Abstract
The conformational pathways and the free energy variations for base opening into the major and minor grooves of a B-DNA duplex are studied using umbrella sampling molecular dynamics simulations. We compare both GC and AT base pair opening within a double-stranded d(GAGAGAGAGAGAG)· d(CTCTCTCTCTCTC) oligomer, and we are also able to study the impact of opening on the conformational and dynamic properties of DNA and on the surrounding solvent. The results indicate a two-stage opening process with an initial coupling of the movements of the bases within the perturbed base pair. Major and minor groove pathways are energetically comparable in the case of the pyrimidine bases, but the major groove pathway is favored for the larger purine bases. Base opening is coupled to changes in specific backbone dihedrals and certain helical distortions, including untwisting and bending, although all these effects are dependent on the particular base involved. Partial opening also leads to well defined water bridging sites, which may play a role in stabilizing the perturbed base pairs.
INTRODUCTION
Nucleic acids undergo a wide variety of thermally induced fluctuations that occur on timescales ranging from picoseconds to milliseconds and with spatial extents ranging from fractions of an angstrom to tens of angstroms. These fluctuations can play important roles in the biological functioning of the nucleic acids. In the case of DNA, fluctuations in local helical conformation, which occur on the picosecond to nanosecond timescale, play a significant role in specific protein–DNA binding by enabling proteins to indirectly probe the base sequence via local changes in mechanical and dynamic behavior.
Amongst the larger deformations of the double helix, base pair disruption is a necessary step to making reactive sites on the bases accessible for chemical attack. Base opening, by which we imply the destruction of Watson–Crick hydrogen bonding within a base pair and movement of at least one base out of helical stack, is an intrinsic part of enzyme-catalyzed DNA modifications, such as selective methylation (1). It is also the first step in the larger scale disruption of the double helix necessary for both replication and transcription. Since the bases are held within the double helix both by Watson–Crick hydrogen bonding and by base stacking, base opening involves much higher activation energies than simple helical deformations, typically of the order of 10–20 kcal mol–1, and consequently it occurs on much longer timescales, typically of the order of tens of milliseconds (2).
Base pair opening can be studied indirectly by following the exchange of labile base protons with the surrounding solvent (3). NMR spectroscopy has proved to be an excellent tool for such purposes (2,4–6). In the case of the imino protons of thymine and guanine, NMR studies, as a function of catalyst concentration and of temperature, have enabled the kinetic and thermodynamic parameters of base opening to be obtained for a number of base sequences. Within canonical B-DNA, adenine–thymine (AT) and guanine–cytosine (GC) pairs are typically associated with opening times of 1–10 and 5–50 ms, respectively, while open bases have lifetimes of the order of a few nanoseconds (2). Certain sequences, such as A tracts (AnTm, where n ≥ 3, m ≥ 0), can lead to an order of magnitude slower exchange rates for thymine (4), while runs of GC pairs appear to accelerate guanine imino exchange (7).
Experimental studies are however unable to define the conformational changes involved in opening and thus the exact nature of an ‘open’ state. Although conformations with bases trapped out of helical stack have been observed crystallographically within DNA (8) and within DNA– protein complexes (1), it is not clear whether these states are directly related to spontaneous base opening in solution. A number of theoretical studies of opening have thus been carried out by molecular mechanics (9–13), molecular dynamics (MD) (14,15) and Brownian dynamics (16,17). However, it has only recently become possible to calculate the free energy pathways associated with base opening. This progress has been achieved by the combination of increased computer power, improvements in force fields (18,19) and, notably, improved treatment of long-range electrostatic interactions (20,21), which enable stable, multi-nanosecond simulations of DNA to be obtained. We made a first step in this direction using a geometrical opening restraint developed for molecular mechanics studies (11) associated with umbrella sampling MD simulations (22–24). Compared to earlier approaches, which either forced the bases of a pair apart with a distance restraint (14,15) or used simplified models assuming the dominant role of given backbone dihedrals (12,13), our restraint enables each base of the pair to be controlled individually, without any prior assumptions as to the conformational consequences of opening. We used this approach to study AT base pair opening in a B-DNA oligomer with a repeating d(GAGAGAGAGAGA) sequence (25), and later extended this to flipping a cytosine completely out of stack within the target sequence of the HhaI methyltransferase enzyme (26). A similar study of a related sequence was also undertaken by Banavali and MacKerell (27) using a different opening restraint, a different force field and a different sampling protocol, but with qualitatively similar results. In the present paper, we compare both AT and GC opening within a single repeating sequence B-DNA oligomer, and extend our earlier studies to a more thorough investigation of the consequences of base opening on neighboring base pairs, the phosphodiester backbones, local and global helical conformations and the environment of solvent and counterions.
MATERIALS AND METHODS
All simulations were performed with the AMBER program version 6.0 (28) using the Parm99 force field (29). A double-stranded DNA 13mer with the sequence d(GAGAGAGAGAGAG)·d(CTCTCTCTCTCTC) was first constructed in a canonical B-DNA conformation. The molecule was then neutralized by the addition of 24 Na+ counterions positioned using the electrostatic potential of the solute. The system was then solvated with 5940 TIP3P water molecules, corresponding to a solvent layer of at least 10 Å, within a truncated octahedral box, which, after equilibration, had a face-to-face dimension of 62 Å. All calculations were carried out using periodic boundary conditions. Long range electrostatic interactions were treated using the particle-mesh Ewald approach (20,21) with a 9 Å direct space cutoff. The simulations were performed at constant temperature and pressure using the Berendsen algorithm (30), and the center of mass motion of the system was removed every 10 ps. Bond lengths involving hydrogen atoms were constrained using SHAKE (31). The equations of motion were integrated using the Verlet algorithm and a 2 fs time step.
