Abstract
RecQ5 is one of five RecQ helicase homologs identified in humans. Three of the human RecQ homologs (BLM, WRN and RTS) have been linked to autosomal recessive human genetic disorders (Bloom syndrome, Werner syndrome and Rothmund–Thomson syndrome, respectively) that display increased genomic instability and cause elevated levels of cancers in addition to other symptoms. To understand the role of RecQ helicases in maintaining genomic stability, the WRN, BLM and Escherichia coli RecQ helicases have been characterized in terms of their DNA substrate specificity. However, little is known about other members of the RecQ family. Here we show that Drosophila RECQ5 helicase is a structure-specific DNA helicase like the other RecQ helicases biochemically characterized so far, although the substrate specificity is not identical to that of WRN and BLM helicases. Drosophila RECQ5 helicase is capable of unwinding 3′ Flap, three-way junction, fork and three-strand junction substrates at lower protein concentrations compared to 5′ Flap, 12 nt bubble and synthetic Holliday junction structures, which can be unwound efficiently by WRN and BLM.
INTRODUCTION
Bloom syndrome, Werner syndrome and a subset of Rothmund–Thomson syndrome are caused by mutations in genes that encode DNA helicases belonging to the RecQ family (BLM, WRN and RTS, respectively) (1–3). The individuals affected by these genetic disorders are predisposed to cancer and, as a general hallmark, display high genomic instability on a cellular level (4). In addition to the RecQ homologs mentioned above, two additional RecQ family genes have been identified in humans, RECQL and RECQ5 (5,6). To date, no human genetic disorder has been associated with these genes.
Members of the RecQ helicase family are present in organisms ranging from bacteria to humans although the number of RecQ homologs present in each organism varies. RECQ5 orthologs have been identified in humans, mice, Caenorhabditis elegans and Drosophila melanogaster (7–9) and alternative splicing of the RECQ5 transcript has been reported in humans and Drosophila (9,10). In both cases at least three protein isoforms are produced as a result of alternative splicing. At present, nothing is known regarding the differential functions of the three RECQ5 isoforms in flies and humans. However, there is some information on the localization of the proteins and transcripts. Northern blot analysis revealed that human RecQ5 transcripts are present in all tissues examined. It has been demonstrated that the large isoform of human RECQ5, RECQ5β, coimmunoprecipitates and colocalizes with topoisomerase 3α and 3β (10) suggesting that these proteins interact. However, the region on RECQ5β required for this interaction has not been mapped.
To begin to understand how the phenotypes in Bloom syndrome and Werner syndrome arise, and to describe the biological role of the RecQ helicases, several laboratories have characterized the biochemical function of these proteins. Many of the biochemical studies have focused on Escherichia coli RecQ, the founding member of the RecQ helicase family (11–15), while others have focused on the BLM helicase (16–21) and the WRN helicase (21–27). All three proteins catalyze a 3′ to 5′ DNA helicase reaction and each enzyme is capable of unwinding a wide spectrum of substrates (28–30). For example, E.coli RecQ helicase can unwind blunt-ended duplex substrates, a three-strand junction, a synthetic Holliday junction, fork structures and internal regions on a double-stranded DNA (13,14). BLM and WRN can unwind many of these same DNA structures (synthetic Holliday junctions, G4-quadruplex DNA, 12 nt bubble structures, triple helix structures and fork structures) but not blunt-ended substrates (21,27,30). All three proteins have been shown to facilitate branch migration over long stretches of DNA (13,18,26). BLM protein can also unwind D-loop structures and has a higher affinity for D-loops with a 5′ end invasion compared to D-loops with a 3′ end invasion (20). Based upon the phenotypes of Bloom syndrome and Werner syndrome cell lines, the proteins they interact with, and the substrates they can unwind, BLM and WRN have been proposed to play roles in the recovery of stalled replication forks and/or recombinational repair (28).
In a previous study, a small isoform of the Drosophila RECQ5 protein (DmRECQ5) was purified and characterized (31). These initial investigations used partial duplex DNA substrates with varying duplex regions (20, 42 and 93 bp) to characterize the helicase activity of DmRECQ5. Although the partial duplex substrates were unwound by the purified protein, a high protein concentration (≥1 µM) was required to reach maximum unwinding of the 42 and 93 bp partial duplex substrates. Unwinding of a blunt duplex substrate was not detected. These results suggested that DmRECQ5, like several other RecQ helicases, might prefer specific DNA structures for unwinding.
Here we present evidence that DmRECQ5 preferentially unwinds specific DNA structures including a 3′ Flap, a three-strand junction and a three-way junction. These substrates were unwound at significantly lower protein concentrations than the substrates utilized previously, probably due to binding of DmRECQ5 to the fork structure present in each substrate. Interestingly, unwinding of a Holliday junction, 5′ Flap and 12 nt bubble structures, which can be unwound by other RecQ proteins (WRN, BLM and/or E.coli RecQ), could not be detected or required significantly higher protein concentrations. The implications of these results are discussed in terms of a potential cellular function for RECQ5.
MATERIALS AND METHODS
Protein purification
A small isoform of DmRECQ5 was overexpressed and purified from E.coli ER2566 using the IMPACT system (New England Biolabs) as previously described (31).
Construction of helicase substrates
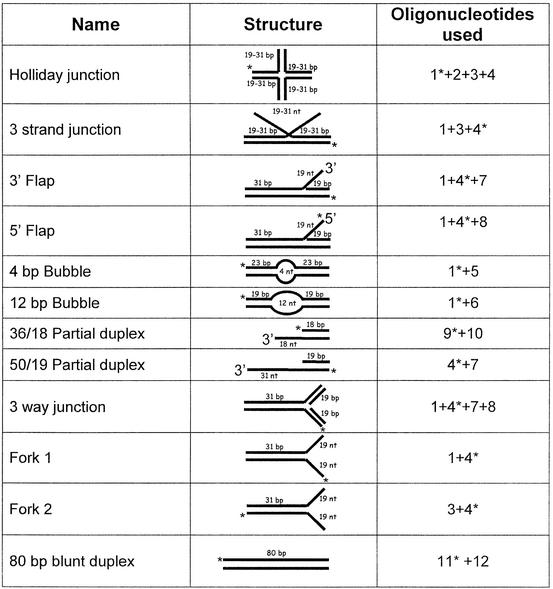
Oligonucleotides were synthesized by IDT Inc., and are listed in Table 1. Each oligonucleotide was purified on a 20% denaturing polyacrylamide gel. To prepare each substrate, one of the oligonucleotides was labeled on the 5′ end using T4 polynucleotide kinase and [γ-32P]ATP. The annealing mixtures (150 µl) contained 7 mM Tris–HCl (pH 7.5), 6.6 mM MgCl2, 50 mM NaCl, 1 mM DTT and the other oligonucleotide(s) at a 4:1 molar ratio compared to the labeled strand. The annealing mixtures were boiled for 5 min in a beaker containing 300 ml of ddH2O and allowed to cool over several hours to room temperature. Substrates were purified on a native 10% polyacrylamide gel using 0.5× TBE (44.5 mM Tris base, 44.5 mM borate, 1 mM EDTA) as the running buffer, electroeluted from the gel slice in 1× TBE + 10 mM MgCl2 and dialyzed against TEN Buffer [10 mM Tris–HCl (pH 8.0), 50 mM NaCl, 10 mM EDTA]. The substrates are shown in Table 2.
