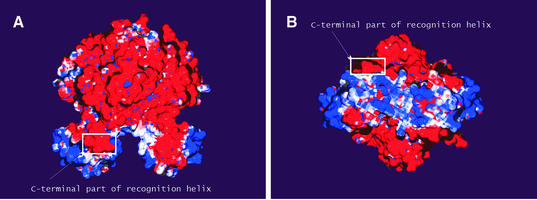
Figure 4 shows the electrostatic surface potential of the OxyR dimer computed by numerical solution of the Poisson–Boltzmann equation. The C-terminal domain is negatively charged. In contrast, a positive charge is found in regions of the surface directly involved in protein–DNA interaction. The wing of the winged HTH motif is the most positively charged part of the DBD. This is in accordance with the postulated role of the wing in the interaction with the phosphate backbone of DNA. A positive charge is also mapped to the N-terminal end of the recognition helix which, according to our model, is positioned within the major groove of the DNA. The C-terminal part of this helix is negatively charged. This supports our result that this part of the recognition helix is positioned outside of the major groove.
Figure 4.
The electrostatic surface potential of the OxyR dimer. Electrostatic surface representation of the reduced form of the OxyR dimer with blue and red regions indicating positive and negative electrostatic regions, respectively. (A) The molecule shown in the same orientation as in Figure 2D. (B) The molecule is rotated by 90° around its long axis showing the face turned towards the DNA. Note the highly basic nature of the parts of the DBD which directly interact with the DNA. The negatively charged C-terminal part of recognition helix is marked by the square on both pictures.
The distribution of charges on the surface of the OxyR molecule suggests the involvement of an electrostatic steering effect during the formation of the OxyR–DNA complex. The positively charged DBD will be attracted toward the DNA molecule while the negatively charged C-terminal domain is repelled. It should be noted that the distribution of charges on the surface of OxyR is not a general property of LTTRs. For example, the cofactor binding domain of Klebsiella pneumoniae CysB protein is not predominantly negatively charged (data not shown). However, the positive electrostatic potential mapped to the surface of the DBD is a common feature since positively charged residues located in this domain are conserved among LTTRs.
All the experimental work regarding mutational studies discussed above and in the following section are listed in Table 1.
Table 1. Mutational studies discussed in this work.
| Proteina | Mutation | Localization in the structureb | Function affected | Ref. |
|---|---|---|---|---|
| OXYR_ECOLI | L32F | Hydrophobic core of the DBD | DNA binding | 5 |
| GCVA_ECOLI | V32I/A | Hydrophobic core of the DBD | DNA binding | 29 |
| NAHR_PSEPU | A27T | Hydrophobic core of the DBD | DNA binding | 30 |
| CYSB_ECOLI | I48T | Hydrophobic core of the DBD | DNA binding | 6 |
| CYSB_ECOLI | I33N | Hydrophobic core of the DBD | DNA binding | 31 |
| OXYR_ECOLI | T31M | Recognition helix | DNA binding | 5 |
| OXYR_ECOLI | S33N | Recognition helix | DNA binding | 56 |
| CYSB_SALTY | S34R | Recognition helix | DNA binding | 31 |
| GCVA_ECOLI | S38P | Recognition helix | DNA binding | 29 |
| NAHR_PSEPU | R43H | Recognition helix | DNA binding | 30 |
| NAHR_PSEPU | P35S | Recognition helix | DNA binding | 30 |
| OXYR_ECOLI | R50W | Wing | DNA binding | 5 |
| OXYR_ECOLI | R4C | Exposed residue in helix α1 | DNA binding | 5 |
| GCVA_ECOLI | L30A | Turn of HTH | Activator function | 29 |
| GCVA_ECOLI | F31A/L | Turn of HTH | Activator function | 29 |
| CYSB_ECOLI | Y27G | Turn of HTH | Activator function | 6 |
| CYSB_ECOLI | N309Ter | C-terminus | Tetramer formation | 6 |
| OXYR_ECOLI | E225K | C-terminus | Tetramer formation | 5 |
aProteins are named by SwissProt database identifiers.
bMutations in the proteins other than OxyR were mapped to the model structure according to the sequence alignment shown in Figure 1.
