Abstract
Primary, secondary and higher-order structures of downstream elements of mammalian pre-mRNA polyadenylation signals [poly(A) signals] are re viewed. We have carried out a detailed analysis on our database of 244 human pre-mRNA poly(A) signals in order to characterize elements in their downstream regions. We suggest that the downstream region of the mammalian pre-mRNA poly(A) signal consists of various simple elements located at different distances from each other. Thus, the downstream region is not described by any precise consensus. Searching our database, we found that ∼80% of pre-mRNAs with the AAUAAA or AUUAAA core upstream elements contain simple downstream elements, consisting of U-rich and/or 2GU/U tracts, the former occurring ∼2-fold more often than the latter. Approximately one-third of the pre-mRNAs analyzed here contain sequences that may form G-quadruplexes. A substantial number of these sequences are located immediately downstream of the poly(A) signal. A possible role of G-rich sequences in the polyadenylation process is discussed. A model of the secondary structure of the SV40 late pre-mRNA poly(A) signal downstream region is presented.
INTRODUCTION
Poly(A) tails are essential structural and functional elements of eukaryotic mRNAs. They are important for the regulation of mRNA stability, mRNA export from the nucleus to the cytoplasm, translation initiation and, possibly, for other cellular events (1–5). Moreover, use of alternative polyadenylation sites has been shown to be important for the regulation of gene expression (6). It has been suggested that polyadenylation signals [poly(A) signals] are required for effective splicing, transcription termination and, possibly, even for translation termination (7–10). Thus, further study of polyadenylation mechanisms is of great importance.
Polyadenylation of pre-mRNA is a universal modification, but different organisms use different mechanisms to carry it out (11–14). The polyadenylation mechanism of mammalian pre-mRNAs has been well studied (12,14–17). The reaction is performed by a large set of proteins and is dependent on a minimum of two elements in the pre-mRNA molecule. There is an upstream element consisting of a highly conserved AAUAAA hexamer, and a downstream element often described as a poorly conserved GU- or U-rich sequence (for example, see 18–20).
This review is devoted to analyzing the downstream elements of higher eukaryotic pre-mRNA poly(A) signals. After analyzing data from the literature, we conclude that these elements usually consist of collections of short U-rich and/or GU-rich sequences.
PRIMARY STRUCTURE OF DOWNSTREAM ELEMENTS
The polyadenylation reaction of mammalian pre-mRNAs proceeds in two stages: the endonucleolytic cleavage of pre-mRNA and the subsequent addition of poly(A) sequence to the newly formed 3′ end (12,14–17). The cleavage reaction requires the cleavage and polyadenylation specificity factor (CPSF), the cleavage stimulation factor (CstF), the cleavage factors I and II (CF I and CF II), and, in most cases, poly(A) polymerase (PAP). The addition of poly(A) requires CPSF, PAP and a third protein, poly(A) binding protein 2. The assembly of the cleavage complex, which contains most or all of the processing factors and the substrate RNA, occurs cooperatively (16). CPSF binds to the AAUAAA hexamer, CstF interacts with the U/GU-rich element located downstream of the hexamer, and these two proteins interact with each other. Pre-mRNA is cleaved at a site located between the hexamer and the U/GU-rich element. The interaction of CPSF and CF I with pre-mRNA is apparently one of the earliest events of the cleavage complex assembly. CF II has been only partially purified so far, and its role is not completely clear (21).
The characteristic feature of the AAUAAA element is that most single base substitutions significantly reduce the cleavage and polyadenylation efficiencies. The only exception is AUUAAA, which directs cleavage in vitro with an efficiency of 66%, relative to the wild-type level (22). Polyadenylation of precleaved pre-mRNAs proceeds with an efficiency of >10% in the presence of eight out of the possible 18 singly substituted variants of the AAUAAA hexamer. Recent statistical analysis of higher eukaryotic DNA databases (23–25) has revealed that the AAUAAA element is not as universal as it has been considered to be previously (15–17). According to recent data (23–25), only ∼70% of human 3′ expressed sequence tags (ESTs) contain one of the two optimal sequences (AAUAAA or AUUAAA). By in silico detection of poly(A) signals, Graber et al. (23) showed that the occurrence of AAUAAA variants with single base substitutions correlates with their respective processing efficiencies measured in vitro. The most efficiently processed variants appeared to be elements of poly(A) signals of eukaryotic pre-mRNAs (24). They are described by an NNUANA consensus, where N is any nucleotide. Thus, nucleotides at positions 3, 4 and 6 are the most conserved.
The use of suboptimal AAUAAA-like elements in vivo could be explained by the cooperative mode of pre-mRNA sequences recognition by the polyadenylation factors, when a weak interaction of one protein factor with the binding site can be compensated by a stronger interaction of another factor (20,23,26,27).
An exact consensus sequence for the downstream element of the poly(A) signal has not been determined. Here we suggest that no general consensus exists, but rather the downstream element consists of various simple elements located at different distances from each other.
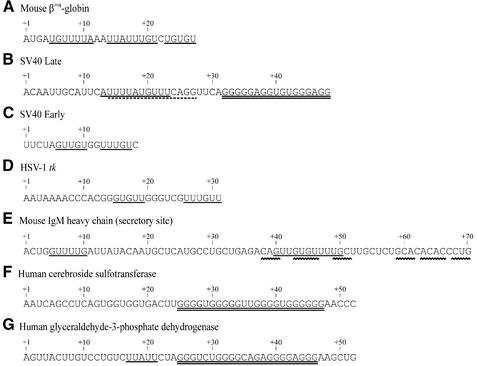
Results of Chen and Nordstrom (28) provide a good illustration of some special features of downstream elements. The efficiency of polyadenylation of chimeric constructs containing the poly(A) signal of the mouse βmaj-globin pre-mRNA was examined. Some parts of the downstream element (Fig. 1A) were replaced with random CA-containing tracts in this signal. This replacement reduced the efficiency of 3′ end processing. In particular, replacements of +5/+10, +11/+16, +17/+22 and +23/+27 tracts (Fig. 1A) decreased the processing efficiency approximately by 27, 7, 50 and 10%, respectively. This led to a conclusion that the downstream element of the poly(A) signal of the mouse βmaj-globin pre-mRNA has two major U/G-rich functional components (+5/+10 and +17/+22), and that the portions of the element are functionally redundant. The signals of other pre-mRNAs examined had similar features (26).
Figure 1.
The sequences of downstream regions of pre-mRNA poly(A) signals. The four out of five base UREs and 2GU/U elements are indicated by a single line, G-rich tracts are indicated by a double line. The binding site for CstF is indicated by a dotted line. The segments which form the double-helical structure are indicated by a wavy line.
U-rich downstream elements have been investigated in more depth than G/U-rich tracts. Wilusz and co-workers (29–32) have shown that an oligo(U) tract composed of 5 nt could functionally substitute for the downstream region of the poly(A) signal of SV40 late (SV40 L), SV40 early (SV40 E) and Ad5 L3 pre-mRNAs (29). Moreover, pentamers consisting of four U residues and any other residue within this pentamer [a four out of five base U-rich element (URE)], were proven to be sufficient for polyadenylation of SV40 L pre-mRNA (31,32). The authors also characterized the positional requirements for both the AAUAAA and URE motifs in the polyadenylation reaction. The cleavage was found to occur within a region 11–23 nt downstream of the AAUAAA element (with a few exceptions). The optimal position for the URE is 10–30 nt downstream of the cleavage site, but this element can be located closer than 10 nt to the cleavage site. The authors suggested that the spacing requirements for the AAUAA and URE motifs reflect the spatial requirements for a stable interaction between CPSF and CstF (32).