Table 1. Oligonucleotides.
| Oligo number | Oligo name | Sequence |
|---|---|---|
| 1 | X12–1 | 5′-GACGCTGCCGAATTCTGGCTTGCTAGGACATCTTTGCCCACGTTGACCCG-3′ |
| 2 | X12–2 | 5′-CGGGTCAACGTGGGCAAAGATGTCCTAGCAATGTAATCGTCTATGACGTC-3′ |
| 3 | X12–3 | 5′-GACGTCATAGACGATTACATTGCTAGGACATGCTGTCTAGAGACTATCGC-3′ |
| 4 | X12–4 | 5′-GCGATAGTCTCTAGACAGCATGTCCTAGCAAGCCAGAATTCGGCAGCGTC-3′ |
| 5 | BUB4 | 5′-CGGGTCAACGTGGGCAAAGATAGAATAGCAAGCCAGAATTCGGCAGCGTC-3′ |
| 6 | BUB12 | 5′-CGGGTCAACGTGGGCAAAGTCTATGCTTAGCGCCAGAATTCGGCAGCGTC-3′ |
| 7 | 3′ Flap | 5′-GCTGTCTAGAGACTATCGC-3′ |
| 8 | 5′ Flap | 5′-CGGGTCAACGTGGGCAAAG-3′ |
| 9 | BF18 | 5′-GCCTCGCTGCCGTCGCCA-3′ |
| 10 | BF36 | 5′-TGGCGACGGCAGCGAGGCTTTTTTTTTTTTTTTTTT-3′ |
| 11 | JS-80 | 5′-CCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTTTCTTTATAAATAGAGTGTTATGAAAAATTAGTTTC-3′ |
| 12 | JS-80nic | 5′-GAAACTAATTTTTCATAACACTCTATTTATAAAGAAAAATCAGCAAAAACTTGTTTTTGCGTGGGGTGTGGTGCTTTTGG-3′ |
Table 2. Structures of substrates.
*Radioactive label.
Helicase assays
Helicase reaction mixtures (20 µl) contained 25 mM Tris–HCl (pH 8.0), 20 mM NaCl, 5 mM MgCl2, 50 µg/ml BSA, 1 mM DTT, 3 mM dATP, ∼54 pM substrate and the indicated amount of DmRECQ5. dATP was used in these reactions because it supports somewhat higher levels of unwinding than does ATP (31). Incubation was at 30°C for 10 min. The reactions were quenched with 10 µl of stop solution (37.5% glycerol, 50 mM EDTA, 0.05% xylene cyanol, 0.05% bromophenol blue, 0.3% SDS). The reaction products were resolved on a non-denaturing polyacrylamide gel (10%, 20:1 cross-linking ratio) run at 100 V for 16 h. The results were visualized using a phosphorimager and analyzed with ImageQuant software (Molecular Dynamics). Percent [32P]DNA displaced for each substrate is the sum of all DNA molecules that are formed by the unwinding of that particular substrate. Percent of each DNA molecule formed was calculated using the following equation:
%[32P]DNA = (P – Bp) / [(S – Bs) + (P – Bp)] × 100
where P is the unwound product, S is the intact substrate, and Bp and Bs represent the background associated with the product and substrate, respectively.
Electrophoretic mobility shift assays (EMSA)
Reaction mixtures (20 µl) contained 25 mM Tris–HCl (pH 8.0), 20 mM NaCl, 2 mM MgCl2, 100 µg/ml BSA, 1 mM DTT, 1 mM ATPγS, ∼54 pM substrate and the indicated amount of DmRECQ5. The reactions were incubated at 30°C for 10 min after the addition of protein. After addition of 3 µl of loading dye (74% glycerol, 0.01% xylene cyanol and bromophenol blue) to each reaction, they were placed on ice until loading on the gel. The bound DNA was resolved from the unbound DNA on a native 4% polyacrylamide gel (15:1 crosslinking ratio) run at 200 V for 2.5 h at 4°C using 50 mM Tris, 380 mM glycine and 2 mM EDTA as the running buffer. The results were visualized using a phosphorimager and analyzed with ImageQuant software (Molecular Dynamics).
DNase I footprinting
DNase I footprinting reactions were performed under the same conditions as helicase assays with the following changes: the reaction volume was 10 µl and ATPγS was substituted for dATP. After a 10 min incubation at 30°C to allow protein binding to the DNA substrate, CaCl2 (1 mM final concentration) and DNase I (10 ng) were added to each reaction and incubation was continued for 4 min. Each reaction was quenched with 28 µl of gel loading solution (85% formamide, 25 mM EDTA, 0.01% each of xylene cyanol and bromophenol blue) and boiled for 3 min. The reaction products were resolved on a denaturing polyacrylamide gel (16%, 20:1 cross-linking ratio) run at 25 W for 3 h.
A DNA ladder was prepared by digesting 1.1 pmol of a 30 nt 5′-end labeled oligonucleotide with 0.08 U of phosphodiesterase I (US Biochemical Corp.) in 40 mM Tris–HCl (pH 9) and 2 mM MgCl2. The reaction (100 µl) was assembled on ice and incubated at 30°C. Aliquots (20 µl) were removed at 2.5, 5, 10, 15 and 20 min, and stopped by the addition of 80 µl of 100 mM NaCl, 10 mM Tris–HCl (pH 7.5), 1 mM EDTA and 100 µl of phenol/chloroform. After phenol/chloroform extraction, 12 µl samples were resolved on a 16% polyacrylamide, 8 M urea denaturing gel to determine the progress of the reaction at each time point. Typically, the 10 and 15 min time points were pooled to use as markers.