Interaction of LTTRs with the C-terminal domain of the α subunit of RNA polymerase
It has been postulated that many prokaryotic transcription factors, including LTTRs, activate transcription by interaction with the C-terminal domain of the α subunit of RNA polymerase (αCTD). For example, mutational studies have shown that lysine 271 of the E.coli αCTD may interact with proteins regulating the transcription of cysA, melAB, araBAD and araE promoters (38). The αCTD K271E mutation significantly reduced the expression levels of reporter genes. Mutation of lysine 271 to alanine (K271A) reduces Fis-activated transcription from proP P2, rrnB P1 and rrnE P1 promoters both in vivo and in vitro (39,40). On the basis of the known structure of the αCTD it was postulated that residues 271–273 form a ridge on the protein surface which takes part in the interaction with the Fis transcription factor (39). Alanine scanning mutational studies of Fis have shown that residues Q68, R71 and G72 are responsible for interaction with the αCTD (39).
Biochemical studies of LTTRs have revealed a few mutants in which transcriptional activator function was impaired while repressor activity remained unaffected. Examples are the L30A and F31A/L mutations in GcvA (29) and Y27G in CysB (6). It was proposed that these residues may interact with the αCTD. It was also shown that the rate of CysB activated transcription from the cysA promoter is significantly decreased by mutations at position K271 of αCTD (38). This suggests that LTTRs may interact with the same region of RNA polymerase as the Fis transcription factor. According to the alignment shown in Figure 1, residues F31 of GcvA and Y27 of CysB occupy equivalent positions. Both aromatic side chains are exposed on the surface, do not directly interact with DNA and are therefore capable of interaction with the αCTD. However, the aforementioned residues are located in different parts of the conserved HTH structure compared with residues Q68, R71 and G72 of the Fis protein implicated in the interaction with the αCTD. In the case of GcvA and CysB, both aromatic residues are located in the first position of the turn in HTH (Fig. 5A). Residues Q68, R71 and G72 of Fis occupy the loop at the C-terminal end of the HTH motif. This means that there is a significant difference between the location of the residues implicated in the interaction with the K271 region of the αCTD in Fis and LTTRs. The turn following the recognition helix in Fis is oriented in such a way that Fis and the αCTD may be positioned ‘side-by-side’ on the DNA molecule (39,40). In contrast, the turn in the HTH motif of LTTRs is directed toward the part of DNA molecule positioned between the two DBDs of the dimer which makes ‘side-by-side’ interaction impossible.
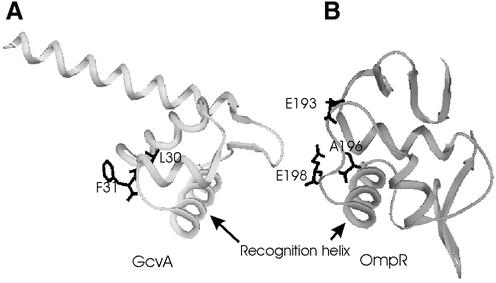
Figure 5.
Location of the residues which may act as contacts with αCTD. (A) Model of the DBD of the GcvA transcription factor. (B) Crystal structure of the DBD of the OmpR transcription factor. Note that in both structures the residues implicated in the contacts with αCTD are located in the turn of the HTH motif.
Location of a putative RNA polymerase interaction region in the turn of the HTH motif is not unique to LTTRs. The structure of the OmpR transcriptional regulator (41) revealed that residues identified by mutational studies as being involved in the interaction with RNA polymerase are located within the turn region extended to 11 residues (Fig. 5B). Kondo et al. (41) postulated that as the turn region faces one side of the DNA, the αCTD may bind the DNA bases covered by OmpR. According to our model, a similar mechanism may be valid for LTTRs; the turn region faces one side of the DNA thus allowing interaction with the αCTD within the region covered by the DBD.