Chen et al. (32) analyzed 131 poly(A) signals for mammalian pre-mRNAs available in the GenBank database. The four out of five base UREs, located within 30 nt downstream of the cleavage site, were present in 74% of natural signals. A UV cross-linking study showed that such elements serve as the binding sites for the 64 kDa subunit of CstF (30) (as an illustration, the CstF binding site in the SV40 L pre-mRNA is shown in Fig. 1B). Proceeding from these facts, Chen et al. (32) concluded that the URE is a major downstream element of poly(A) signals.
In certain instances, G/U-rich tracts are important compo nents of the downstream element of the poly(A) signal. McDevitt et al. (33) showed that the GUUGUGGU sequence, which is a fragment of a natural poly(A) signal of SV40 E pre-mRNA (Fig. 1C), could substitute for the intact downstream element of this mRNA. In this case, the efficiency of the cleavage reaction was ∼35% of wild type. Different single base substitutions in this fragment led both to increases and decreases in processing efficiency. In particular, an increase in efficiency occurred when either G residue in the GG dimer was substituted by U. The GUUGUUGU and GUUGUGUU variants were approximately three times more effective than the original fragment.
These results are interesting in the light of data of Takagaki and Manley (20). Using a SELEX method, they established that the isolated RNA binding domain of the 64 kDa subunit of CstF selected GU-rich sequences containing GU- and U-repeats, but not G-repeats. They also demonstrated that such sequences were specifically recognized by full-length CstF and served as downstream elements in pre-mRNA cleavage assays. These experimental data (20) allow to assume that the functional part of the GUUGUGGU fragment of the downstream element of the SV40 E pre-mRNA poly(A) signal (33) is the GUUGU portion, which does not contain G-repeats.
As mentioned above, the UGUGU fragment of mouse βmaj-globin pre-mRNA downstream element (+23/+27 tract, Fig. 1A), although not essential for polyadenylation, contributes to maximal efficiency (28). Since the replacement of this fragment with a CA tract does not affect the +5/+10 and +17/+22 fragments containing the four out of five base UREs (Fig. 1A), a true role of UGUGU as a downstream element may not be adequately reflected in this experiment. Moreover, this element is more essential in another assay where a partially mutated poly(A) signal of βmaj-globin pre-mRNA was used. In this context, deletion of the fragments containing the UGUGU element (+22/+27) or URE (+16/+22) (Fig. 1A) led to a ∼20% decrease in the cleavage efficiency (28).
Other short G/U-rich sequences in the downstream region of the poly(A) signal were found in Herpes Simplex Virus type 1 thymidine kinase (HSV-1tk) pre-mRNA (34). The replacement of a CGGGUGUU tract (Fig. 1D) in the downstream region of this pre-mRNA with a linker sequence led to a 69% reduction in polyadenylation efficiency. Based on results with other pre-mRNAs, we suggest that the functional portion of this tract may be the sequence GUGUU. The three above-mentioned tracts (GUUGU, UGUGU and GUGUU) are all possible variants of a pentamer consisting of GU dimers and a U residue. Since all these sequences play some role in the polyadenylation reaction, we propose that another simple downstream element is present along with the well known element four out of five base URE. We term it the ‘two GU and one U’ (2GU/U) element (12).
McLauchlan et al. (35) studied sequences at the 3′ termini of 95 pre-mRNAs from higher eukaryotes and their viruses. They proposed that a YGUGUUYY sequence (where Y is pyrimidine), located ∼30 nt downstream of AAUAAA, is a consensus for the downstream element of the poly(A) signal, since 67% of the examined pre-mRNAs contained this or similar sequences. Other consensuses were also proposed, for example, UUGANNNUUUUUU (36). Nevertheless, a convincing consensus for the downstream element was not achieved (16). It may be that there is no general consensus sequence for the downstream region of the pre-mRNA poly(A) signal, but rather this region contains a different number of various simple elements (each 5 nt long, as a minimum) located at different distances from each other.
To analyze further the structure of poly(A) signals, we collected a database of human poly(A) signals (37), which includes 244 DNA sequences randomly selected from GenBank (National Center for Biotechnology Information, USA). The sequences correspond to regions of pre-mRNAs extending 200 nt upstream of the cleavage site (or up to the 5′ end of the last exon) and 200 nt downstream. The sequences downstream of the cleavage site were identified by comparing the genomic DNAs with mRNA sequences from GenBank. The sequences in our database belong to different chromosomes and are expressed in various tissues. A portion of the sequences in the database, representing 70 nt upstream and 70 nt downstream of the poly(A) site, is presented in the Supplementary Material. The full database containing the 400 nt long sequences is available from the authors upon request.
Results of our analysis are presented in Tables 1–3. We found that 69% of the pre-mRNAs analyzed contained the canonical AAUAAA hexamer. This percentage is higher than that obtained using 3′ EST analysis (50–60%), but lower than the percentage obtained in earlier studies using cDNA (80–90%) (25 and references therein). Our value is similar to that obtained by Tabaska and Zhang (∼74%) who used both 3′ ESTs and sequences from GenBank to build a database of poly(A) signals that contains 280 mRNA and 136 DNA sequences (38). MacDonald and Redondo (25) provided some explanations why 3′ EST data differ from conventional cDNA data. In particular, they noted that when a cDNA is submitted to GenBank, the most common variant of the 3′ end sequence is reported, whereas the EST approach gives equal weight to every instance of a 3′ end sequence, including both rarely expressed and more common variant forms of mRNAs. Thus, our database is probably biased toward more common variants of poly(A) signals. Even so, 17% of our pre-mRNAs contained neither AAUAAA nor AUUAAA hexamers. We therefore classified pre-mRNAs in our database by the type of core upstream element (Table 1). We defined the AAUAAA and AUUAAA hexamers as type I and II, respectively. The occurrence of the AUUAAA element is ∼12–15% in both our database (Table 1) and in other studies (25,38). Some pre-mRNAs in our database contain the AAUAAA hexamer with a single base substitution at the 1st, 2nd and 5th positions (type III). As was already mentioned, these variants function in natural pre-mRNAs.
Table 1. Core upstream elements of 244 human pre-mRNA poly(A) signals studieda.
| Element type | Element | Portion of pre-mRNAs containing the element (%) |
|---|---|---|
| I | AAUAAA | 69 |
| II | AUUAAA | 14 |
| III | NAUAAA | 9 |
| ANUAAA | ||
| AAUANA | ||
| IV | AANAAA | 2 |
| AAUNAA | ||
| AAUAAN | ||
| V | Substitutions of any two bases in AAUAAA | 3 |
| VI | Other than I–V | 3 |
aThe database of human poly(A) signals is reported in Zarudnaya et al. (37) and available as Supplementary Material (Table S1).
Table 3. Downstream regions of human poly(A) signals of pre-mRNAs studieda.
| Element | Variant | Portion of pre-mRNAs containing downstream elements (%)b |
|---|---|---|
| URE | NUUUU | 66–70 |
| UNUUU | ||
| UUNUU | ||
| UUUNU | ||
| UUUUN | ||
| 2GU/U | GUUGU | 27–31 |
| UGUGU | ||
| GUGUU | ||
| Two or more UREs and 2GU/U elements | – | 46 |
| Core downstream elements other than URE and 2GU/U | – | 20 |
aThe database of human poly(A) signals is reported in Zarudnaya et al. (37) and available as Supplementary Material (Table S1).
b204 pre-mRNAs with core upstream elements of types I and II were surveyed. Core downstream elements are located within the +1/+32 region downstream of the cleavage site.
Hexamers of types I–III were found to occur as far as 16–35 nt upstream of the cleavage site (counting from the first nucleotide of the element). The distribution of distances between the hexamer and the cleavage site is skewed, with a maximum at +21. In contrast to the core upstream elements, the downstream elements were found over a much wider region, throughout the downstream region (+1/+70) (see Table S1). However, they occurred 3-fold more frequently in the +2/+22 than in the +36/+70 region. Thus, the distance between the upstream and the corresponding downstream elements observed in the most pre-mRNAs studied covers 25–50 nt.