The solvent and counterions were relaxed by energy minimization and allowed to equilibrate around a static DNA molecule during 100 ps at 300 K and constant volume. This simulation was then continued for 50 ps, partially relaxing the DNA by applying harmonic restraints to each atom of 5 kcal mol–1 Å–2. These restraints were finally relaxed over a period of 250 ps at constant pressure, and an unrestrained simulation of 2 ns was performed before base opening trajectories were begun.
Base opening was controlled using a geometrical restraint developed during our earlier molecular mechanics studies (11). This restraint, which has been added to the AMBER program, first involves projecting the glycosidic bond of the opening base (C1′–N1 for pyrimidines or C1′–N9 for purines) into a plane containing the C1′–C1′ vector and normal to a local helical axis vector U. U is determined by calculating the mean of the vectors joining the C1′ atoms of the nucleotides on either side of the opening base pair in each strand and then calculating the perpendicular projection of this vector with respect to the C1′–C1′ vector of the opening base pair. The opening angle is then calculated between the projected glycosidic bond and the C1′–C1′ vector. This ensures that the opening angle indeed measures base opening towards the grooves and not base buckling within the helical stack of base pairs (11). In canonical B-DNA, the opening angle θ has a value of ∼55° for the bases of a Watson–Crick pair. If we define the sign of the rotation of base using an axis pointing in the 5′–3′ direction of the corresponding strand, then positive (right-handed) rotation around this axis leads to increasing θ and base opening into the major groove. Negative (left-handed) rotation leads to decreasing θ and base opening into the minor groove. It is remarked that Banavali and MacKerell (27) controlled base opening using a pseudo-dihedral angle between four center of mass coordinates corresponding to the opening base, its attached sugar, the adjacent sugar (5′ for C opening and 3′ for G opening) and its associated base pair. In contrast with our approach, this restraint exerts forces only on one side of the opening base, but the consequences of this are not easy to judge.
After equilibration of the relaxed oligomer, the opening of the bases belonging to the AT and GC pairs closest to the center of the molecule (A8:T19 and G7:C20, numbering each strand of the oligomer in the 5′–3′ direction and starting with the GA strand) was studied using umbrella sampling (22–24). This consisted of a series of simulation ‘windows’ for steadily increasing or decreasing values of θ. For each window i, θ was modified by 5° and restrained to the new value using a harmonic biasing potential Vi(θ) with a force constant of k = 0.05 kcal mol–1 degree–2. We then carried out 50 ps of simulation to equilibrate this conformation, followed by 150 ps of further simulation in order to sample the fluctuations of θ. The conformation obtained at the end of this sampling was used as the starting point for the following window. It is again remarked that this contrasts with the work of Banavali and MacKerell (27), who generated the starting conformations for the entire opening pathway before beginning sampling, using 5 ps of simulation per window and a strong force constant. Our opening pathways required a total of 41 windows for each base, starting with the window corresponding to the relaxed base pair and adding 20 windows in the direction of each groove (changing θ by a total of ±100°). During the sampling phase of each window, the values of θ were stored in bins with a width of 0.5° in order to generate a biased probability histogram, P*i(θ). It was verified that adjacent histograms showed significant overlaps, indicating that all values of θ within the desired range had been appropriately sampled. [Note that sampling was carried out with a 50% smaller restraining force constant than in our earlier simulations (25).]
These histograms were used to obtain the potential of mean force (PMF) or free energy associated with base opening either by a simple splicing algorithm or by the constant temperature weighted histogram analysis method (WHAM) (32–34). In the first case, the free energy of the ith window Wi(θ) is given by:
Wi(θ) = –kBT ln[P*i(θ)] – Vi(θ) + Ci1
where kB is the Boltzmann constant, T is the constant temperature and Ci is a coefficient that can be adjusted to ensure the continuity of the free energy curve at the points of maximum overlap between successive window probability distributions. Note that the splicing method only takes into account non-overlapping sections of the probability distributions. In contrast, in the WHAM approach, the unbiased probability distribution P(θ) is calculated from the complete biased probability distribution as:
P(θ) = [∑ P*i(θ)] / [∑ ni exp {[Fi – Vi(θ)] / kBT}]2
where ni is the number of data points in window i and the coefficients Fi are defined by:
Fi = –kBT · ∫ln P(θ) exp [–Vi(θ) / kBT] dθ3
All sums run over the total number of windows sampled. Once equations 2 and 3 have been iterated to achieve self-consistency (using a tolerance of 10–4), the relative free energy at a given opening angle θ is obtained as:
W(θ) = –kBT ln P(θ)4
Convergence of the free energy results was checked by comparing the results obtained with 50 and 100 ps of sampling to those with 150 ps, and also by repeating many of the opening pathways with different initial atomic velocities. Both these methods suggest that the results obtained are precise to ∼1–2 kcal mol–1 (25,26).
We analyzed the structure of DNA along the base opening pathways using the helicoidal and backbone parameters calculated with the CURVES program (35). Global helicoidal parameters were employed and the helical axis was generated without reference to the opening base pair or the terminal base pairs in order to avoid the perturbations this would cause. Axial bending was calculated as the angle between the ends of the CURVES helical axis using only four base pairs on either side of the opening base. The direction of bending was obtained by calculating where the end-to-end vector of the overall helical axis intersected a plane perpendicular to the helical axis at the level of the opening base pair. The reference for calculating the bending direction (0°) was taken to be the major groove dyad in the central plane and its sign was defined using right-hand rotation around an axis pointing in the 5′–3′ direction of the GA strand. Note that histograms with respect to bending direction only include conformations for which the bending amplitude was >15°.