Biochemical studies of mutated CysB and GcvA proteins and our model suggest that the K271 region on the αCTD surface may take a part in the interaction with LTTR regulators, but the relative orientations of the interacting proteins on the DNA appear to be different than in the case of the αCTD–Fis complex. This may be caused by the presence of the wing in LTTRs that may hinder access of the αCTD to the end of the recognition helix. A detailed description of the interaction between the αCTD and LTTRs requires further studies that may be assisted by the model presented in this work.
CONCLUSIONS
Three well established and independent sequence analysis methods show that the N-terminal DBDs of the LTTR and ModE families of transcriptional regulators share remote, but significant, sequence similarity. As the structure of the E.coli ModE protein is known, the sequence alignment of the ModE and LTTR protein families allows reliable homology modeling of the LTTR family DBD for which no structural insight is currently available. The model of the E.coli OxyR DBD indicates that the sequence conservation between the two protein families occurs mainly in the hydrophobic core of the DBD. Several amino acids located within the HTH motifs and implicated in DNA binding are also conserved in comparison with other HTH proteins. Conservation of the hydrophobic core and functionally important residues further supports the sequence alignment presented in this work.
We show by computer simulation that the homology model of the DBD dimer and the crystal structure of the C-terminal domain dimer determine their relative orientation in the case of both reduced and oxidized forms of the OxyR. There is a unique (RMS deviation of non-hydrogen atom positions of ∼1.3 Å) arrangement of the DBD and C-terminal domain dimers in which the ends of polypeptide chains can be joined by flexible loops without introducing steric clashes and the distorted conformations of a polypeptide chain.
Our model has permitted structural interpretation of biochemical results collected from studies on several mutated LTTRs. According to the model, even subtle alterations in the hydrophobic core of the DBD may result in severely impaired DNA binding activity. Other mutations influencing DNA binding are mapped to residues directly interacting with the DNA molecule. Most of these direct protein–DNA interactions involve the phosphate backbone of DNA. We also applied our model to study residues implicated by mutational studies in the interaction with the α subunit of RNA polymerase. These residues mapped to the turn region of the conserved HTH motif and were oriented towards one side of the DNA molecule. This suggest that similarly to the OmpR transcription factor, the RNA polymerase may contact the LTTR factors within the DNA region occupied by the regulator. We believe that the model presented in this work may be of great value for directing further biochemical studies of the LysR-type family of transcriptional regulators.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr Monika Hryniewicz for valuable discussions and the reading of our manuscript. We are also grateful to Dr Jaroslaw Poznanski for the help with constructing the XPLOR software simulation protocol. We thank Dr Piotr Zielenkiewicz for critical comments on the manuscript.