RESULTS
In the initial biochemical characterization of a small isoform of DmRECQ5, partial duplex substrates with duplex regions of varying lengths (20, 42 and 93 bp) were used to characterize the helicase reaction (31). Although unwinding of all three substrates was observed, high DmRECQ5 concentrations were required to measure unwinding of the longer duplex regions and the fraction of the substrate unwound was not increased by the addition of a single-stranded DNA (ssDNA) binding protein (E.coli ssDNA binding protein and yeast replication protein A). Unwinding of a blunt duplex substrate of 89 bp, on the other hand, was not detected even at 3.4 µM DmRECQ5, indicating that the protein cannot initiate unwinding from blunt ends. One of the explanations proposed for the high protein concentration requirement was that the substrates tested were not ‘preferred’ substrates of DmRECQ5. To test this possibility the DmRECQ5-catalyzed unwinding of a series of DNA substrates with a variety of structures was examined.
DmRECQ5 unwinds specific DNA structures
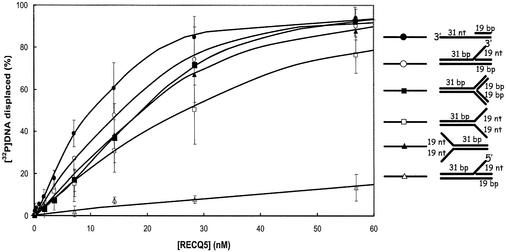
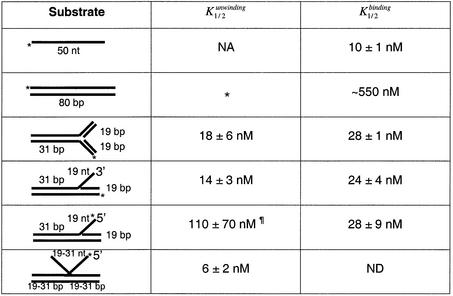
The oligonucleotides and the specific DNA structures constructed using these oligonucleotides are shown in Tables 1 and 2, respectively. The unwinding of each of these substrates was measured in the presence of increasing concentrations of DmRECQ5 and the results of these experiments are presented in Figure 1 and Table 3. The 3′ Flap substrate, fork substrates, 50/19 partial duplex substrate and the three-way junction substrate were all unwound at significantly lower protein concentrations (Fig. 1) compared to what had been observed with the 20 bp partial duplex substrate (31). For example, half maximal unwinding (Kunwinding1/2) of the 20 bp partial duplex substrate was reached at a DmRECQ5 concentration of ∼100 nM compared to the 50/19 partial duplex substrate where the Kunwinding1/2 was 9 nM (Table 3). Similarly, the Kunwinding1/2 for the three-way junction substrate was 18 nM and the Kunwinding1/2 for the 3′ Flap substrate was 14 nM. Both Fork 1 and Fork 2 were unwound with Kunwinding1/2 values (16 and 20 nM) similar to those measured for the 3′ Flap, the three-way junction and the 50/19 partial duplex substrate. Another substrate that was efficiently unwound by DmRECQ5 was a three-strand junction substrate. The Kunwinding1/2 value for this substrate was 6 nM (Table 3). The fact that each of these substrates was unwound efficiently by RECQ5 in vitro may suggest that RECQ5 encounters and unwinds similar structures in vivo.
Figure 1.
Unwinding of DNA substrates by DmRECQ5. Helicase assays using the 3′ Flap (open circles), 5′ Flap (open triangles), three-way junction (filled squares), 50/19 partial duplex (filled circles) and fork (fork 1, open squares; fork 2, filled triangles) substrates were as described in Materials and Methods using the indicated amount of DmRECQ5. Unwinding reactions were initiated by the addition of enzyme and incubated for 10 min at 30°C. Each data point is the average of at least three experiments. Error bars represent the standard deviation about the mean.
Table 3. Unwinding and binding parameters.
NA, not applicable; ND, not determined.
*No unwinding detected.
¶<40% unwinding detected.
It is also significant that DmRECQ5 unwound several of the substrates tested very poorly or not at all. For example, the unwinding of the 5′ Flap substrate required significantly higher DmRECQ5 concentrations than unwinding of the 3′ Flap substrate (Fig. 1 and Table 3). Unwinding of a 5′ Flap substrate could be detected at higher protein concentrations. However, the extent of unwinding did not exceed 40% even at 910 nM RECQ5 after reaching a plateau at ∼300 nM RECQ5 (data not shown). In addition, unwinding of a 4 nt bubble structure was not detected (data not shown). The failure to unwind the 4 nt bubble structure is consistent with the previous finding that DmRECQ5 cannot unwind blunt-ended DNA substrates (31). It is also apparent that the protein cannot load at the ssDNA–duplex DNA junction provided by the 4 nt bubble. Increasing the size of the bubble structure to 12 nt did not dramatically improve the use of this structure as a substrate for unwinding. At high protein concentrations, low but detectable unwinding of the 12 nt bubble structure was observed (∼30% at 910 nM protein). Similarly, under these reaction conditions no significant unwinding of a synthetic Holliday junction substrate with a 12 bp complementary core region was detected. In contrast, WRN helicase can unwind a 5′ Flap substrate (30) and both WRN and BLM are capable of unwinding a 12 nt bubble structure and a synthetic Holliday junction (21). Thus, several structures unwound by other RecQ helicases did not serve as substrates for DmRECQ5.
Unwinding of the synthetic Holliday junction substrate
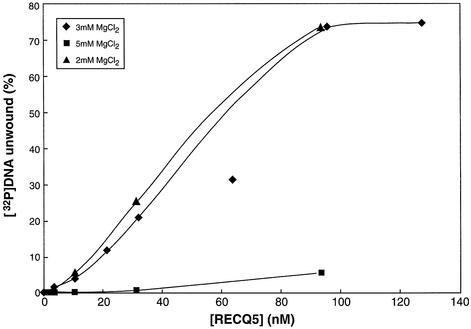
The failure to unwind a Holliday junction was surprising since several other RecQ helicases including BLM, WRN and E.coli RecQ have been demonstrated to unwind this structure (13,21). The helicase reactions contained 3 mM dATP and 5 mM MgCl2 resulting a free Mg++ concentration of ∼2 mM. However, the magnesium concentration in the helicase reactions with BLM, WRN and E.coli RecQ was the same as or similar to the ATP concentration, resulting in no or low concentrations of free Mg++. Intriguingly, it has been reported that free magnesium has inhibitory effects on the unwinding of synthetic Holliday junctions by two other helicases, RecG and MER3p (32,33). For this reason unwinding of a synthetic Holliday junction by DmRECQ5 was tested at different Mg++ concentrations. As shown in Figure 2, DmRECQ5 is indeed capable of unwinding a synthetic Holliday junction at low free Mg++ concentrations. However, even at 3 mM MgCl2, the Kunwinding1/2 was higher compared to unwinding of the 50/19 partial duplex substrate, fork substrates, three-way junction substrate and 3′ Flap substrate (40 versus 16 nM). This property of DmRECQ5 distinguishes it from WRN and BLM, that are capable of unwinding synthetic Holliday junctions at higher rates than fork structures.