A very small portion of the pre-mRNAs in our database (2%) contained AAUAAA hexamers with single base substitutions at the 3rd, 4th or 6th positions (type IV, Table 1). Some of these elements, for example AAGAAA or AAUGAA, are functional in some pre-mRNAs and deleterious in others (24 and references therein). The portion of pre-mRNAs containing the AAUAAA hexamer with single base substitutions other than AUUAAA (elements of types III and IV) made up 11% of the transcripts analyzed (Table 1), which is close to the value reported by Tabaska and Zhang (9%) (38).
Elements with substitutions at any two bases in the AAUAAA consensus were grouped as type V (Table 1). Information on elements of this type is very sparse in the literature. Only UAUAUA has been shown to be functional. This element functions weakly in papilloma virus pre-mRNA (24 and references therein). The remaining 3% of pre-mRNAs were found to contain neither the canonical hexamer nor its variants in the –16/–35 region (type VI, Table 1), but the majority of these pre-mRNAs contain hexamers of types I–III (true signal elements) in the –38/–200 region. It could be that hexamers located in the –36/–54 region can be functional elements if the corresponding downstream elements are located very close to the cleavage site, so that the distance between the core upstream and downstream elements does not exceed 50–55 nt.
Some hexamers located far from the cleavage site may be functional if they are brought together with the downstream element by formation of a stem–loop structure, as was shown to occur in case of the Human T-Cell Leukemia Virus Type I (HTLV-1) poly(A) signal (39). In addition, hexamers of types I–III located outside the –16/–35 region may be upstream elements of alternative poly(A) signals.
What is the possible role of non-canonical upstream elements? Hexamers of types III–V or other undetermined elements may be used preferentially in specific tissues or cell types. Wallace et al. (40) and Dass et al. (41) discovered a new form of CstF-64, τCstF-64, which is highly expressed in male germ cells, and to a smaller extent in brain. The authors proposed that τCstF-64 plays a significant role in polyadenylation of pre-mRNAs with non-AAUAAA core upstream elements in germ cells. As a pilot study, MacDonald and Redondo (25) sequenced 3′ ends of 122 cDNAs from a library prepared using mouse male germ cell types and showed that 59% of these RNAs contained AAUAAA and 12% contained AUUAAA, which is similar to the frequencies obtained in other 3′ EST studies (25 and references therein). Thus, the question remains open whether cell types expressing τCstF-64 will show a higher incidence of non-AAUAAA poly(A) signals.
We compared the occurrence of non-AAUAAA core upstream elements of poly(A) signals in 78 human pre-mRNAs from different cells (37) and 39 mammalian germ cell-expressed pre-mRNAs earlier reported by Wallace et al. (40). Surprisingly, the percentage of pre-mRNAs with core upstream elements of types III and V in germ cells appeared to be ∼2-fold higher than in other cells, and the percentage of germ cell pre-mRNAs with hexamers of type II is ∼4-fold lower than in other cells (Table 2). The portions of pre-mRNAs with hexamers IV and VI appeared to be approximately equal. These results suggest that the hexamers III and V are major non-AAUAAA upstream elements of poly(A) signals in male germ cells, which is consistent with the hypothesis of Wallace et al. about the significance of non-canonical upstream elements of poly(A) signals in germ cells. A more extended database of germ cell pre-mRNAs should be analyzed to support this hypothesis.
Table 2. Comparison of non-AAUAAA core upstream elements of poly(A) signals in 78 human pre-mRNAs from different cells (37) and 39 mammalian germ cell-expressed pre-mRNAs (40).
| Element type | Portion of pre-mRNAs containing the element (%) | |
|---|---|---|
| Human cells | Mammalian germ cells | |
| II | 45 | 10 |
| III | 23 | 41 |
| IV | 6 | 8 |
| V | 12 | 26 |
| VI | 14 | 15 |
The elements are located within the –16/–35 region.
Our findings on the structure of the downstream region of poly(A) signals of pre-mRNAs with core upstream elements I and II are presented in Table 3. As shown here, 66–70% of pre-mRNAs contain UREs, in agreement with reported data (32). A substantial portion of pre-mRNAs (27–31%) contain the 2GU/U element. The percentages of transcripts containing UREs and 2GU/U elements are given as a range because some pre-mRNAs contain tracts that cannot be unambiguously interpreted. For example, the UUUGUGU sequence in mouse cystatin 9-like (CST9L) pre-mRNA includes a signal pentamer that may be classified as both a URE (UUUGU) and a 2GU/U element (UGUGU).
Notably, Graber et al. (23), using computer analysis of ∼900 Drosophila 3′ ESTs, revealed that 6 nt ‘words’, such as UGUUUU, UGUGUU and UUUUUU, were the most common words in the region of 10–20 nt downstream of the cleavage/polyadenylation site. Though the authors studied six-letter ‘words’, the UGUGU and GUGUU pentamers are present. We suppose that 2GU/U elements can be significant not only in Drosophila pre-mRNAs but also in mammalian transcripts. The following findings support this suggestion. The AAUAAA hexamer was shown to occur in Drosophila pre-mRNAs approximately as often as in mammalian pre-mRNAs (23). It is therefore likely that the downstream elements of poly(A) signals are also identical in insects and mammals. According to Hatton et al. (42), Drosophila CstF and CPSF recognize mammalian poly(A) signals, since these signals have been successfully used in vectors for expressing proteins in Drosophila cells (42 and references therein).
As many as 20% of analyzed pre-mRNAs with hexamers I and II contain neither UREs nor 2GU/U elements in the region between the cleavage site and the nucleotide +32. However, the majority of these contain tracts that are similar to one of these elements. For example, a sequence in the ficolin 3 pre-mRNA (UCUUC) could be a suboptimal URE. Our database analysis showed that 46% of the transcripts with AAUAAA or AUUAAA hexamer (types I and II) contained two or more UREs or 2GU/U elements. In most of these transcripts the downstream elements are separated by 2 nt or more, as seen in Figure 1D, but in some cases the elements are present in one long tandem sequence, as can be seen in Figure 1B and E. It is reasonable to suppose that the presence of multiple downstream elements increases the probability of CstF binding to pre-mRNA in the vicinity of the upstream element of the poly(A) signal, and thus the overall efficiency of cleavage complex assembly.
The frequency of UREs and 2GU/U elements in pre-mRNAs with upstream elements of types III–VI is comparable with that in pre-mRNAs with hexamers of types I and II (see Table S1 in the Supplementary Material). However, these data are excluded from Table 3, because the pools of these pre-mRNAs in our database are not sufficient for a reliable estimation.
Earlier in our review (12) we discussed the analysis of downstream poly(A) signals preformed by McLauchlan et al. (35) using different eukaryotic species and we also evaluated the frequency of UREs and 2GU/U elements in database of McLauchlan. Remarkably, the YGUGUUYY consensus proposed by McLauchlan contains some variants of UREs and 2GU/U elements. The frequency of these elements in the database of McLauchlan is similar to that which we obtained with human poly(A) signals, which provides additional evidence that the structures of the downstream region of poly(A) signals in different species are not drastically different, at least in the case of pre-mRNAs with the canonical AAUAAA hexamer. Further investigations will be required to elucidate the sequences of the downstream elements in non-human mammalian pre-mRNAs with core upstream elements of different types.