Atomic accessibilities were calculated using a Korobov grid of 610 points per atom, Pauling atomic radii and a probe sphere of 1.4 Å radius (36). Solvent structure was studied by accumulating the positions of the water molecules surrounding the perturbed base pairs using 1 ps snapshots during the 150 ps of trajectory at each opening angle. In order to define an axis system around the opening base pair, we used the C1′–C1′ vector of the perturbed base pair as the x-axis, the local helical axis U as the z-axis and major groove dyad, defined by the vector product of the two former vectors, as the y-axis. Water molecules were only considered if their oxygen atoms fell with x = y = ±8 Å and z = ±1 Å.
Excluding both repeated opening pathways, and the extended simulations carried out for certain partially open states, ∼35 ns of simulation were necessary to complete this study. Calculations were performed partially on a Linux PC cluster using two 1.2 GHz processors for each run and requiring ∼1 week per nanosecond of simulation.
RESULTS AND DISCUSSION
We divide the discussion of our umbrella sampling simulations into three sections dealing respectively with the energetics of base opening, its structural consequences for the DNA oligomer and the associated changes in solvent distribution around the oligomer.
Free energy profile and base movements
The opening free energy profiles for the four bases are shown in Figure 1. As in our earlier publications (25,26), we describe opening with the relative opening angle Δθ. This angle is defined as θ–θ0, where θ0 is the value of θ in the unperturbed DNA duplex. The average structures obtained from our initial dynamic simulations indicate that θ0 is 55 ± 2° for all the bases studied. Following the definitions given in the preceding section, positive Δθ values correspond to base opening into the major groove and negative values to opening into the minor groove.
Figure 1.
Free energy curves as a function of the relative opening angle Δθ with respect to the relaxed position of each base within the central AT and GC pairs of a B-DNA 13mer with an alternating GA sequence. Purine pathways are shown as solid lines and pyrimidine pathways as dotted lines.
Figure 1 shows that the free energy profiles for opening each of the four bases can be divided into two regimes: a quadratic zone for –20° < Δθ < +25° and a roughly linear zone beyond these values. Reaching the limit of the quadratic zone requires free energy increases of ∼7–11 kcal mol–1, with slightly lower values on the major groove side (Δθ > 0) for all bases except guanine. Opening until Δθ = ±100° requires a total free energy input of 13–23 kcal mol–1. Although all four bases can be opened into both the major and minor grooves, the free energy profile is only roughly symmetric for the pyrimidines, while the purines show a clear energetic preference of 7–9 kcal mol–1 for opening into the major groove. It is nevertheless remarked that cytosine also has lower free energy values for intermediate opening angles along the major groove pathway.
These results are very similar to those presented in our earlier publication (25) concerning AT opening, despite minor changes in the force field used and in the simulation protocol. They are also similar to the results of Banavali and Mackerell (27), for small opening angles. However, these authors find virtually no energy increase beyond the quadratic zone. In setting up the protocol for simulating base opening, we repeated most of the opening pathways shown in Figure 1 more than once. These results confirmed the findings we have described and showed that the free energy curves could be reproduced to within ∼2 kcal mol–1. These studies however also revealed some rarer events which are discussed shortly.
As previously seen for the AT base pair (25), opening either base leads to coupled movements of the paired partner. This is illustrated in Figure 2. In each diagram, the horizontal axis refers to the restrained opening base and the vertical axis to the paired partner, whose freely varying opening angle is plotted. The basic behavior in each case is that for small opening angles, the partner follows the opening base, before springing back, somewhat like the behavior of saloon doors. This coupling is strongest for controlled thymine opening and weakest for cytosine. For larger opening angles, the partner base may again move, towards either of the grooves, presumably to reinforce its stacking with the neighboring base pairs.
Figure 2.
Coupling movements within base pairs during opening. The solid curve (Δθ vertical axis) shows movement of the paired base during opening of the restrained base (Δθ horizontal axis). In the case of cytosine opening (bottom right) the dotted curve shows the movement of thymine on the 3′ side of the restrained base.
If we look at the effect of base opening on the neighboring base pairs, no significant perturbation is seen when opening either adenine or thymine, in line with our earlier results (11,25). However, a striking change occurs for opening cytosine into the minor groove, when the adjacent 3′-thymine was found to open in a coupled manner, maintaining stacking with the restrained cytosine. This effect is illustrated by the dotted line in the bottom right-hand diagram of Figure 2, which plots the progress of the thymine opening angle. In order to verify that cytosine opening significantly weakens the adjacent AT pair, we re-calculated the free energy profile for thymine opening into the minor groove, with the 5′-cytosine held at Δθ = –55°. The result (Fig. 3) shows that thymine can now reach an opening angle of –50° for a free energy cost of 4 kcal mol–1 (b) compared to almost 12 kcal mol–1 when the adjacent GC pair is unperturbed (a). No such coupling of adenine to guanine opening was seen, although one of the pathways calculated for guanine displacement into the minor groove did show disruption of the AT pair. This coupling requires further studies, but suggests that the strictly local nature of base pair opening (37) may not hold for all base sequences.
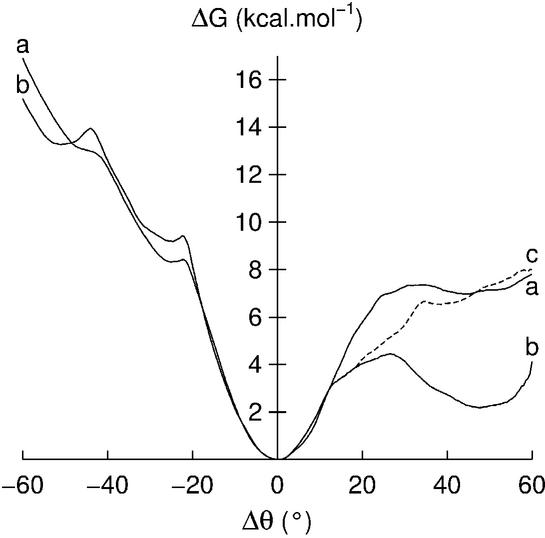
Figure 3.
