Abstract
The human SRC gene encodes pp60c–src, a non-receptor tyrosine kinase involved in numerous signaling pathways. Activation or overexpression of c-Src has also been linked to a number of important human cancers. Transcription of the SRC gene is complex and regulated by two closely linked but highly dissimilar promoters, each associated with its own distinct non-coding exon. In many tissues SRC expression is regulated by the housekeeping-like SRC1A promoter. In addition to other regulatory elements, three substantial polypurine:polypyrimidine (TC) tracts within this promoter are required for full transcriptional activity. Previously, we described an unusual factor called SRC pyrimidine-binding protein (SPy) that could bind to two of these TC tracts in their double-stranded form, but was also capable of interacting with higher affinity to all three pyrimidine tracts in their single-stranded form. Mutations in the TC tracts, which abolished the ability of SPy to interact with its double-stranded DNA target, significantly reduced SRC1A promoter activity, especially in concert with mutations in critical Sp1 binding sites. Here we expand upon our characterization of this interesting factor and describe the purification of SPy from human SW620 colon cancer cells using a DNA affinity-based approach. Subsequent in-gel tryptic digestion of purified SPy followed by MALDI-TOF mass spectrometric analysis identified SPy as heterogeneous nuclear ribonucleoprotein K (hnRNP K), a known nucleic-acid binding protein implicated in various aspects of gene expression including transcription. These data provide new insights into the double- and single-stranded DNA-binding specificity, as well as functional properties of hnRNP K, and suggest that hnRNP K is a critical component of SRC1A transcriptional processes.
INTRODUCTION
The human SRC proto-oncogene encodes the 60 kDa non-receptor tyrosine kinase pp60c–src, which has been implicated in many diverse cellular processes including differentiation, focal adhesion dynamics, regulation of cell–cell contact, bone remodeling, mitosis and others (1–4). Numerous findings throughout the past two decades have also pointed towards a role for SRC in the process of cellular transformation and cancer (2). For example, activation and/or overexpression of pp60c–src has been observed in many colon (5–8) as well as breast (9–12) carcinomas, and activating mutations in the SRC gene have been detected in a limited number of advanced colon tumors (13). Further evidence from our own laboratory suggests that in certain colon cancer cell lines, increased c-Src protein levels can be attributed to the transcriptional activation of the SRC gene (14). Therefore, to elucidate the mechanisms responsible for regulating SRC transcription we have been studying the regulatory motifs within the 5′ region of the SRC gene and have identified two promoters: SRC1α and SRC1A. The SRC1α promoter, located ∼1 kb upstream of the SRC1A promoter, is regulated by the liver-enriched hepatocyte nuclear factor 1 family of transcription factors, and transcripts originating from this promoter show a restricted tissue-specific expression pattern (15). Transcripts arising from the SRC1A promoter, on the other hand, show a ubiquitous expression pattern more typical of housekeeping genes. Consistent with this classification, the SRC1A promoter is GC rich and uses multiple transcription start sites; however, it also contains three perfect polypurine:polypyrimidine (TC) tracts, TC1, TC2 and TC3. We have previously shown that SRC1A promoter activity was dependent upon two critical Sp1 sites, GC1 and GA2, as well as the binding of a factor called SRC pyrimidine-binding protein (SPy) (16). SPy was originally described as a factor capable of binding specifically to TC1 and TC2 (but not TC3) in their double-stranded form, and to all of the TC tract pyrimidine sequences in single-stranded form. TC1 and TC2 were found to share a common motif (CTTCC), which was absent from TC3. Mutation of this sequence to CTTTC within either TC tract abolished SPy double-stranded binding and compromised SRC1A promoter activity; however, this mutation failed to inhibit SPy single-stranded pyrimidine binding (16). Therefore, it appeared that SPy had differential binding specificities for both double- and single-stranded DNA. In addition, the apparent affinity of SPy for single-stranded DNA was observed to be greater than for the double-stranded counterparts (16). Lastly, SPy was shown to regulate the SRC1A promoter cooperatively with the transcription factor Sp1 (16).
Given the highly unusual DNA-binding characteristics of SPy and its requirement for maximum SRC1A transcription, we decided to study this factor in greater detail and describe here its purification using a DNA-affinity based approach. Subsequent matches between experimental tryptic peptide masses determined by matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry and in silico tryptic digests derived from protein sequence databases identified SPy as heterogeneous nuclear ribonucleoprotein K (hnRNP K). This highly modular protein interacts with many biomolecules including RNA, single-, double- and triple-stranded DNA (17–19), as well as a number of important transcriptional regulators and signaling molecules (17,20). Interestingly, these include pp60c–src itself as well as other c-Src family members (21). The functions of hnRNP K, while generally poorly defined, appear to be numerous, and the protein has been implicated in multiple levels of gene expression including transcription, translation and the processing of RNA (22–25). Our data show that hnRNP K also plays an important role in the regulation of the SRC1A promoter and provides new insights into the DNA-binding specificity and biological activity of hnRNP K.