Using a SELEX method to determine CstF-binding sites, Beyer et al. (43) showed that purified CstF from calf thymus and HeLa cells selected RNA ligands with one of three conserved elements: element 1 (AUGCGUUCCUCGUCC), element 2a (YGUGUN0-4UUYAYUGYGU) or element 2b (UUGYUN0-4AUUUACU(U/G)N0-2YCU). These tracts and their fragments could serve as downstream elements of poly(A) signals in chimeric constructs. Computer-assisted analysis of the EMBL library data showed the majority of the element 2a-like sequences to be located downstream of the coding region, and the authors postulate that this element may represent a novel consensus sequence for downstream elements. The authors also showed that CstF factors purified from different organisms preferentially selected different RNA ligands (43). The factors extracted from HeLa cells and a calf thymus predominantly selected element 1 and elements 2a/2b, respectively.
As seen from an analysis of the novel consensus 2a, it contains simple elements, four out of five base UREs or 2GU/U tracts, the number of which, as a rule, is not less than two. This lends support to our model where the downstream poly(A) signal consists of different numbers of various simple elements located at different distances from one another. The majority of variants of element 1 reported in Beyer et al. (43) do not contain UREs or 2GU/U pentamers, although most of them contain the GCGUU or UUCCU tracts which are homologous to GUGUU and UREs, respectively.
To conclude this section, we would like to emphasize that the precise sequence of the downstream element is, apparently, not crucial for the polyadenylation reaction. In some cases (for example, see 20,28,43) the cleavage reaction proceeds in the absence of any known downstream elements, though less efficiently. This could be explained by the properties of the RNA-binding domain of the 64 kDa subunit of CstF, which is of the RNP2/RNP1 type (44). Proteins with this domain are known to specifically recognize many different RNA sequences (44–47). Wang and Hall (47) studied the crystal structures of the first two RBDs (of the RNP2/RNP1 type) of the HuD protein in complex with fragments of AU-rich elements (AREs) which control the stability of short-lived mRNAs. The authors show that mutations at some positions in ARE severely reduce HuD binding to the ARE, while mutations at other positions are better tolerated. By analogy, such position dependence is expected upon CstF binding to the U/GU-rich downstream elements. We suppose that in this case the key contacts occur between RBD of the 64 kDa subunit of CstF and two or three U residues located in the downstream element. The conformation of these U residues is probably optimal when they are located in the most frequently occurring elements: UGUUUU, UGUGUU and UUUUUU. In the absence of UREs or 2GU/U-pentamers, CstF, which cooperatively interacts with other polyadenylation factors, is likely to bind to other U-rich tracts, where the arrangement of U residues is less optimal.
SECONDARY STRUCTURE OF PRE-mRNA POLY(A) SIGNALS
Studies of the role of RNA secondary structure in the recognition of the AAUAAA hexamer reveal that this element is recognized in a single-stranded form (16,48,49). Effective binding of CPSF to pre-mRNA (in nuclear extracts or in vivo) is also possible when a part of the hexamer is involved in a double-helical structure, provided the stability is not very high (18,50).
Not many studies on the secondary structure of poly(A) signal downstream elements are known in the literature, and the conclusions drawn from them are ambiguous. Chen and Wilusz (48) showed that chimeric RNAs containing any of the main regions of poly(A) signal (the AAUAAA hexamer, the cleavage site or the URE) in a secondary structure were cleaved very inefficiently as compared to pre-mRNAs containing a wild-type signal.
The structure of the poly(A) signal of adenovirus-2 L4 pre-mRNA, the downstream element of which contains neither UREs nor 2GU/U pentamers, was studied by Sittler et al. (49). The authors failed to find any clear correlation between the secondary structure of this element and its function.
Results obtained by Phillips et al. (19) support the conclusion that one of two downstream elements of mouse IgM secretory poly(A) signal participates in a stem–loop structure with two asymmetric internal loops. Both the stem and the internal loops are important for efficient polyadenylation. In Figure 1E we show the sequences involved in the formation of the double-stranded segments of this hairpin. As seen here, the element consisting of GU/U repeats is partially involved in the double-stranded structure, although four U residues of this element are single stranded. The GU/U element is located 39 nt, and the second element (URE) only 4 nt downstream of the cleavage site (51,52). Thus, both elements are located suboptimally: the first is rather far from the cleavage site, and the second is rather close. The authors suggest that the participation of the distal downstream element in a hairpin structure may result in better recognition of this element by polyadenylation factors, compensating for its suboptimal location.
How can CstF interact with downstream elements that participate in secondary structures? Does it interact, for example, at the moment when double-helical segments of the hairpin are opened in the process of RNA ‘breathing’, and one of the signal pentamers (UREs or 2GU/U tracts) becomes accessible for the polyadenylation factor? Are downstream hairpins a target for structure modifying enzymes (RNA helicases etc.) as Phillips et al. (19) have suggested? The answers to these questions are still unknown. In this connection, we refer to recent results of Bléoo et al. (53), which suggest a functional and a spatial relationship between CstF-64 and human DEAD box protein DDX1, a putative RNA helicase.
Phillips et al. (19) suggest that involvement of the downstream element in a hairpin structure is not peculiar to the mouse IgM poly(A) signal. They demonstrate that the downstream regions of the hamster and the human IgM secretory poly(A) signals can also form hairpin structures with internal loops. The sequences forming the descending arms of the hairpins are highly conserved in all three cases [fig. 8 in Phillips et al. (19)]. The proximal segments are similar in the mouse and the hamster hairpins, while the proximal segment of the human hairpin is G-rich. The role of the highly conserved sequence in the distal segment of the hairpins is unknown. The authors noted (19) that there is another obvious candidate, besides CstF, for the role of protein binding specifically to the hairpin. This is a protein with a molecular mass of 30 kDa. The binding of this protein to mouse IgM secretory pre-mRNA depends on a 55 nt sequence encompassing the distal downstream element (51). Induction of this 30 kDa protein correlates with the increase of secretory poly(A) site usage.
The poly(A) signal of SV40 L pre-mRNA contains an auxiliary downstream element (AUX DSE) in addition to the core element (URE) (54). This element is a G-rich sequence (GRS) GGGGGAGGUGUGGG (containing neither URE nor 2GU/U). The GRS serves as the binding site for hnRNP H/H′ protein (55). Interaction between this protein and the GRS stimulates polyadenylation of SV40 L pre-mRNA. It has to be noted that the full GRS in the downstream region of SV40 L pre-mRNA (Fig. 1B) is 3 nt longer than the element GRS which binds hnRNP H/H′ protein.
Hans and Alwine (56) used nuclease sensitivity structure analysis techniques to determine the secondary structure of the SV40 L poly(A) signal. They found that a large part of the U-rich downstream element and a part of the AAUAAA hexamer are involved in a secondary structure, while the GRS is primarily in a single-stranded form. By replacing portions of the downstream region with unrelated sequences, they showed that the ability of this region (particularly nucleotides 2696–2700) to form double-stranded structures correlates with cleavage efficiency. This indicates that the secondary structure of the region downstream of AAUAAA has a functional significance. The mechanism by which pre-mRNA secondary structure influences the polyadenylation process is still unknown. The authors propose that the secondary structure may aid CstF interaction with the downstream element. They also make an intriguing suggestion that the RNA structure in the downstream region may have a catalytic role in the cleavage process.
Though the latter suggestion cannot be excluded, it is generally assumed that the 30 kDa subunit of CPSF is the endonuclease that cleaves pre-mRNA in the polyadenylation process, since this protein is a homolog of Drosophila CLP protein, which possesses endoribonucleolytic activity (17,57–60). Others suggest that the cleavage is performed by a predicted nuclease in the metallo-β-lactamase fold of the 73 kDa subunit of CPSF (61). In both these cases catalytic function is attributed to a protein, not to RNA.