Free energy curves for thymine opening into the minor groove: (a) reference curve, as shown in Figure 1; (b) curve obtained when the adjacent 5′-cytosine is pre-opened into the minor groove by 55°.
We now turn to another specific event that involved one of the profiles calculated for adenine opening. This profile (Fig. 4b) is very similar to the ‘normal’ profile (Fig. 4a and already presented in Fig. 1) on the minor groove side, but exhibits an energy minimum for Δθ = ∼50° on the major groove side. Analysis of the solvent and ion environment in this simulation showed that two sodium counterions were close to the opening base, whereas this was not the case for the normal profile. We consequently re-sampled the opening profile, starting from Δθ = 10°, after having moved the sodium ions in question further away from DNA, by exchanging their positions with those of two distant water molecules and re-equilibrating the system. The resulting profile (Fig. 4c) now closely resembles the original free energy pathway. This demonstrates a general problem with slowly moving counterions for nanosecond scale simulations, which we are currently trying to characterize more quantitatively.
Figure 4.
Free energy curves for adenine opening: (a) reference curve, as shown in Figure 1; (b) curve perturbed by the proximity of two sodium counterions; (c) curve recalculated after moving the two counterions away from the opening base.
Conformational changes
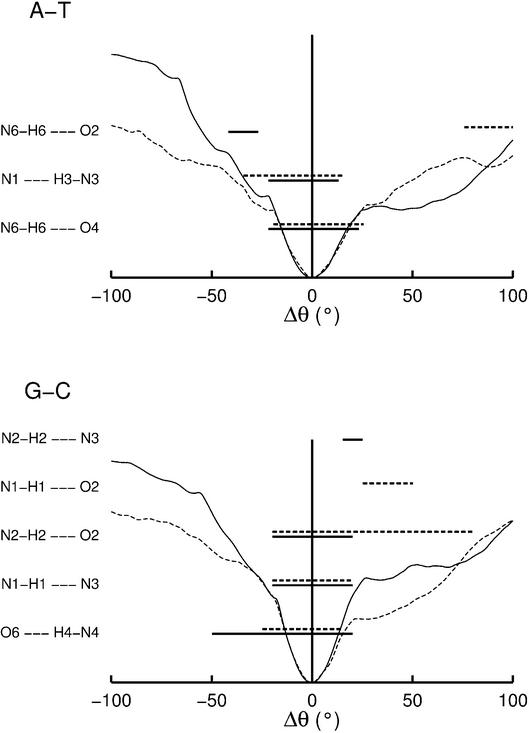
Hydrogen bonding, accessibility and base orientation. We now look at the conformational consequences of base opening. First, it is interesting to ask when the base pair hydrogen bonds break along the base opening profiles. The results are shown in Figure 5 as horizontal bars. Bars are plotted when the corresponding hydrogen bond donor–acceptor distance (i.e. between the heavy atoms) is <3.5 Å and deviates from linearity by <30°. Solid lines refer to purine opening and dotted lines to pyrimidine opening. The free energy profiles, which are also plotted in Figure 5, show that their quadratic zones indeed refer to elastic distortions of the base pairs, since the Watson–Crick hydrogen bonds almost always break at the limits of these zones. It is however interesting to note that, for the GC pair, the O6–N4 bond, which lies on the major groove side of this base pair, persists for minor groove opening until Δθ = –50°. Conversely, the N2–O2 bond on the minor groove side persists for major groove opening until Δθ = 80°. For larger opening angles, both the GC and AT pairs form unconventional hydrogen bonds, due to the proximity of appropriate acceptor and donor groups. The zones corresponding to these new interactions, G(N1)–C(O2), G(N2)– C(N3) and A(N6)–T(O2), are also illustrated in Figure 5.
Figure 5.
Hydrogen bonds formed within the AT and GC base pairs as a function of base opening. The horizontal lines indicate hydrogen bond with distances <3.5 Å and <30° departure from linearity. The opening free energy profiles are shown for reference. Solid lines indicate purine opening and dotted lines indicate pyrimidine opening.
We can attempt to determine which value of Δθ is likely to correspond to ‘opening’ as defined experimentally by the observation of imino proton exchange with the surrounding solvent (2). This can be measured by following the surface accessibility of the imino protons of thymine and guanine as these bases open. Figure 6 shows that these protons only become significantly accessible once the base pair hydrogen bonds have been broken and the opening bases have moved out of the helical stack. This only occurs for Δθ values of ∼±50°. It is of note that, in Figure 5, thymine has a lower accessibility for major groove opening compared to our earlier simulations (25). This is related to stronger overall bending and positive roll angles between the base pairs flanking the opening base (see discussion below).
Figure 6.
Percentage surface accessibilities (with respect to the isolated bases) for the imino protons of guanine N1 (solid line) and thymine N3 (dotted line) as a function of the opening angle Δθ. Values are calculated from the averaged conformation after 150 ps of sampling at the specified opening angle using a probe sphere representing a water molecule (radius 1.4 Å).
It is also noted that accessibility does not necessarily increase monotonically for increased opening. This is because once the opening base escapes from the helical stack, it need not remain in a plane more or less perpendicular to the DNA axis. Base rotation is strongest for minor groove opening and, as a result, the imino proton of thymine at Δθ = –80° becomes practically inaccessible (Fig. 6). Base rotation into the groove appears to be advantageous both because it enables the relatively hydrophobic base faces to be shielded from aqueous solution and also because it allows hydrogen bond formation between the open base and either base or backbone sites within the grooves. We do not detail these interactions since they are strongly dependent on the nature of the opening base and on the surrounding DNA sequence. It should however be noted that rotation into the minor groove places the opening base on the 5′ side, with respect to its own strand.