Figure 2.
Unwinding of a synthetic Holliday junction substrate. Helicase assays using a synthetic Holliday junction substrate were as described in Materials and Methods using the indicated amount of DmRECQ5 and MgCl2. Unwinding reactions were initiated by the addition of enzyme and incubated for 10 min at 30°C. Each data point is the average of at least two experiments.
DmRECQ5 unwinds the lagging strand in a ‘stalled replication fork’
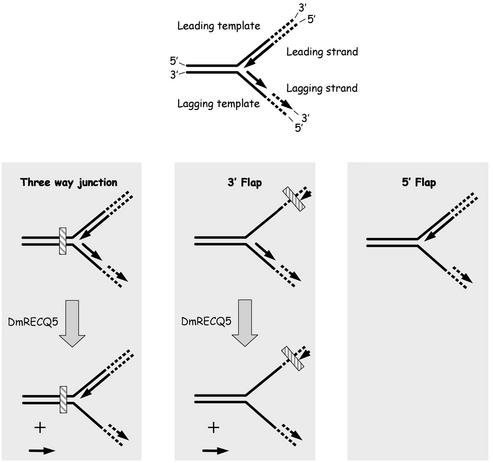
Three of the substrates used in this study resemble structures proposed for a stalled replication fork (see Fig. 7). The three-way junction substrate resembles a stalled replication fork where both the leading and the lagging arms have been converted to duplex DNA. The 3′ Flap substrate can be envisioned as a replication fork where the synthesis of the leading strand has been blocked while synthesis of the lagging strand has continued. The 5′ Flap substrate resembles the structure proposed for a stalled replication fork where synthesis of the lagging strand has been blocked or has not yet reached the point of the leading strand.
Figure 7.
The three-way junction, 3′ Flap and 5′ Flap substrates resemble a stalled replication fork. A replication fork is illustrated at the top to describe all the strands involved and their polarities. The three panels illustrate the observed unwinding pathway for each substrate. The hatched box on the three-way junction and the 3′ Flap structures denotes a lesion that blocks replication. The solid lines represent the oligonucleotides present in each substrate used. Dashed lines are drawn to further illustrate the proposed replication fork. The three-way junction mimics a fork where a lesion blocks synthesis on both sides. The 3′ Flap mimics the fork where DNA synthesis of the leading strand is blocked. The 5′ Flap mimics the fork where DNA synthesis of the lagging strand has been blocked or has not yet reached the point of the leading strand, due to uncoupling of the leading and lagging strand synthesis. The resulting structures from the RECQ5-catalyzed unwinding reaction are depicted on the bottom. DmRECQ5 removes the lagging strand in the three-way junction and 3′ Flap substrates.
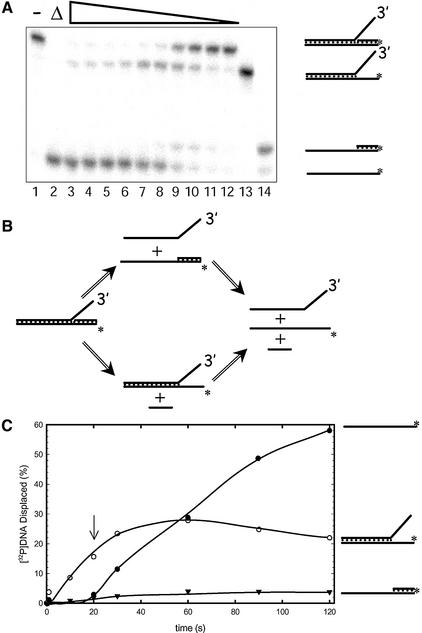
In unwinding reactions using the 3′ Flap substrate the appearance of both a fork structure and a partial duplex structure as intermediates was observed (Fig. 3A). The appearance of the partial duplex structure was not surprising since DmRECQ5 exhibits a 3′ to 5′ polarity like other RecQ helicases. If the protein loads on the 3′ ssDNA tail, and then translocates in a 3′ to 5′ direction, it would displace the top strand from the bottom strand resulting in the production of the partial duplex structure (Fig. 3B; upper pathway). Production of a fork structure could be achieved if the protein recognized and bound the junction, and then translocated in a 3′ to 5′ direction along the bottom strand removing the ‘lagging’ strand (Fig. 3B; lower pathway). Because DmRECQ5 will inevitably act upon the intermediates described above, and unwind these structures to produce single-stranded oligonucleotides, determination of which pathway occurred more frequently required further analysis. For this purpose, time points early in the reaction (<2 min) were analyzed and the fork structure was determined to be the first intermediate produced (Fig. 3C). In fact, the appearance of the 50mer product succeeds the appearance of the fork suggesting that the 50mer is the product of unwinding of the fork structure rather than a product of unwinding the partial duplex substrate. The partial duplex structure detected during this unwinding reaction never exceeded 4% of the total DNA molecules in the reaction suggesting that the majority, if not all, of the DmRECQ5 protein preferred to initiate unwinding at the junction to produce the fork structure.
Figure 3.