A secondary-structure scheme of the SV40 L poly(A) signal was not presented in Hans and Alwine (56) because of discrepancy between the computer-predicted structures based on programs available at that time and the nuclease sensitivity data. Inasmuch as computer programs to predict RNA secondary structure are constantly improving, we used a more recent version of the mfold program, together with the latest free energy rules (62,63) (http://www.bioinfo.rpi.edu/applications/mfold/) to predict the folding of the SV40 L pre-mRNA fragments. We then compared the predicted structures with the experimental results of Hans and Alwine (56).
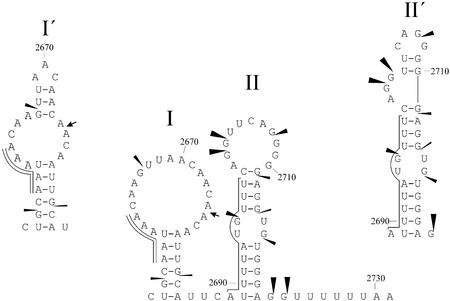
Figure 2 shows a computer-predicted secondary structure of the SV40 L pre-mRNA fragment from nucleotides 2653 to 2731, which includes the AAUAAA hexamer, URE and GRS. This structure has two hairpins. Hairpin I includes AAUAAA and hairpin II includes the URE and the GRS. Variants of hairpins I and II (hairpins I′ and II′), which have a less favorable free energy of folding, are also shown in Figure 2.
Figure 2.
The secondary structure scheme of the core poly(A) signal of the SV40 L pre-mRNA. The secondary structures of the pre-mRNA fragments were determined using the mfold version 3.1 program for RNA folding by Zuker and Turner (http://www.bioinfo.rpi.edu/applications/mfold/). The U-rich downstream element and the AAUAAA hexamer are marked by single and double lines, respectively. The cleavage site is marked by an arrow. The sites attacked by RNase T1 (56) are marked by triangles. Thermodynamic stabilities of the hairpins I, I′, II and II′ are –3.1, –2.5, –4.3 and –2.4 kcal/mol, respectively.
We also predicted the folding of longer fragment of the SV40 L pre-mRNA (nucleotides 2634–2731), which along with AAUAAA, the URE and the GRS contained one of three auxiliary upstream elements of the SV40 L poly(A) signal. The auxiliary upstream elements specifically interact with U1A protein (64). Hairpins I and II are also formed in the 2634–2731 nucleotide fragment. However, the formation of hairpin I was not predicted on folding the 2603–2731 nucleotide fragment containing all three auxiliary upstream elements and also the full substrate RNA (nucleotides 2531–2731) studied in Hans and Alwine (56). In both cases the AAUAAA hexamer was involved in other double-stranded structures. Hairpin II was predicted to form in one of the thermodynamically favorable variants of the secondary structure of the full substrate RNA. We suppose hairpin II may also form in much longer RNA sequences, including the full-length SV40 L pre-mRNA.
Hans and Alwine (56) used the following nucleases: RNase V1, which cleaves double-helical RNAs non-specifically; RNase T1, which cleaves at single-stranded G residues, and RNase PhyM, which cleaves at single-stranded A and U residues. The reported data (56) are consistent with the secondary structure model presented in Figure 2, provided that the variants of RNA hairpins coexist in dynamic equilibrium. As an example of nuclease sensitivity data, all the sites cleaved by RNase T1 more or less efficiently (56) are marked in Figure 2 with triangles, the sizes of which approximately correlate with the intensity of the RNA digestion.
The only discrepancy between the experimental data and the secondary structure presented in Figure 2 is the upper part of the descending arm of the hairpin II stem (nucleotides 2711–2714), which is insensitive to RNase V1. But it should be noted that insensitivity of some RNA segments to this nuclease does not necessarily mean that they are in a single-stranded form. According to Lowman and Draper (65), RNase V1 recognizes a 4–6 nt segment of sugar–phosphate backbone with an approximately helical conformation and does not require paired bases. The conformation of the GAGG segment in the upper part of the hairpin II may be suboptimal for the nuclease V1 in contrast to the conformation of the complementary UUUC segment. Consistent with our model, the GAGG tract is poorly cleaved by both single-stranded-specific nucleases.
Interestingly, some previous computer programs predicted the formation of the hairpin II without the internal loop, i.e. with all the nucleotides in the stem being paired, but as can be seen in Figure 2, this contradicts the experimental results.
Thus, our computer predictions of SV40 L poly(A) signal secondary structure and the experimental results of Hans and Alwine (56) enable us to propose that URE and the auxiliary downstream element (GRS) may be present in the ascending and descending arms of hairpin II structure, respectively. Remarkably, such functional arrangement of hairpin II of the SV40 L poly(A) signal is similar to that of the hairpin with the distant downstream element of the IgM poly(A) signal, where the GU/U-rich downstream element and the highly conserved sequence (putative auxiliary downstream element) are located in ascending and descending arms of the hairpin, respectively. It would be interesting to determine how widespread this pattern may be.
Recent studies of Arhin et al. (66) showed that G-rich auxiliary elements are not limited to SV40 L pre-mRNA. According to their data, ∼34% of mammalian poly(A) signals contain short G-tract(s) in the region downstream of the core elements. All the tested G-rich elements bound hnRNP H/H′ protein, which resulted in stimulation of polyadenylation. The affinity of hnRNP H/H′ binding for various poly(A) signals was significantly different, which affected the abilities to stimulate 3′-end processing. These authors suggested that downstream G-rich tracts are common auxiliary elements of poly(A) signals (66).
Protein hnRNP H/H′, which binds to GRSs, belongs to the H subfamily of hnRNP proteins, which also includes hnRNP F and hnRNP 2H9 (67,68). HnRNP H and H′, which are 96% identical to each other (67), and hnRNP F, contain three RNA-binding domains of the quasi-RNP2/RNP1 type, while hnRNP 2H9 contains two such domains. Caputi and Zahler (69) showed that all the members of the H group specifically interact with RNA sequences containing a GGGA tract, whereas only hnRNP H/H′ recognizes a GGGGGC sequence. HnRNP H/H′ is likely to recognize the GGGU sequence as well (70).
Considering these facts, it can be assumed that the other members of the H group of hnRNP proteins may also bind to G-rich auxiliary downstream elements, since most of auxiliary downstream elements, which are reported in Arhin et al. (66), contain the GGGA tract(s) and the rest contain the GGGU or GGGGGC tracts. The role of hnRNP 2H9 binding to the G-tracts in the polyadenylation process is currently unclear. HnRNP F was shown recently to be able to diminish pre-mRNA 3′-end processing (71). The authors supposed that the binding of hnRNP F or H′/F heterodimer to a poly(A) signal near the core downstream element may inhibit the association of CstF with pre-mRNA and/or the cleavage reaction.
All the members of the H group of hnRNP proteins also participate in splicing regulation (68–70,72–75), the mechanism of their functioning in this process is not yet determined. In some cases, hnRNP H may sterically hinder the binding of SR proteins to splicing regulatory sequences, thereby inhibiting their functions (70,73). In other cases (72,74,75) the interaction of hnRNP H protein with its binding site was required for assembly of other proteins onto splicing regulator elements, and the protein was essential for function of these elements. HnRNP H/H′ was suggested to play a similar role in the polyadenylation process. Arhin et al. (66) suggested that it directly interacts with CstF to stimulate assembly of general polyadenylation factors on the core poly(A) signal. Alternatively, the authors suggested that the protein may alter the structure of the nascent transcript to present the core signal elements to the general polyadenylation factors in a productive fashion. In this connection, interestingly, the RNA-binding sites for hnRNP H/H′/F proteins may form G-quadruplexes, which we discuss in the next section.