REFERENCES
- 1.Henikoff S., Haughn,G.W., Calvo,J.M. and Wallace,J.C. (1988) A large family of bacterial activator proteins. Proc. Natl Acad. Sci. USA, 85, 6602–6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schell M.A. (1993) Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol., 47, 597–626. [DOI] [PubMed] [Google Scholar]
- 3.Perez-Rueda E. and Collado-Vides,J. (2000) The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res., 28, 1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kullik I., Stevens,J., Toledano,M.B. and Storz,G. (1995) Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for DNA binding and multimerization. J. Bacteriol., 177, 1285–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lochowska A., Iwanicka-Nowicka,R., Plochocka,D. and Hryniewicz,M.M. (2001) Functional dissection of the LysR-type CysB transcriptional regulator. Regions important for DNA binding, inducer response, oligomerization and positive control. J. Biol. Chem., 276, 2098–2107. [DOI] [PubMed] [Google Scholar]
- 7.Tyrrell R., Verschueren,K.H., Dodson,E.J., Murshudov,G.N., Addy,C. and Wilkinson,A.J. (1997) The structure of the cofactor-binding fragment of the LysR family member, CysB: a familiar fold with a surprising subunit arrangement. Structure, 5, 1017–1032. [DOI] [PubMed] [Google Scholar]
- 8.Choi H., Kim,S., Mukhopadhyay,P., Cho,S., Woo,J., Storz,G. and Ryu,S. (2001) Structural basis of the redox switch in the OxyR transcription factor. Cell, 105, 103–113. [DOI] [PubMed] [Google Scholar]
- 9.Lo Conte L., Brenner,S.E., Hubbard,T.J., Chothia,C. and Murzin,A.G. (2002) SCOP database in 2002: refinements accommodate structural genomics. Nucleic Acids Res., 30, 264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gough J., Karplus,K., Hughey,R. and Chothia,C. (2001) Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol., 313, 903–919. [DOI] [PubMed] [Google Scholar]
- 11.Pieper U., Eswar,N., Stuart,A.C., Ilyin,V.A. and Sali,A. (2002) MODBASE, a database of annotated comparative protein structure models. Nucleic Acids Res., 30, 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall D.R., Gourley,D.G., Leonard,G.A., Duke,E.M., Anderson,L.A., Boxer,D.H. and Hunter,W.N. (1999) The high-resolution crystal structure of the molybdate-dependent transcriptional regulator (ModE) from Escherichia coli: a novel combination of domain folds. EMBO J., 18, 1435–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoesser G., Baker,W., van den Broek,A., Camon,E., Garcia-Pastor,M., Kanz,C., Kulikova,T., Lombard,V., Lopez,R., Parkinson,H., Redaschi,N., Sterk,P., Stoehr,P. and Tuli,M.A. (2001) The EMBL nucleotide sequence database. Nucleic Acids Res., 29, 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- 17.Brunger A.T. (1992) X-PLOR Version 3.1. A Aystem for X-ray Crystallography and NMR. Yale University Press, New Haven, CT.
- 18.Nilges M.A. (1993) A calculation strategy for the structure determination of symmetric dimers by 1H NMR. Proteins, 17, 297–309. [DOI] [PubMed] [Google Scholar]
- 19.Nilges M., Clore,G.M. and Gronenborn,A.M. (1988). Determination of three-dimensional structures of proteins from interproton distance data by dynamical simulated annealing from a random array of atoms. FEBS Lett., 239, 129–136. [DOI] [PubMed] [Google Scholar]
- 20.Nilges M., Kuszewski,J. and Brunger,A.T. (1991) In Hoch,J.C. (ed.), Computational Aspects of the Study of Biological Macromolecules by NMR. Plenum Press, New York.