Unwinding of the 3′ Flap substrate. (A) A DmRECQ5 titration using the 3′ Flap substrate. Lanes 1 and 2 contained no enzyme. The reaction shown in lane 2 was boiled to denature the substrate. Lane 3, 910 nM DmRECQ5; lane 4, 455 nM DmRECQ5; lane 5, 226 nM DmRECQ5; lane 6, 113 nM DmRECQ5, lane 7, 57 nM DmRECQ5; lane 8, 28 nM DmRECQ5; lane 9, 14 nM DmRECQ5; lane 10, 7 nM DmRECQ5; lane 11, 3.6 nM DmRECQ5; lane 12, 1.8 nM DmRECQ5. Lanes 13 and 14 are markers to indicate where the fork and the partial duplex structures migrate on the gel, respectively. The substrate and each intermediate/product are shown schematically on the right. (B) A schematic representation of unwinding of the 3′ Flap substrate. Production of two intermediates can be explained by two pathways. The partial duplex intermediate is produced by the upper pathway when the protein loads on the ssDNA 3′ flap and translocates along this strand displacing the top strand from the bottom strand. The fork intermediate is produced by the lower pathway when the protein loads on the bottom strand at the junction translocating 3′ to 5′ along this strand unwinding the annealed oligonucleotide. (C) Time course of 3′ Flap unwinding. The reaction mixture (100 µl) was as described in Materials and Methods and the reaction was initiated by the addition of RECQ5 to a final concentration of 53 nM. Aliquots (10 µl) of the reaction were withdrawn at the indicated time points, quenched and analyzed on a native polyacrylamide gel. Lines correspond to fraction of fork (open circles), partial duplex (filled triangles) and 50mer (filled circles) present during the time course. The arrow points to the 20 s time point when the appearance of the final product (50mer) is first observed.
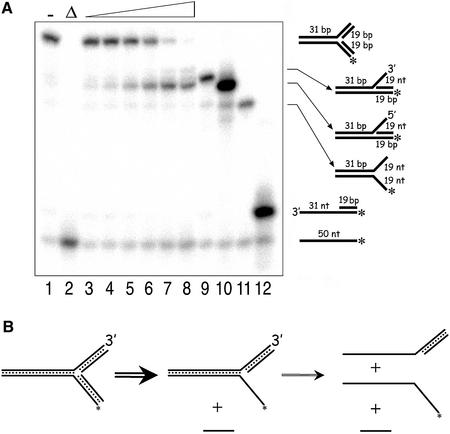
In the unwinding reactions using the three-way junction substrate, the 5′ Flap structure was the predominant intermediate observed (Fig. 4A). Production of a 5′ Flap substrate from a three-way junction substrate can be achieved by removal of the ‘lagging’ strand as in the case of the 3′ Flap substrate (Fig. 4B). The fact that the appearance of a 3′ Flap structure, or intermediates derived from unwinding of a 3′ Flap substrate were not detected suggests that in this substrate the ‘leading’ strand is not unwound by DmRECQ5. Thus, at high concentrations of DmRECQ5 essentially all of the substrate had been converted to a 5′ Flap intermediate but only a fraction of this intermediate was converted to fully unwound 50mer. The latter result is consistent with the poor unwinding of the 5′ Flap substrate described above.
Figure 4.
Unwinding of the three-way junction substrate. (A) A DmRECQ5 titration using the three-way junction substrate. Lanes 1 and 2 contained no enzyme. The reaction shown in lane 2 was boiled prior to loading on the gel to denature the substrate. Lanes 3–8 show a protein titration (1.8, 3.5, 7, 14, 28, 57 nM, respectively). Lanes 9–12 are markers to indicate where the 3′ Flap, 5′ Flap, Fork1 and the 50/19 partial duplex structures migrate on the gel, repectively. (B) A schematic representation of unwinding of the three-way junction substrate. Production of the 5′ Flap structure from the three-way junction substrate can be achieved if RECQ5 loads on the bottom strand at the junction and translocates along this strand displacing the annealed oligonucleotide. At high protein concentrations some of the 5′ Flap intermediate is unwound to release the labeled 50mer. This activity is shown by a thin arrow compared to the thicker arrow to produce the 5′ Flap intermediate from the three-way junction substrate.
The unwinding of these substrates suggested that DmRECQ5 recognized the junction, loaded on the bottom strand and translocated 3′ to 5′ along this strand preferentially unwinding the ‘lagging’ strand in a ‘stalled replication fork’ whether the ‘leading’ strand was present or not. These results may infer a role for this protein in the processing of stalled replication forks (see Discussion).
RECQ5 binds at the junction
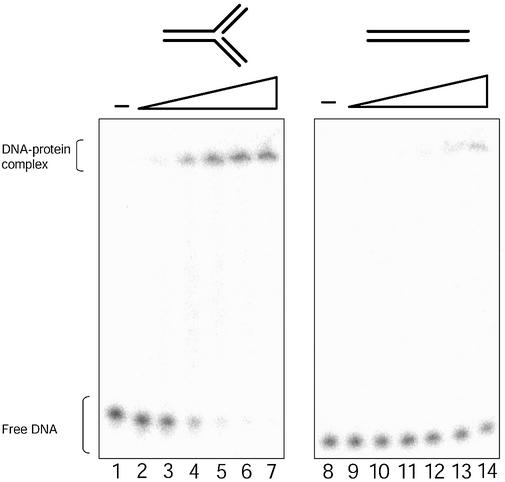
The appearance of specific unwinding intermediates with defined kinetics strongly suggests that DmRECQ5 binds and initiates unwinding from the junction on these substrates. In addition, unwinding of the three-way junction, which contains three blunt ends that cannot serve as initiation sites for unwinding, suggests that unwinding initiates at the junction. To confirm this interpretation, EMSA were performed (Fig. 5 and Table 3). The binding of DmRECQ5 to a 50 base ssDNA oligonucleotide, the 3′ Flap substrate, the 5′ Flap substrate, the three-way junction substrate and an 80 bp blunt duplex substrate was measured. No significant binding to the 80 bp blunt duplex substrate was detected (Kbinding1/2 ∼ 550 nM). This result is in agreement with the lack of unwinding of this structure. The three-way junction substrate is also a double-stranded, blunt-ended structure. However, DmRECQ5 bound the three-way junction substrate (Kbinding1/2 = 28 nM). The difference between the two substrates is the presence of the fork structure in the three-way junction. This result suggests that DmRECQ5 recognizes and binds the fork structure within the three-way junction, which is absent in the 80 bp blunt duplex substrate.
Figure 5.
EMSA of the three-way junction substrate. DNA binding reactions were as described in Materials and Methods. Lanes 1–7, reactions contained the three-way junction substrate; lanes 8–14, reactions contained the 80 bp blunt duplex substrate. Lanes 1 and 8 contained no DmRECQ5; lanes 2 and 9 contained 7.3 nM DmRECQ5; lanes 3 and 10 contained 14.5 nM DmRECQ5; lanes 4 and 11 contained 29 nM DmRECQ5; lanes 5 and 12 contained 58 nM DmRECQ5; lanes 6 and 13 contained 116 nM DmRECQ5; lanes 7 and 14 contained 232 nM DmRECQ5. Schematics of each substrate are shown at the top of the corresponding gel.