G-QUADRUPLEXES AS POSSIBLE AUXILIARY ELEMENTS OF POLY(A) SIGNALS
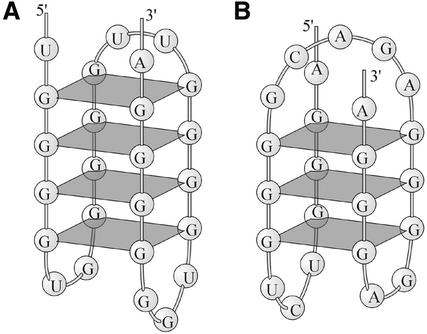
DNA or RNA sequences consisting of clusters of G-repeats may form four-stranded structures composed of stacked G-tetrads, so called G-quadruplexes (76). The thermodynamic stability of G-quadruplexes depends on the number of G-tetrads, the sequence of the loops connecting G-tetrads, and the size of the loops (77–79). Based on these data and other experimental findings on the formation of G-quadruplexes, we previously reported that ∼27% of pre-mRNA sequences in our database of human poly(A) signals have a potential to form such structures (37). A portion of the article (37) is presented as Supplementary Material. In particular, this article discusses G-quadruplex formation and shows that the G-rich region of the SV40 L poly(A) signal is likely to be capable of forming G-quadruplexes. The schematic structures of G-quadruplexes formed by the full GRS of SV40 L pre-mRNA and possible quadruplexes adopted by GRSs of several human pre-mRNAs available in our database are also presented as Supplementary Material (Fig. S1). Here we present the schematic illustrations of two other putative quadruplexes of human pre-mRNAs. The first is formed by a GRS located 24 nt downstream of the cleavage site of human cerebroside sulfotransferase pre-mRNA (Figs 1F and 3A). The second quadruplex is formed by the fragment located 3 nt downstream of URE of human glyceraldehyde-3-phosphate dehydrogenase pre-mRNA (Figs 1G and 3B).
Figure 3.
Patterns of pre-mRNA fragments folding into G-quadruplexes. (A) Fragment of human cerebroside sulfotransferase pre-mRNA. (B) Fragment of human glyceraldehyde-3-phosphate dehydrogenase pre-mRNA.
Interestingly, the sequences with a potential to form G-quadruplexes occur in pre-mRNAs of our database ∼5-fold more often in the +1/+70 region (11% of pre-mRNAs) than in the –1/–70 region (2%), while in the +71/+200 and –71/–200 regions they occur approximately equally (8 and 7%, respectively). The frequency of the clusters of G-repeats in the region located immediately downstream of the cleavage site is unlikely to be fortuitous and may be evidence that G-quadruplexes play some role in the polyadenylation process.
It is also notable that hnRNP H, H′ and F, besides binding to different G-rich tracts, are capable of binding to poly(G) at 2 M NaCl (80). At this ionic strength (under equilibrium conditions), poly(G) is a completely four-stranded macromolecule (81). Certain proteins are known to recognize G-quadruplexes, facilitate their formation, or disintegrate them (82). Some of them can bind specifically both to four-stranded and single-stranded forms of polynucleotides (83). It is possible that H, H′ and F proteins can also recognize both single-stranded G-rich tracts and G-quadruplexes and/or promote the formation of four-stranded structures.
What functions could G-quadruplexes fulfill in the polyadenylation process? Consideration of how auxiliary elements may function helps to answer this question. Chen and Wilusz (48) have proposed three possible mechanisms of stimulation of the polyadenylation reaction by auxiliary DSEs. First, they suggest that some of these elements may promote processing efficiency by maintaining the elements of the core poly(A) signal in an unstructured form, which enables the general polyadenylation factors to assemble efficiently on the pre-mRNA molecule. Secondly, the authors suggest that auxiliary DSEs may form stable structures which prevent CstF from sliding along the pre-mRNA, and thereby limit the region of interaction with the transcript to the downstream region of the poly(A) signal only. It was shown that a pseudoknot from a viral RNA can functionally substitute for the undetermined auxiliary DSE of the Ad5 L3 pre-mRNA poly(A) signal. A mutated pseudoknot sequence had no effect on polyadenylation efficiency. Thirdly, the authors propose that the interaction of specific proteins with auxiliary DSEs, as in the case of hnRNP H/H′, may enhance the efficiency of 3′-end processing, probably by stimulating an assembly of the general polyadenylation factors on the pre-mRNA.
Four-stranded structures stabilized by G-tetrads could promote all three above-mentioned functions of auxiliary DSEs. First, a G-quadruplex, like a pseudoknot, could prevent CstF migration from the U/GU-rich downstream element. This could stimulate the polyadenylation process. In addition, G-quadruplexes could maintain the core poly(A) signal in an accessible conformation. Some GRSs form highly stable intramolecular quadruplexes that can limit a number of possible conformations adopted by RNA and thereby maintain the poly(A) signal in an optimal conformation. Finally, G-quadruplexes could serve as the binding sites for proteins that influence the efficiency of the polyadenylation process. The GRSs in regions of pre-mRNAs upstream of the cleavage site may function in the same manner as GRS located downstream, in particular, by interacting with auxiliary proteins.
It should be noted that the formation of the G-quadruplex in the SV40 L poly(A) signal may ensure opening of the hairpin with the URE (Fig. 2) and access of the CstF to the downstream element.
In conclusion, we would like to mention other possible functions of GRSs. They may function as specific signals not only in pre-mRNAs, but also in the corresponding DNAs that encode these GRS-containing pre-mRNAs. It has been shown that tandemly arranged GRSs …CTGGCCTTGGGGG AGGGGGAGGC… (which are the binding sites for the transcription factor MAZ), located downstream of the poly(A) signal in chimeric constructs, specifically paused RNA polymerase II and stimulated polyadenylation in vitro (84). We note that the binding site for MAZ being in the single-stranded form (for example, in the transcription bubble) might form quadruplex structures with two G-tetrads and this structure could induce pausing of RNA polymerase.
SUPPLEMENTARY MATERIAL
With permission of the journal Biopolymery i Kletka, a portion of the text of the article (37), one table and one figure are available as Supplementary Material at NAR Online. The portion of the article reprinted here concerns some trends in G-quadruplex formation. Table S1 contains the database of poly(A) signals of 244 human pre-mRNAs. Figure S1 displays the schematic structures of possible quadruplexes for several human pre-mRNAs.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs O. Yo. Cherepenko, T. I. Chausovs’kyi and S. S. Syzenko for assistance and helpful discussion of the manuscript. We are very grateful to three anonymous reviewers for critical comments on the manuscript.