- 21.Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- 22.Sippl M.J. (1993) Recognition of errors in three-dimensional structures of proteins. Proteins, 17, 355–362. [DOI] [PubMed] [Google Scholar]
- 23.Godzik A., Kolinski,A. and Skolnick,J. (1992) Topology fingerprint approach to the inverse protein folding problem. J. Mol. Biol., 8, 409–416. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez R. and Sali,A. (1998) Large-scale protein structure modeling of the Saccharomyces cerevisiae genome. Proc. Natl Acad. Sci. USA, 95, 13597–13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eddy S.R. (1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 26.Sali A. and Blundell,T.L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol., 234, 779–815. [DOI] [PubMed] [Google Scholar]
- 27.Zheng M., Aslund,F. and Storz.,G. (1998) Activation of the OxyR transcription factor by reversible disulfide bond formation. Science, 279, 1718–1721. [DOI] [PubMed] [Google Scholar]
- 28.Toledano M.B., Kullik,I., Trinh,F., Baird,P.T., Schneider,T.D. and Storz,G. (1994) Redox-dependent shift of OxyR–DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell, 78, 897–909. [DOI] [PubMed] [Google Scholar]
- 29.Jourdan A.D. and Stauffer,G.V. (1998) Mutational analysis of the transcriptional regulator GcvA: amino acids important for activation, repression and DNA binding. J. Bacteriol., 180, 4865–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schell M.A., Brown,P.H. and Raju,S. (1990) Use of saturation mutagenesis to localize probable functional domains in the NahR protein, a LysR-type transcription activator. J. Biol. Chem., 265, 3844–3850. [PubMed] [Google Scholar]
- 31.Colyer T.E. and Kredich,N.M. (1994) Residue threonine-149 of the Salmonella typhimurium CysB transcription activator: mutations causing constitutive expression of positively regulated genes of the cysteine regulon. Mol. Microbiol., 13, 797–805. [DOI] [PubMed] [Google Scholar]
- 32.Chen C.S., White,A., Love,J., Murphy,J.R. and Ringe,D. (2000) Methyl groups of thymine bases are important for nucleic acid recognition by DtxR. Biochemistry, 39, 10397–10407. [DOI] [PubMed] [Google Scholar]
- 33.Kwon H.J., Bennik,M.H., Demple,B. and Ellenberger,T. (2000) Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nature Struct. Biol., 7, 424–430. [DOI] [PubMed] [Google Scholar]
- 34.Parkinson G., Wilson,C., Gunasekera,A., Ebright,Y.W., Ebright,R.E. and Berman,H.M. (1996) Structure of the CAP–DNA complex at 2.5 angstroms resolution: a complete picture of the protein–DNA interface. J. Mol. Biol., 260, 395–408. [DOI] [PubMed] [Google Scholar]
- 35.Yuan H.S., Finkel,S.E., Feng,J.A., Kaczor-Grzeskowiak,M., Johnson,R.C. and Dickerson,R.E. (1991) The molecular structure of wild-type and a mutant Fis protein: relationship between mutational changes and recombinational enhancer function or DNA binding. Proc. Natl Acad. Sci. USA, 88, 9558–9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gajiwala K.S., Chen,H., Cornille,F., Roques,B.P., Reith,W., Mach,B. and Burley,S.K. (2000) Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature, 403, 916–921. [DOI] [PubMed] [Google Scholar]
- 37.Littlefield O. and Nelson,H.C. (1999) A new use for the ‘wing’ of the ‘winged’ helix–turn–helix motif in the HSF–DNA cocrystal. Nature Struct. Biol., 6, 464–470. [DOI] [PubMed] [Google Scholar]
- 38.Giffard P.M. and Booth,I.R. (1988) The rpoA341 allele of Escherichia coli specifically impairs the transcription of a group of positively-regulated operons. Mol. Gen. Genet., 214, 148–152. [DOI] [PubMed] [Google Scholar]
- 39.McLeod S.M., Aiyar,S.E., Gourse,R.L. and Johnson,R.C. (2002) The C-terminal domains of the RNA polymerase alpha subunits: contact site with Fis and localization during co-activation with CRP at the Escherichia coli proP P2 promoter. J. Mol. Biol., 316, 517–529. [DOI] [PubMed] [Google Scholar]
- 40.Aiyar S.E., McLeod,S.M., Ross,W., Hirvonen,C.A., Thomas,M.S., Johnson,R.C. and Gourse,R.L. (2002) Architecture of Fis-activated transcription complexes at the Escherichia coli rrnB P1 and rrnE P1 promoters. J. Mol. Biol., 316, 501–516. [DOI] [PubMed] [Google Scholar]
- 41.Kondo H., Nakagawa,A., Nishihira,J., Nishimura,Y., Mizuno,T. and Tanaka,I. (1997) Escherichia coli positive regulator OmpR has a large loop structure at the putative RNA polymerase interaction site. Nature Struct. Biol., 4, 28–31. [DOI] [PubMed] [Google Scholar]