The binding parameter Kbinding1/2, defined as the protein concentration required to observe one-half of the maximum substrate in the bound form, was virtually the same for the 3′ Flap, the 5′ Flap and the three-way junction substrates (Kbinding1/2 = 24, 28, 28 nM, respectively). The Kbinding1/2 of the 3′ Flap substrate and the three-way junction substrates were very similar to the Kunwinding1/2 measured for these substrates (Kunwinding1/2 = 14 and 18 nM versus Kbinding1/2 = 24 and 28 nM). However, this was not the case for the 5′ Flap substrate. DmRECQ5 bound to the 5′ Flap substrate efficiently but could not unwind more than 40% of this substrate even at high protein concentrations. This result can be explained if DmRECQ5 recognizes and binds the fork structure but then translocates in a 3′ to 5′ direction along the ssDNA (i.e. outward from the fork, on the template strand for the lagging strand). Furthermore, DmRECQ5 bound the 50 nt ssDNA oligonucleotide with higher affinity (Kbinding1/2 = 10 nM) than all the other substrates tested. Therefore, the ssDNA region in the 3′ Flap and the 5′ Flap substrates, in addition to the fork structure, may also contribute to the binding by DmRECQ5.
DNase I footprinting experiments were performed to detect the protein binding site on the three-way junction substrate (Fig. 6) and the 3′ Flap substrate (data not shown). The reactions were performed in the presence of ATPγS, a poorly hydrolyzable analog of ATP, to capture the protein/DNA complexes at the initiation of the unwinding reaction. The three-way junction substrate with the radioactive label in the bottom strand was used for the experiment shown in Figure 6. The footprint is detectable at 15 nM DmRECQ5 and becomes apparent at 30 nM DmRECQ5 (Fig. 6B, lanes 5 and 6), which is in agreement with the binding parameter (Kbinding1/2 = 28 nM) for this substrate. Nearly all the protected bases correspond to the region surrounding the junction, indicative of protein binding in this area. We also note an alteration in the protection pattern near one end of the DNA substrate. This may reflect an unstable interaction of the protein with the duplex DNA end that is detectable in a DNase I protection assay but not detected using EMSA (see Fig. 5). If this is the case, the interaction is apparently never sufficiently stable to allow an unwinding reaction to initiate since DNA substrates with only duplex ends lacking junctions are not unwound by DmRECQ5 helicase (31). Finally, the overall size of the observed footprint appears to increase as the protein concentration increases. This may be due to the binding of additional molecules of DmRECQ5 protein. This result, and the fact that the assembly state of bound DmRECQ5 (i.e. monomer versus oligomer) is not known, precludes any conclusion regarding a minimal binding site size.
Figure 6.
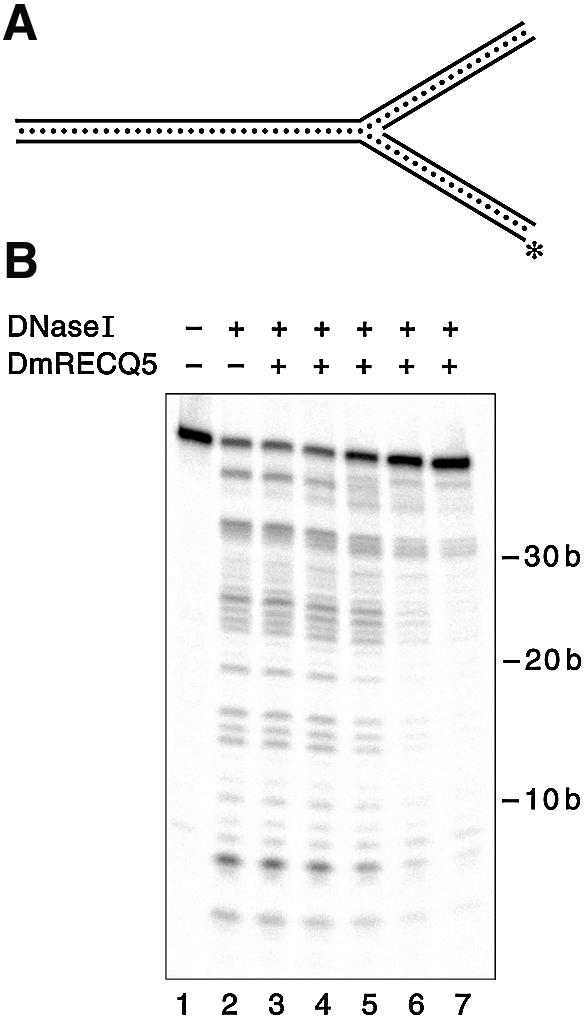
DNase I footprinting on the three-way junction substrate. DNase I footprinting reaction mixtures were prepared and analyzed as described in Materials and Methods. (A) Schematic view of the DNA substrate. The labeled strand is marked with the asterisk. (B) The reaction in lane 1 contained no enzymes, the reaction in lane 2 contained no DmRECQ5 and lanes 3–7 contained DmRECQ5 at 3.8, 7.3, 14.6, 29.1 and 58.3 nM, respectively.
DISCUSSION
DNA helicases belonging to the RecQ helicase family have been shown to unwind a wide range of DNA structures (13,20,21,27,30). For example, E.coli RecQ has been shown to unwind the following substrates: blunt-ended duplex DNA, three-strand junctions, Holliday junctions and covalently closed plasmids. The BLM and WRN gene products, two human RecQ helicases, cannot initiate unwinding from blunt ends or from internal regions of a duplex molecule like the E.coli RecQ helicase, but they are capable of unwinding synthetic Holliday junctions, 12 nt bubble structures, triplex DNA, G-quadruplex DNA and fork structures. In fact, the set of substrates unwound by WRN and BLM overlap although the rate of unwinding of individual substrates may differ from one enzyme to the other.