Backbone conformation. A detailed analysis of the changes in backbone conformations as base opening occurs show several general features. First, the dihedral angles which constitute the dinucleotide junctions adjacent to the opening base, or belonging to the equivalent junctions in the complementary strand, often show an increase in their oscillations or undergo full transitions, including: α: g– → g+; β: t → g+; γ: g+ → t or, more rarely, g+ → g–; and εζ: BI (tg–) → BII (g–t). These changes are likely to be the consequence of an increase in conformational freedom corresponding to the destruction of base stacking on either side of the opening base.
Several trends in the dihedral angle changes can be observed: transitions in the strand containing the opening base tend to occur on the 5′ side of this base for opening into the minor groove and on the 3′ side for opening into the major groove. The complementary strand shows an inverse tendency and also exhibits a high proportion of εζ: BI → BII transitions. None of these effects, however, appears to be indispensable for base opening as their occurrence depends on which base is involved and on which pathway is studied. Indeed, all these changes are likely to be coupled to the base–groove interactions discussed above, and are thus also sensitive to the nature of the opening base and to its surrounding sequence.
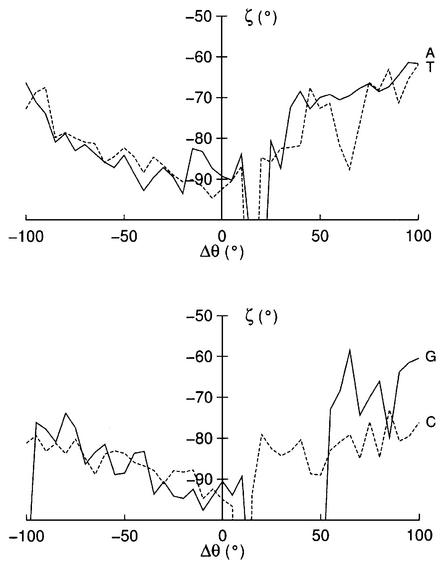
If we now turn to finer conformational changes, it is possible to find more direct coupling with base opening. This is the case for ζ on the 3′ side of the opening base (Fig. 7), which moves from its average value of ∼–90° to values of ∼–70° as opening occurs into either groove. This coupling has already been invoked by Chen et al. (12,13) in a series of reduced (and constrained) coordinate molecular mechanics studies as a means of provoking base opening. The ζ dihedral seems to be a good candidate for this purpose since its central O3′–P bond is almost perpendicular to the base pair plane. We carried out preliminary MD simulations to test this mechanism (data not shown), but were unable to produce any base opening. We thus conclude that although ζ and opening are coupled, the changes in ζ are a consequence of opening and not a driving force. It is of note that, in addition to the fine changes in the 3′ ζ angle, Figure 7 also shows temporary g– → t transitions, which coincide with breaking the hydrogen bonds of the base pairs along the major groove opening pathway for each of the bases studied. It is reasonable to assume that these changes may also be part of the opening mechanism.
Figure 7.
Changes in ζ (C3′–O3′–P–O5′) on the 3′ side of the opening base as a function of Δθ. Solid lines correspond to purines and dotted lines to pyrimidines.
Sugar phase also appears to show quite strong coupling to base opening, moving from typical south (around C2′-endo) to north (C3′-endo) puckers as opening occurs. For the minor groove pathway, these changes generally involve the nucleotide on the 5′ side of the opening base, whereas for the major groove pathway it is the sugar of the opening base itself that transits. Similar pucker changes occur in the complementary strand, but can involve either the facing nucleotide or its 5′ or 3′ neighbors. Lastly, significant changes in the glycosidic angle χ also occur, but these are directly related to the rotation of the opening base into the groove of the duplex which have been discussed above.
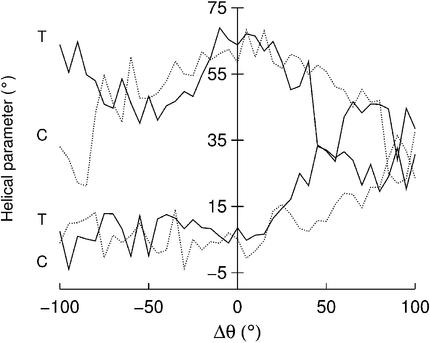
Helical conformation. We have looked at the influence of base opening both on the helical conformation surrounding the perturbed base pair and on the overall structure of the duplex. The first study involved calculating the CURVES (35) inter-base pair parameters (shift, slide, rise, tilt, roll, twist) between the base pairs that flank the opening base. The results show variations of ∼1 Å for the translational parameters and of ∼10° (tilt) or ∼20° (twist and roll) for the rotational parameters as base opening occurs towards either groove. Few of these parameters, however, show clear trends linked to opening. The main exception to this rule involves the pyrimidine opening pathways. For both cytosine and thymine, twist decreases regularly as these bases are opened into either groove (as shown by the upper two curves in Fig. 8). In the case of the major groove pathway, unwinding is accompanied by an increase in roll (lower two curves in Fig. 8), which closes down the distance between the flanking base pairs on the major groove side and results in strong bending into the major groove (see below). The trend to increasingly positive roll for major groove opening is also visible for the purines (data not shown), although the changes are smaller than those seen for pyrimidine opening.
Figure 8.
Changes in the helical parameters twist (upper two curves) and roll (lower two curves) between the base pairs flanking the opening base as a function of Δθ. Solid lines correspond to thymine opening and dotted lines to cytosine opening.