REFERENCES
- 1.Wang Z., Day,N., Trifillis,P. and Kiledjian,M. (1999) An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell. Biol., 19, 4552–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Decker C.J. and Parker,R. (1993) A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev., 7, 1632–1643. [DOI] [PubMed] [Google Scholar]
- 3.Chen Z., Li,Y. and Krug,R.M. (1999) Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J., 18, 2273–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craig A.W.B., Haghighat,A. and Yu,A.T.K. (1998) Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature, 392, 520–523. [DOI] [PubMed] [Google Scholar]
- 5.Zarudnaya M.I. and Hovorun,D.M. (1999) Hypothetical double-helical poly(A) formation in a cell and its possible biological significance. IUBMB Life, 48, 581–584. [DOI] [PubMed] [Google Scholar]
- 6.Edwalds-Gilbert G., Veraldi,K.L. and Milcarek,C. (1997) Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res., 25, 2547–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooke C., Hans,H. and Alwine,J.C. (1999) Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal. Mol. Cell. Biol., 19, 4971–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeung G., Choi,L.M., Chao,L.C., Park,N.J., Liu,D., Jamil,A. and Martinson,H.G. (1998) Poly(A)-driven and poly(A)-assisted termination: two different modes of poly(A)-dependent transcription termination. Mol. Cell. Biol., 18, 276–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zarudnaya M.I., Potyahaylo,A.L., Dzerzhyns’kyi,M.E. and Hovorun,D.M. (2001) Molecular mechanisms of coupling of the transcription termination and the pre-mRNA polyadenylation. Ukr. Biokhim. Zh., 73, 28–32 (in Russian). [PubMed] [Google Scholar]
- 10.Hilleren P. and Parker,R. (1999) mRNA surveillance in eukaryotes: kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA, 5, 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manley J.L. (1995) Messenger RNA polyadenylation: a universal modification. Proc. Natl Acad. Sci. USA, 92, 1800–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zarudnaya M.I. (2001) mRNA polyadenylation. 1. 3′-end formation of vertebrates’ mRNAs. Biopolimery i kletka, 17, 93–108 (in Russian). [Google Scholar]
- 13.Zarudnaya M.I. (2001) mRNA polyadenylation. 2. Formation of poly(A) tails in yeast, plant, prokaryote and virus mRNAs. Biopolimery i kletka, 17, 185–202 (in Russian). [Google Scholar]
- 14.Edmonds M. (2002) A history of poly(A) sequences: from formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol., 71, 285–389. [DOI] [PubMed] [Google Scholar]
- 15.Colgan D.F. and Manley,J.L. (1997) Mechanism and regulation of mRNA polyadenylation. Genes Dev., 11, 2755–2766. [DOI] [PubMed] [Google Scholar]
- 16.Wahle E. and Rüegsegger,U. (1999) 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev., 23, 277–295. [DOI] [PubMed] [Google Scholar]
- 17.Zhao J., Hyman,L. and Moore,C. (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev., 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klasens B.I.F., Thiesen,M., Virtanen,A. and Berkhout,B. (1999) The ability of the HIV-1 AAUAAA signal to bind polyadenylation factors is controlled by local RNA structure. Nucleic Acids Res., 27, 446–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillips C., Kyriakopoulou,C.B. and Virtanen,A. (1999) Identification of a stem–loop structure important for polyadenylation at the murine IgM secretory poly(A) site. Nucleic Acids Res., 27, 429–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takagaki Y. and Manley,J.L. (1997) RNA recognition by the human polyadenylation factor CstF. Mol. Cell. Biol., 17, 3907–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Vries H., Rüegsegger,U., Hübner,W., Friedlein,A., Langen,H. and Keller,W. (2000) Human pre-mRNA cleavage factor IIm contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J., 19, 5895–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheets M.D., Ogg,S.C. and Wickens,M.P. (1990) Point mutations in AAUAAA and the poly(A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res., 18, 5799–5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graber J.H., Cantor,C.R., Mohr,S.C. and Smith,T.F. (1999) In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl Acad. Sci. USA, 96, 14055–14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beaudoing E., Freier,S., Wyatt,J.R., Claverie,J.-M. and Gautheret,D. (2000) Patterns of variant polyadenylation signal usage in human genes. Genome Res., 10, 1001–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacDonald C.C. and Redondo,J.-L. (2002) Reexamining the polyadenylation signal: were we wrong about AAUAAA? Mol. Cell. Endocrinol., 190, 1–8. [DOI] [PubMed] [Google Scholar]
- 26.Wahle E. (1995) 3′-End cleavage and polyadenylation of mRNA precursors. Biochim. Biophys. Acta, 1261, 183–194. [DOI] [PubMed] [Google Scholar]
- 27.Graber J.H., Cantor,C.R., Mohr,S.C. and Smith,T.F. (1999) Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res., 27, 888–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J.S. and Nordstrom,J.L. (1992) Bipartite structure of the downstream element of the mouse beta globin (major) poly(A) signal. Nucleic Acids Res., 20, 2565–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilusz J. and Shenk,T. (1990) A uridylate tract mediates efficient heterogeneous nuclear ribonucleoprotein C protein–RNA cross-linking and functionally substitutes for the downstream element of the polyadenylation signal. Mol. Cell. Biol., 10, 6397–6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacDonald C.C., Wilusz,J. and Shenk,T. (1994) The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol. Cell. Biol., 14, 6647–6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou Z.-F., Chen,F. and Wilusz,J. (1994) Sequence and position requirements for uridylate-rich downstream elements of polyadenylation signals. Nucleic Acids Res., 22, 2525–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen F., MacDonald,C.C. and Wilusz,J. (1995) Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res., 23, 2614–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDevitt M.A., Hart,R.P., Wong,W.W. and Nevins,J.R. (1986) Sequences capable of restoring poly(A) site function define two distinct downstream elements. EMBO J., 5, 2907–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang F., Denome,R.M. and Cole,C.N. (1986) Fine-structure analysis of the processing and polyadenylation region of the herpes simplex virus type 1 thymidine kinase gene by using linker scanning, internal deletion and insertion mutations. Mol. Cell. Biol., 6, 4611–4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLauchlan J., Gaffney,D., Whitton,J.L. and Clements,J.B. (1985) The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3′ termini. Nucleic Acids Res., 13, 1347–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Renan M.J. (1987) Conserved 12-bp element downstream from mRNA polyadenylation sites. Gene, 60, 245–254. [DOI] [PubMed] [Google Scholar]
- 37.Zarudnaya M.I., Potyahaylo,A.L., Kolomiets,I.M. and Hovorun,D.M. (2002) Auxiliary elements of mammalian pre-mRNAs polyadenylation signals. Biopolimery i kletka, 18, 500–517. [Google Scholar]
- 38.Tabaska J.E. and Zhang,M.Q. (1999) Detection of polyadenylation signals in human DNA sequences. Gene, 231, 77–86. [DOI] [PubMed] [Google Scholar]
- 39.Proudfoot N. (1991) Poly(A) signals. Cell, 64, 671–674. [DOI] [PubMed] [Google Scholar]
- 40.Wallace A.M., Dass,B., Ravnik,S.E., Tonk,V., Jenkins,N.A., Gilbert,D.J., Copeland,N.G. and MacDonald,C.C. (1999) Two distinct forms of the 64,000 Mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. Proc. Natl Acad. Sci. USA, 96, 6763–6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dass B., McMahon,K.W., Jenkins,N.C., Gilbert,D.J., Copeland,N.G. and MacDonald,C.C. (2001) The gene for a variant form of the polyadenylation protein CstF-64 is on chromosome 19 and is expressed in pachytene spermatocytes in mice. J. Biol. Chem., 276, 8044–8050. [DOI] [PubMed] [Google Scholar]
- 42.Hatton L.S., Eloranta,J.J., Figueiredo,L.M., Takagaki,Y., Manley,J.L. and O’Hare,K. (2000) The Drosophila homologue of the 64 kDa subunit of cleavage stimulation factor interacts with the 77 kDa subunit encoded by the suppressor of forked gene. Nucleic Acids Res., 28, 520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beyer K., Dandekar,T. and Keller,W. (1997) RNA ligands selected by cleavage stimulation factor contain distinct sequence motifs that function as downstream elements in 3′-end processing of pre-mRNA. J. Biol. Chem., 272, 26769–26779. [DOI] [PubMed] [Google Scholar]