Another member of the RecQ family, RECQ5 from Drosophila was recently purified and shown to possess DNA helicase activity (31). In the initial study it was reported that the DmRECQ5 protein can unwind partial duplex substrates but it cannot initiate unwinding from blunt ends, a property it shares with the BLM and WRN proteins. Here the DmRECQ5 is reported to be a structure specific helicase that can unwind 3′ Flap, three-way junction, three-strand junction and fork structures. This result suggests that DmRECQ5 may bind and initiate unwinding on these DNA structures specifically. Interestingly, the structures unwound by DmRECQ5 show little overlap with the structures unwound by BLM and WRN. Importantly, a synthetic Holliday junction substrate that can be unwound by BLM and WRN was not a preferred substrate for DmRECQ5. In addition, a 12 nt bubble structure was very poorly unwound by DmRECQ5 while the rate of unwinding of this structure by WRN was comparable to the WRN-catalyzed unwinding rate of G4-quadruplex substrates, where WRN displayed the highest unwinding rate (21). The 5′ Flap substrate, which was unwound poorly by RECQ5, was unwound efficiently by the WRN protein (30). Both WRN and DmRECQ5 can unwind a three-way junction and a 3′ Flap substrate, but the product of the unwinding reaction is different for the two proteins. The WRN helicase unwinds toward the fork, separating the two ‘template’ strands, and DmRECQ5 unwinds outward from the fork, separating the ‘lagging’ strand from its template. These results demonstrate that even though RecQ helicases are structure specific, the specificity is not the same for all members of the family. Thus, it seems likely there is a division of labor among the RecQ helicases in terms of unwinding substrates.
Further investigation of the DmRECQ5-catalyzed unwinding of these substrates revealed a preference for binding at the fork junction present in each of the structures. The intermediates formed during the unwinding of the 3′ Flap substrate, and especially the three-way junction substrate which does not contain ssDNA, the site where most helicases are thought to load, suggest that RECQ5 recognizes the junction in these substrates. Moreover, the oligonucleotide unwound by RECQ5 can be envisioned as the ‘lagging’ strand of a stalled replication fork (Fig. 7). Unwinding of this specific duplex can be achieved by loading on and then translocating along the bottom strand (the template for lagging strand synthesis). Both EMSA and DNase I footprinting experiments demonstrated RECQ5 binding to the fork junction structure. The fact that the three-way junction, which lacks any ssDNA, was bound by DmRECQ5 provided strong support for the idea that the protein can recognize the junction. The DNase I footprint on both the 3′ Flap substrate (data not shown) and the three-way junction substrate also demonstrates that the protein can bind at the junction and protect a region around the junction on the substrate.
A surprising result was that the unwinding of the oligonucleotide based 50/19 partial duplex substrate was achieved at much lower DmRECQ5 concentrations compared to the partial duplex substrates tested previously (31). For example, the Kunwinding1/2 of the 20 bp partial duplex substrate was determined to be ∼100 nM, significantly higher than the Kunwinding1/2 value for the 50/19 partial duplex substrate (9 nM). This discrepancy may be the result of one or more factors as explained below. First, in the reactions containing M13 ssDNA circle based substrates the substrate concentration was higher than the reactions containing the oligonucleotide based substrates (230 versus 54 pM substrate). This difference could contribute to the difference of the Kunwinding1/2 values of the partial duplex substrates. In addition, the large amount of ssDNA and secondary structures present in the M13 circle of the 20 bp partial duplex substrate may constitute multiple non-specific binding sites for DmREQ5, thus inhibiting recognition and unwinding of the duplex region.
Assuming that the human RECQ5 protein will demonstrate similar substrate specificity, these data suggest that structures arising in recombinational repair pathways and/or during the recovery of stalled replication forks, which are poor substrates for WRN or BLM, may be acted upon by RECQ5. The idea of having several helicases that can recognize and unwind structures that are similar, yet different, may explain why the deletion of a single RecQ helicase is not lethal. A structure that may arise during the recovery of a stalled replication fork may be recognized and unwound by more than one RecQ homolog. Depending on which helicase is recruited to the site, the process may continue along one pathway rather than another ultimately resolving the recombination intermediate.
Based on the phenotypes of mutant cell lines, localization in the nucleus and colocalization/interaction with other proteins, BLM and WRN have been proposed to function in recombinational repair pathways involved in maintaining genomic stability. Although their specific molecular roles are far from being determined, both have been proposed to play roles in replication, specifically in the recovery of stalled replication forks. In addition, BLM may be responsible for termination of non-productive 5′ end invading D-loop structures and BLM (and WRN) may play a role in double-stranded break repair.
To date, there is no information regarding the phenotype exhibited by an organism with a mutation in RECQ5. This makes it particularly difficult to pinpoint a role for RECQ5 in the cell. However, since two of the substrates unwound by RECQ5 resemble stalled replication forks, and a specific duplex region is unwound in each case, it is tempting to speculate that RECQ5 may play a role in the processing of stalled replication forks that is complementary to the roles played by WRN and BLM. It is intriguing that the E.coli RecG helicase, which has been shown to play a role in the recovery of stalled replication forks (34), removes the same strand in the 3′ Flap and three-way junction substrates as DmRECQ5. Perhaps RecG helicase in E.coli and RECQ5 helicase in metazoans have similar roles in recombinational repair in the cell.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Susan Whitfield for help with artwork preparation and members of the Matson and Sekelsky laboratories for helpful discussions. This work was supported by US National Institutes of Health Grant GM 33476 (to S.W.M.).