The second study looked for evidence of coupling between base opening and bending, as found in our earlier work (9–11). Figure 9 shows histograms of bending amplitude and direction for each opening base. In order to accumulate sufficient data, we have divided the opening pathway into three segments: Δθ < –50° corresponding to significant minor groove opening (shown in blue in Fig. 9), Δθ > 50° corresponding to significant major groove opening (shown in green) and –50° < Δθ < 50° corresponding to only moderately perturbed base pairing (shown in red). These results can be contrasted with 5 ns of data on the relaxed DNA oligomer, shown by the black lines in each diagram. Before base opening, the helix has a mean bending amplitude of ∼10°. This bending is rather anisotropic and clearly favoring bending towards the major groove direction in the center of the oligomer. This result is in line with other MD results (38) and with observations from crystallographic data (39). Once the base pair is disrupted (red curves), a small increase in bending amplitude occurs, associated, in the case of A and T, with a sharpening of the bending direction histogram around the major groove dyad. This trend continues for significant opening into the minor groove (blue curves), where all bases show an increase in bending amplitude. The bending direction is once again focused around the major groove dyad, with the exception of thymine which shows a bias towards the TC strand. Significant opening into the major groove (green curves) provokes even stronger bending for all bases except adenine, although, once again, the favored bending direction is into the major groove (although cytosine also shows some of bending towards the minor groove). We have tested these results, which were accumulated during the opening process, with a number of longer simulations where the open base is held at a fixed Δθ angle. An example of this can be seen in the plots for thymine, where the blue and green dotted curves correspond to results obtained over 1 ns at Δθ = –65° and Δθ = +65°, respectively. These curves show similar results to the data accumulated during the opening process, although the bias of bending towards the TC strand for minor groove thymine opening is enhanced. We can conclude that opening and bending are indeed coupled, although, in contrast to the predictions of our earlier molecular mechanics studies (9–11), enhanced bending is dominantly in the major groove direction, rather than in the direction opposite to base opening.
Figure 9.
Changes in overall helical bending amplitude and direction as a function of base opening. For each base, histograms are shown for the relaxed duplex (black), small opening angles, –50° < Δθ < 50° (red), significant minor groove opening, Δθ < –50° (blue) and significant major groove opening, Δθ > 50° (green). For thymine, the dotted curves refer to 1 ns simulations at fixed opening angles: Δθ = –65° (blue), Δθ = 65° (green).
Solvation
We have looked at the impact of base pair opening on the distribution of water around the double helix by sampling water positions within a 16 × 16 × 2 Å box centered on the opening base pair (see Materials and Methods). Water oxygen positions, which were sampled every 1 ps for 150 ps in each window, are indicated by colored spheres. Each water molecule is assign a different color, although, given the limited number of colors which can be visually distinguished, geometrically distant waters may be allocated the same colors. This graphic enables persistently occupied hydration sites to be easily visualized. The results in Figure 10 refer to the unperturbed AT and GC base pairs and show that well defined hydration sites have water residency times of the order of tens of picoseconds, for example around the pyrimidine O2 groups. This is compatible with current values deduced from NMR experiments (40). During disruption of the base pairs, the coupled base movements discussed in Backbone Conformation lead to the creation of a number of new bidentate water binding positions between the disrupted Watson–Crick partners. As Figure 11 shows, for the case of Δθ = ±45°, this is associated with a striking increase in long-lived water binding sites. The majority of these sites are occupied for >50% of the 150 ps sampling at the specified opening angle.
Figure 10.
Water distributions sampled during 150 ps around the relaxed AT and GC base pairs (Δθ = 0°). Colored spheres indicate water oxygen atoms lying within a 16 × 16 × 2 Å box centered on the base pair. Each water molecule has a fixed color during the sampling, but, due to a limited palette, distant waters may be assigned identical colors. Molecular images were generated with VMD (43).
Figure 11.
Water distributions sampled during 150 ps around partially opened AT and GC base pairs. Minor groove openings are shown on the left (Δθ = –45°) and major groove openings on the right (Δθ = 45°). Colored spheres indicate water oxygen atoms (see caption to Fig. 10).
Analysis of these sites has enabled us to define the average bound water positions, which are shown in Figure 12. It is of note that the presence of such a bridging water molecule has also been seen in semiempirical quantum mechanical studies (41) and in crystallographic studies of an open GC base pair (8). Their existence has also been deduced from NMR studies of base pair opening to explain exchange rates in the absence of added catalyst by allowing an indirect proton transfer from T(N3) to A(N1) (37). These waters, which bridge acceptor and donor sites on the opening base pair, may be expected to play a role in stabilizing partially open base pair at rather specific opening angles, and it is tempting to associate some of the depressions in the free energy curves with such water bridges, although we have not attempted to investigate this further. Work is presently underway in the group of C. Griesinger (Max Planck Institut für Biophysikalische Chemie, Göttingen) and H. Schwalbe (University of Frankfurt) to determine whether such structures correspond to intermediate open states (before imino proton exchange occurs), which explain observed changes in the width of imino proton NMR resonances.
Figure 12.
Bridging sites between the bases of partially open base pairs, derived from the water clusters visible in Figure 12. Minor groove openings are shown on the left (Δθ = –45°) and major groove openings on the right (Δθ = 45°). Each water shown is that lying closest to the center of the corresponding cluster.
Lastly, it is worth remarking that opening beyond ∼80° leads to the formation of a water channel linking the two sides of the double helix, filling the gap left in the base stack. It is of note that a similar effect has recently been observed in molecular dynamics simulations of DNA containing an abasic site (42).
CONCLUSIONS
Base opening can be studied theoretically in a controlled manner using a simple angular restraint coupled with an MD umbrella sampling approach. This method allows the free energy and conformational changes linked to opening to be calculated for a DNA oligomer while taking into account explicit solvent and counterions.
The results for a repeating GA sequence oligomer show that both AT and GC base pairs begin to open with a quadratic energy regime corresponding to elastic deformation, followed by a roughly linear regime once the Watson–Crick hydrogen bonds rupture and progressive base unstacking occurs. Accessibility calculations show that base opening, as measured experimentally by guanine or thymine imino proton exchange, requires ∼50° of opening into either the minor or major grooves. Although both routes are energetically accessible, major groove opening is clearly favored for the larger purine bases.