- 44.Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- 45.Deo R.C., Bonanno,J.B., Sonenberg,N. and Burley,S.K. (1999) Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell, 98, 835–845. [DOI] [PubMed] [Google Scholar]
- 46.Varani G. and Nagai,K. (1998) RNA recognition by RNP proteins during RNA processing. Annu. Rev. Biophys. Biomol. Struct., 27, 407–445. [DOI] [PubMed] [Google Scholar]
- 47.Wang X. and Hall,T.M.T. (2001) Structural basis for recognition of AU-rich element RNA by the HuD protein. Nature Struct. Biol., 8, 141–145. [DOI] [PubMed] [Google Scholar]
- 48.Chen F. and Wilusz,J. (1998) Auxiliary downstream elements are required for efficient polyadenylation of mammalian pre-mRNAs. Nucleic Acids Res., 26, 2891–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sittler A., Gallinaro,H. and Jacob,M. (1995) The secondary structure of the adenovirus-2 L4 polyadenylation domain: evidence for a hairpin structure exposing the AAUAAA signal in its loop. J. Mol. Biol., 248, 525–540. [DOI] [PubMed] [Google Scholar]
- 50.Klasens B.I.F., Das,A.T. and Berkhout,B. (1998) Inhibition of polyadenylation by stable RNA secondary structure. Nucleic Acids Res., 26, 1870–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phillips C., Schimpl,A., Dietrich-Goetz,W., Clements,J.B. and Virtanen,A. (1996) Inducible nuclear factors binding the IgM heavy chain pre-mRNA secretory poly(A) site. Eur. J. Immunol., 26, 3144–3152. [DOI] [PubMed] [Google Scholar]
- 52.Phillips C. and Virtanen,A. (1997) The murine IgM secretory poly(A) site contains dual upstream and downstream elements which affect polyadenylation. Nucleic Acids Res., 25, 2344–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bléoo S., Sun,X., Hendzel,M.J., Rowe,J.M., Packer,M. and Godbout,R. (2001) Association of human DEAD box protein DDX1 with a cleavage stimulation factor involved in 3′-end processing of pre-mRNA. Mol. Biol. Cell, 12, 3046–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bagga P.S., Ford,L.P., Chen,F. and Wilusz,J. (1995) The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res., 23, 1625–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bagga P.S., Arhin,G.K. and Wilusz,J. (1998) DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res., 26, 5343–5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hans H. and Alwine,J.C. (2000) Functionally significant secondary structure of the simian virus 40 late polyadenylation signal. Mol. Cell. Biol., 20, 2926–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barabino S.M.L., Hübner,W., Jenny,A., Minvielle-Sebastia,L. and Keller,W. (1997) The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev., 11, 1703–1716. [DOI] [PubMed] [Google Scholar]
- 58.Bai C. and Tolias,P.P. (1998) Drosophila clipper/CPSF 30 K is a post-transcriptionally regulated nuclear protein that binds RNA containing GC clusters. Nucleic Acids Res., 26, 1597–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barabino S.M.L., Ohnacker,M. and Keller,W. (2000) Distinct roles of two Yth 1p domains in 3′-end cleavage and polyadenylation of yeast pre-mRNAs. EMBO J., 19, 3778–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zarudnaya M.I., Kolomiets’,I.M. and Hovorun,D.M. (2002) What nuclease cleaves pre-mRNA in the process of polyadenylation? IUBMB Life, 54, 27–31. [DOI] [PubMed] [Google Scholar]
- 61.Anantharaman V., Koonin,E.V. and Aravind,L. (2002) Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res., 30, 1427–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zuker M., Mathews,D.H. and Turner,D.H. (1999) Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In Barciszewsky,J. and Clark,B.F.C. (eds), RNA Biochemistry and Biotechnology. NATO ASI Series. Kluwer Academic Publishers, Dordrecht, Boston, and London, pp. 11–43.
- 63.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 64.Lutz C.S. and Alwine,J.C. (1994) Direct interaction of the U1 snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev., 8, 576–586. [DOI] [PubMed] [Google Scholar]
- 65.Lowman H.B. and Draper,D.E. (1986) On recognition of helical RNA by cobra venom V1 nuclease. J. Biol. Chem., 261, 5396–5403. [PubMed] [Google Scholar]
- 66.Arhin G.K., Boots,M., Bagga,P.S., Milcarek,C. and Wilusz,J. (2002) Downstream sequence elements with different affinities for the hnRNP H/H′ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res., 30, 1842–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Honoré B., Rasmussen,H.H., Vorum,H., Dejgaard,K., Liu,X., Gromov,P., Madsen,P., Gesser,B., Tommerup,N. and Celis,J.E. (1995) Heterogeneous nuclear ribonucleoproteins H, H′ and F are members of a ubiquitously expressed subfamily of related but distinct proteins encoded by genes mapping to different chromosomes. J. Biol. Chem., 270, 28780–28789. [DOI] [PubMed] [Google Scholar]
- 68.Mahé D., Mähl,P., Gattoni,R., Fischer,N., Mattei,M.-G., Stévenin,J. and Fuchs,J.-P. (1997) Cloning of human 2H9 heterogeneous nuclear ribonucleoproteins. Relation with splicing and early heat shock-induced splicing arrest. J. Biol. Chem., 272, 1827–1836. [DOI] [PubMed] [Google Scholar]
- 69.Caputi M. and Zahler,A.M. (2001) Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H′/F/2H9 family. J. Biol. Chem., 276, 43850–43859. [DOI] [PubMed] [Google Scholar]
- 70.Fogel B.L., McNally,L.M. and McNally,M.T. (2002) Efficient polyadenylation of Rous sarcoma virus RNA requires the negative regulator of splicing element. Nucleic Acids Res., 30, 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Veraldi K.L., Arhin,G.K., Martincic,K., Chung-Ganster,L.-H., Wilusz,J. and Milcarek,C. (2001) hnRNP F inflluences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell. Biol., 21, 1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chou M.-Y., Rooke,N., Turck,C.W. and Black,D.L. (1999) hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol., 19, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen C.D., Kobayashi,R. and Helfman,D.M. (1999) Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat β-tropomyosin gene. Genes Dev., 13, 593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Markovtsov V., Nikolic,J.M., Goldman,J.A., Turck,C.W., Chou,M.-Y. and Black,D.L. (2000) Cooperative assembly of hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol., 20, 7463–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Caputi M. and Zahler,A.M. (2002) SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J., 21, 845–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Patel D.J., Bouaziz,S., Kettani,A. and Wang,Y. (1999) Structures of guanine-rich and cytosine-rich quadruplexes formed in vitro by telomeric, centromeric and triplet repeat disease DNA sequences. In Neidle,S. (ed.), Oxford Handbook of Nucleic Acid Structure. Oxford University Press, Oxford, pp. 389–453.
- 77.Marathias V. and Bolton,P.H. (1999) Determinants of DNA quadruplex structural type: sequence and potassium binding. Biochemistry, 38, 4355–4364. [DOI] [PubMed] [Google Scholar]
- 78.Jing N., Rando,R.F., Pommier,Y. and Hogan,M.E. (1997) Ion selective folding of loop domains in a potent anti-HIV oligonucleotide. Biochemistry, 36, 12498–12505. [DOI] [PubMed] [Google Scholar]
- 79.Smirnov I. and Shafer,R.H. (2000) Effect of loop sequence and size on DNA aptamer stability. Biochemistry, 39, 1462–1468. [DOI] [PubMed] [Google Scholar]
- 80.Matunis M.J., Xing,J. and Dreyfuss,G. (1994) The hnRNP F protein: unique primary structure, nucleic acid-binding properties and subcellular localization. Nucleic Acids Res., 22, 1059–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Souleil C. and Panijel,J. (1968) Immunochemistry of polyribonucleotides. Study of polyriboinosinic and polyriboguanylic acids. Biochemistry, 7, 7–13. [DOI] [PubMed] [Google Scholar]
- 82.Simonsson T. (2001) G-quadruplex DNA structures—variations on a theme. Biol. Chem., 382, 621–628. [DOI] [PubMed] [Google Scholar]
- 83.Sarig G., Weisman-Shomer,P., Erlitzki,R. and Fry,M. (1997) Purification and characterization of qTBP42, a new single-stranded and quadruplex telomeric DNA-binding protein from Rat hepatocytes. J. Biol. Chem., 272, 4474–4482. [DOI] [PubMed] [Google Scholar]
- 84.Yonaha M. and Proudfoot,N.J. (1999) Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol. Cell, 3, 593–600. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.