REFERENCES
- 1.Ellis N.A., Groden,J., Ye,T.-Z., Straughen,J., Lennon,D.J., Ciocci,S., Proytcheva,M. and German,J. (1995) The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell, 83, 655–666. [DOI] [PubMed] [Google Scholar]
- 2.Yu C.-E., Oshima,J., Fu,Y.-H., Wijsman,E.M., Hisama,F., Alisch,R., Matthews,S., Nakura,J., Miki,T., Ouais,S., Martin,G.M., Mulligan,J. and Schellenberg,G.D. (1996) Positional cloning of the Werner’s syndrome gene. Science, 272, 258–262. [DOI] [PubMed] [Google Scholar]
- 3.Kitao S., Shimamoto,A., Goto,M., Miller,R.W., Smithson,W.A., Lindor,N.M. and Furuichi,Y. (1999) Mutations in RECQL4 cause a subset of cases of Rothmund–Thomson Syndrome. Nature Genet., 22, 82–84. [DOI] [PubMed] [Google Scholar]
- 4.van Brabant A.J., Stan,R. and Ellis,N.A. (2000) DNA helicases, genomic instability and human genetic disease. Annu. Rev. Genom. Hum. Genet., 1, 409–459. [DOI] [PubMed] [Google Scholar]
- 5.Puranam K.L. and Blackshear,P.J. (1994) Cloning and characterization of RECQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J. Biol. Chem., 269, 29838–29845. [PubMed] [Google Scholar]
- 6.Kitao S., Ohsugi,I., Ichikawa,K., Goto,M., Furuichi,Y. and Shimamoto,A. (1998) Cloning of the two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics, 54, 443–452. [DOI] [PubMed] [Google Scholar]
- 7.Sekelsky J.J., Brodsky,M.H. and Burtis,K.C. (2000) DNA repair in Drosophila: insights from the Drosophila genome sequence. J. Cell Biol., 150, F31–F36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohhata T., Araki,R., Fukuruma,R., Kuroiwa,A., Matsuda,Y. and Abe,M. (2001) Cloning, genomic structure and chromosomal localization of the gene encoding mouse DNA helicase RECQL5β. Gene, 280, 59–66. [DOI] [PubMed] [Google Scholar]
- 9.Sekelsky J.J., Brodsky,M.H., Rubin,G.M. and Hawley,R.S. (1999) Drosophila and human RecQ5 exist in different isoforms generated by alternative splicing. Nucleic Acids Res., 27, 3762–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimamoto A., Nishikawa,K., Kitao,S. and Furuichi,Y. (2000) Human RecQ5beta, a large isomer of RecQ5 DNA helicase, localizes in the nucleoplasm and interacts with topoisomerases 3alpha and 3beta. Nucleic Acids Res., 28, 1647–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Umezu K., Nakayama,K. and Nakayama,H. (1990) Escherichia coli RecQ protein is a DNA helicase. Proc. Natl Acad. Sci. USA, 87, 5363–5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umezu K. and Nakayama,H. (1993) RecQ DNA helicase of Escherichia coli. Characterization of the helix-unwinding activity with emphasis on the effect of single-stranded DNA-binding protein. J. Mol. Biol., 230, 1145–1150. [DOI] [PubMed] [Google Scholar]
- 13.Harmon F.G. and Kowalczykowski,S.C. (1998) RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev., 12, 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harmon F.G., DiGate,R.J. and Kowalczykowski,S.C. (1999) RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol. Cell, 3, 611–620. [DOI] [PubMed] [Google Scholar]
- 15.Harmon F.G. and Kowalczykowski,S.C. (2001) Biochemical characterization of the DNA helicase activity of the Escherichia coli RecQ helicase. J. Biol. Chem., 276, 232–243. [DOI] [PubMed] [Google Scholar]
- 16.Karow J.K., Chakraverty,R.K. and Hickson,I.D. (1997) The Bloom’s syndrome gene product is a 3′–5′ helicase. J. Biol. Chem., 272, 30611–30614. [DOI] [PubMed] [Google Scholar]
- 17.Karow J.K., Newman,R.H., Freemont,P.S. and Hickson,I.D. (1999) Oligomeric ring structure of the Bloom’s syndrome helicase. Curr. Biol., 9, 597–600. [DOI] [PubMed] [Google Scholar]
- 18.Karow J.K., Constantinou,A., Li,J.-L., West,S.C. and Hickson,I.D. (2000) The Bloom’s syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl Acad. Sci. USA, 97, 6504–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brosh R.M., Li,J.-L., Kenny,M.K., Karow,J.K., Cooper,M.P., Kureekattil,R.P., Hickson,I.D. and Bohr,V.A. (2000) Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activitiy. J. Biol. Chem., 275, 23500–23508. [DOI] [PubMed] [Google Scholar]
- 20.van Brabant A.J., Ye,T., Sanz,M., German,J.L.,III, Ellis,N.A. and Holloman,W.K. (2000) Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry, 39, 14617–14625. [DOI] [PubMed] [Google Scholar]
- 21.Mohaghegh P., Karow,J.K., Brosh,R.M.,Jr, Bohr,V.A. and Hickson,I.D. (2001) The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res., 29, 2843–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray M.D., Shen,J.-C., Kamath-Loeb,A.S., Blank,A., Sopher,B.L., Martin,G.M., Oshima,J. and Loeb,L.A. (1997) The Werner syndrome protein is a DNA helicase. Nature Genet., 17, 100–103. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki N., Shimamoto,A., Imamura,O., Kuromitsu,J., Kitao,S., Goto,M. and Furuichi,Y. (1997) DNA helicase activity in Werner’s syndrome gene product synthesized in a baculovirus system. Nucleic Acids Res., 25, 2973–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen J.-C., Gray,M.D., Oshima,J. and Loeb,L.A. (1998) Characterization of Werner syndrome protein DNA helicase activity: directionality, substrate dependence and stimulation by replication protein A. Nucleic Acids Res., 26, 2879–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fry M. and Loeb,L.A. (1999) Human Werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem., 274, 12797–12802. [DOI] [PubMed] [Google Scholar]
- 26.Constantinou A., Tarsounas,M., Karow,J.K., Brosh,R.M., Bohr,V.A., Hickson,I.D. and West,S.C. (2000) Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep., 1, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brosh R.M., Majumdar,A., Desai,S., Hickson,I.D., Bohr,V.A. and Seidman,M.M. (2001) Unwinding of a DNA triple helix by the Werner and Bloom syndrome helicases. J. Biol. Chem., 276, 3024–3030. [DOI] [PubMed] [Google Scholar]
- 28.Chakraverty R.K. and Hickson,I.D. (1999) Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. Bioessays, 21, 286–294. [DOI] [PubMed] [Google Scholar]
- 29.Karow J.K., Wu,L. and Hickson,I.D. (2000) RecQ family helicases: roles in cancer and aging. Curr. Opin. Genet. Dev., 10, 32–38. [DOI] [PubMed] [Google Scholar]
- 30.Brosh R.J.M., Waheed,J. and Sommers,J.A. (2002) Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J. Biol. Chem., 277, 23236–23245. [DOI] [PubMed] [Google Scholar]
- 31.Ozsoy A.Z., Sekelsky,J.J. and Matson,S.W. (2001) Biochemical characterization of the small isoform of Drosophila melanogaster RECQ5 helicase. Nucleic Acids Res., 29, 2986–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitby M.C. and Lloyd,R.G. (1998) Targeting Holliday junctions by the RecG branch migration protein of Escherichia coli. J. Biol. Chem., 273, 19729–19739. [DOI] [PubMed] [Google Scholar]
- 33.Nakagawa T. and Kolodner,R.D. (2002) The MER3 DNA helicase catalyzes the unwinding of Holliday junctions. J. Biol. Chem., 277, 28019–28024. [DOI] [PubMed] [Google Scholar]
- 34.McGlynn P. and Lloyd,R.G. (2001) Rescue of stalled replication forks by RecG: Simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc. Natl Acad. Sci. USA, 98, 8227–8234. [DOI] [PMC free article] [PubMed] [Google Scholar]