The movements of both bases within a pair are generally coupled until their hydrogen bonding is broken. Although the effect of opening on neighboring base pairs is minor in the case of adenine or thymine opening, cytosine opening into the minor groove, at least for the sequence studied here, dramatically weakens the adjacent 3′-AT pair and leads to coupled cytosine and thymine rotation.
From a conformational point of view, opening can be produced with only small conformational changes in the DNA backbones. However, once a base is removed from the helical stack, the backbones exhibit increased dihedral and sugar pucker transitions. Only the 3′-ζ dihedral shows consistent coupling to base opening, but this dihedral cannot be used to drive opening. Opening modifies the overall structure of the DNA duplex, leading to enhanced bending into the major groove and, in the case of pyrimidine opening, to local helical untwisting.
Concerning environmental effects, partial opening leads to the creation of long-lived, and potentially stabilizing, water bridging sites between the originally paired bases. The effects of counterions on partially open pairs were also seen to be potentially important, but it is clear that understanding such effects will require much longer simulations or other algorithmic approaches.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to acknowledge the CINES (France) computer center for access to their facilities. P.V. also wishes to thank The Wellcome Trust for a traveling research fellowship (grant reference 060078).
REFERENCES
- 1.Klimasauskas S., Kumar,S., Roberts,R.J. and Cheng,X. (1994) HhaI methyltransferase flips its target base out of the DNA helix. Cell, 76, 357–369. [DOI] [PubMed] [Google Scholar]
- 2.Guéron M. and Leroy,J.L. (1992) Base-pair opening in double-stranded nucleic acids. In Eckstein,F. and Lilley,D.M.J. (eds), Nucleic Acids and Molecular Biology. Springer-Verlag, New York, Vol. 6, pp. 1–22.
- 3.Englander S.W. and Kallenbach,N.R. (1984). Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q. Rev. Biophys., 16, 521–655. [DOI] [PubMed] [Google Scholar]
- 4.Leroy J.L., Charretier,E., Kochoyan,M. and Guéron,M. (1988). Evidence from base-pair kinetics for two types of adenine tract structures in solution: their relation to DNA curvature. Biochemistry, 27, 8894–8898. [DOI] [PubMed] [Google Scholar]
- 5.Folta-Stogniew E. and Russu,I.M. (1994). Sequence dependence of base-pair opening in a DNA dodecamer containing the CACA/GTGT sequence motif. Biochemistry, 33, 11016–11024. [DOI] [PubMed] [Google Scholar]
- 6.Guéron M. and Leroy,J.L. (1995). Studies of base pair kinetics by NMR measurement of porton exchange. Methods Enzymol., 261, 383–413. [DOI] [PubMed] [Google Scholar]
- 7.Dornberger U., Leijon,M. and Fritzsche,H. (1999) High base pair opening rates in tracts of GC base pairs. J. Biol. Chem., 274, 6957–6962. [DOI] [PubMed] [Google Scholar]
- 8.van Aalten D.M.F., Erlanson,D.A., Verdine,G.L. and Joshua-Tor,L. (1999) A structural snapshot of base-pair opening in DNA. Proc. Natl Acad. Sci. USA, 96, 11809–11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramstein J. and Lavery,R. (1988) Energetic coupling between DNA bending and base pair opening. Proc. Natl Acad. Sci. USA, 85, 7231–7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramstein J. and Lavery,R. (1990) Base pair opening pathways in B-DNA. J. Biomol. Struct. Dyn., 7, 915–933. [DOI] [PubMed] [Google Scholar]
- 11.Bernet J., Zakrzewska,K. and Lavery,R. (1997) Modelling base pair opening: the role of helical twist. J. Mol. Struct. (Theochem), 398–399, 473–482. [Google Scholar]
- 12.Chen Y.Z., Mohan,V. and Griffey,R.H. (1998) Effect of backbone ζ torsion angle on low energy single base opening in B-DNA crystal structures. Chem. Phys. Lett., 287, 570–574. [Google Scholar]
- 13.Chen Y.Z., Mohan,V. and Griffey,R.H. (1998) The opening of a single base without perturbations of neighboring nucleotides: a study on crystal B-DNA duplex d(CGCGAATTCGCG)2. J. Biomol. Struct. Dyn., 15, 765–777. [DOI] [PubMed] [Google Scholar]
- 14.Keepers J., Kollman,P.A., Weiner,P.K. and James,T.L. (1982) Molecular mechanical studies of DNA flexibility: coupled backbone torsion angles and base-pair opening. Proc. Natl Acad. Sci. USA, 79, 5337–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keepers J., Kollman,P.A. and James,T.L. (1984) Molecular mechanical studies of base-pair opening in d(CGCGC):d(GCGCGC), dG5.dC5, d(TATAT):d(ATATA) and dA5.dT5 in the B and Z forms of DNA. Biopolymers, 23, 2499–2511. [DOI] [PubMed] [Google Scholar]
- 16.Briki F., Ramstein,J., Lavery,R. and Genest,D. (1991) Evidence for the stochastic nature of base pair opening in DNA: a Brownian dynamics simulation. J. Am. Chem. Soc., 113, 2490–2493. [Google Scholar]
- 17.Briki F. and Genest,D. (1993) Molecular dynamics study of the base pair opening process in the self-complementary octanucleotide d(CTGATCAG). J. Biomol. Struct. Dyn., 11, 43–56. [DOI] [PubMed] [Google Scholar]
- 18.Cheatham T.E. III, Cieplak,P. and Kollman,P.A. (1999) A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn., 16, 845–862. [DOI] [PubMed] [Google Scholar]
- 19.Foloppe N. and MacKerell,A.D.,Jr (2000) All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem., 21, 86–104. [Google Scholar]
- 20.Darden T., York,D. and Pedersen,L. (1993) Particle mesh Ewald: an N*log(N)method for computing Ewald sums. J. Chem. Phys., 98, 10089–10092. [Google Scholar]
- 21.Cheatham T.E. III, Miller,J.L., Fox,T., Darden,T.A. and Kollman,P.A. (1995) Molecular dynamics simulation on solvated biomolecular systems: the particle mesh Ewald method leads to stable trajectories of DNA, RNA and proteins. J. Am. Chem. Soc., 117, 4193–4194. [Google Scholar]
- 22.Torrie G.M. and Valleau,J.P. (1977) Nonphysical sampling distribution in Monte Carlo free-energy estimation: umbrella sampling. J. Comput. Phys., 23, 187–199. [Google Scholar]
- 23.Northrup S.H., Pear,M.R., Lee,C.Y., McCammon,J.A. and Karplus,M. (1982) Dynamical theory of activated processes in globular proteins. Proc. Natl Acad. Sci. USA, 79, 4035–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tobias D.J., Sneddon,S.F. and Brooks,C.L.,III (1990) Reverse turns in blocked dipeptides are intrinsically unstable in water. J. Mol. Biol., 216, 783–796. [DOI] [PubMed] [Google Scholar]
- 25.Giudice E., Varnai,P. and Lavery,R. (2001) Energetic and conformational aspect of A:T base pair opening within the DNA double helix. Chem. Phys. Chem., 2, 673–677. [DOI] [PubMed] [Google Scholar]
- 26.Varnai P. and Lavery,R. (2002) Base flipping in DNA: pathways and energetics studied with molecular dynamics simulations. J. Am. Chem. Soc., 124, 7272–7273. [DOI] [PubMed] [Google Scholar]
- 27.Banavali N.K. and MacKerell,A.D.,Jr (2002) Free energy and structural pathways of base flipping in a DNA GCGC containing sequence. J. Mol. Biol., 319, 141–180. [DOI] [PubMed] [Google Scholar]
- 28.Case D.A., Pearlman,D.A., Caldwell,J.W., Cheatham,T.E.,III, Ross,W.S., Simmerling,C.L., Darden,T.A., Metz,K.M., Stanton,R.V., Cheng,A.L., Vincent,J.J., Crowley,M., Tsui,V., Radmer,R.J., Duan,Y., Pitera,J., Massova,I., Seibel,G.L., Singh,U.C., Weiner,P.K. and Kollman,P.A. (1999) AMBER 6. University of California, San Francisco.
- 29.Wang J., Cieplak,P. and Kollman,P.A. (2000) How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem., 21, 1049–1074. [Google Scholar]
- 30.Berendsen H.J.C., Postma,J.P.M., van Gunsteren,W.F., DiNola,A. and Haak,J.R. (1984) Molecular dynamics with coupling to an external bath. J. Chem. Phys., 81, 3684–3690. [Google Scholar]
- 31.Ryckaert J.P., Ciccotti,G. and Berendsen,H.J.C. (1977) Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys., 23, 327–341. [Google Scholar]
- 32.Kumar S., Bouzida,D., Swendsen,R.H., Kollman,P.A. and Rosenberg,A.J.M. (1992) The weighted histogram analysis method for free energy calculations on biomolecules. I. The method. J. Comput. Chem., 13, 1011–1021. [Google Scholar]
- 33.Boczko E.M. and Brooks,C.L.,III (1993) Constant-temperature free energy surfaces for physical and chemical processes. J. Phys. Chem., 97, 4509–4513. [Google Scholar]
- 34.Roux B. (1995) The calculation of the potential of mean force using computer simulations. Comp. Phys. Commun., 91, 275–282. [Google Scholar]
- 35.Lavery R. and Sklenar,H. (1989) Defining the structure of irregular nucleic acids: convention and principles. J. Biomol. Struct. Dyn., 6, 655–667. [DOI] [PubMed] [Google Scholar]
- 36.Lavery R., Pullman,A. and Pullman,B. (1981) Steric accessibility of reactive centers in B-DNA. Int. J. Quant. Chem., 20, 49–62. [Google Scholar]
- 37.Guéron M., Kochoyan,M. and Leroy,J.L. (1987) A single mode of DNA base-pair opening drives imino proton exchange. Nature, 328, 89–92. [DOI] [PubMed] [Google Scholar]
- 38.McConnell K.J. and Beveridge,D.L. (2001) Molecular dynamics simulations of B′-DNA: sequence effects on A-tract-induced bending and flexibility. J. Mol. Biol., 314, 23–40. [DOI] [PubMed] [Google Scholar]
- 39.Crothers D.M. (1998) DNA curvature and deformation in protein–DNA complexes: a step in the right direction. Proc. Natl Acad. Sci. USA, 95, 15163–15165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phan A.T., Leroy,J.L. and Guéron,M. (1999) Determination of the residence time of water molecules hydrating B′-DNA and B-DNA, by one-dimensional zero-enhancement nuclear Overhauser effect spectroscopy. J. Mol. Biol., 286, 505–519. [DOI] [PubMed] [Google Scholar]
- 41.Kryachko E.S. and Volkov,S.N. (2001) Preopening of the DNA base pairs. Int. J. Quant. Chem., 82, 193–204. [Google Scholar]
- 42.Barsky D., Foloppe,N., Ahmadia,S., Wilson,D.M.,III and MacKerell,A.D.,Jr (2000) New insights into the structure of abasic DNA from molecular dynamics simulations. Nucleic Acids Res., 28, 2613–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Humphrey W., Dalke,A. and Schulten,K. (1996) VMD–Visual Molecular Dynamics. J. Mol. Graph., 14, 33–38. [DOI] [PubMed] [Google Scholar]