MATERIALS AND METHODS
Cells and media
SW480 and SW620 human colon cancer cells were obtained from the American Type Culture Collection and grown at 37°C, 5% CO2 in DMEM containing 10% FCS (Cansera) and 1% PenStrep (Life Technologies).
Oligonucleotides
All oligonucleotides were purchased from Life Technologies. Double-stranded TC1 and mutant TC1 oligonucleotides were generated by annealing with their complementary respective strands (Life Technologies). The TC1 and TC2 wild-type and double-stranded mutant sequences have been previously reported (16). Sequences of the TC1 and TC2 single-stranded mutant oligonucleotides used for electrophoretic mobility shift assays (EMSAs), site-directed mutagenesis and construction of the SPy DNA affinity column are as follows.
EMSA oligonucleotides. TC1ss mut: 5′CTTGCTGCTTG CTGCTCCTCCC; TC2ss mut: 5′CTCCCTCCCTCGTGTTG CCGTCCCC.
Site-directed mutagenesis oligonucleotides. TC1ss mutCT: 5′GTCCCCGCGCGCTTGCTGCTTGCTGCTCCTCCCGG CTGGCCTGCC; TC1ss mutGA: 5′GGCAGGCAGCCGGG AGGAGGAGCAAGCAGCAAGCGCGCGGGGAC; TC2ss mutCT: GGCCCGGGGACGGCAACACGAGGGAGGGA GCG; TC2ss mutGA: CGCTCCCTCCCTCGTGTTGCCG TCCCCGGGCC.
DNA affinity oligonucleotides. TC1 affinity CT: 5′GAT CCTTCCTCCTTCCTCCTCCTCCCGATCCTTCCTCCTT CCTCCTCCTCCC; TC1 affinity GA: 5′GATCGGGAGGA GGAGGAAGGAGGAAGGATCGGGAGGAGGAGGAAGGAGGAAG.
The DNA affinity sequences were then ligated to form concatenated double-stranded multimers and coupled directly to CNBr-activated Sepharose, as previously described (26).
Plasmids and reporter gene constructs
The human SRC promoter-CAT constructs p0.38SRC-CAT and the TC1, TC2 and TC1-TC2 double-stranded mutated forms of p0.38SRC-CAT have been previously described (16). Construction of p0.38SRC-CAT harboring single-stranded SPy-binding mutations in either TC1 or TC2 were generated by the QuickChange site-directed mutagenesis protocol (Stratagene) using either the TC1 or TC2 mutagenic sequences listed in the oligonucleotides section. To generate the construct harboring single-stranded mutations in both TC1 and TC2, p0.38SRC-CAT containing the TC1 single-stranded mutation was used as the template in a QuickChange reaction with the mutant TC2 oligonucleotides. All mutant plasmids generated in the PCR stage were then digested with NarI and SacII to isolate the promoter region, and the fragment ligated back into a wild-type p0.38SRC-CAT construct to ensure the absence of possible mutations elsewhere in the plasmid that could have occurred during the mutagenesis step.
Transient transfections and CAT assays
Transfection-grade supercoiled plasmids were prepared using an EndoFree Plasmid Maxi Kit (Qiagen). SW480 cells (3.75 × 105) in six-well plates were transfected with CAT expression vector (3.0 µg) and pCH110 β-galactosidase expression vector (1.0 µg) using Superfect (Qiagen) according to the manufacturer’s instructions. In the case where single-stranded oligonucleotides were added to titrate out SPy/hnRNP K protein, the oligonucleotides were added at various concentrations (0.5, 1.0, 1.5 and 2.0 µg) to the transfection reagent at the same time as the plasmid. Cells were harvested 48 h after transfection with lysis buffer provided with the CAT ELISA kit (Roche), and protein levels determined using Bradford Protein Reagent (Bio-Rad). β-Galactosidase levels were determined as previously described (27), and CAT protein levels quantified using the CAT ELISA kit (Roche). Reported CAT values were standardized to total protein and β-galactosidase expression and are representative of at least three duplicate experiments.
Preparation of nuclear extracts and EMSA
Nuclear extracts were prepared from human cells according to the method of Andrews and Faller (28). Typically, three 150 mm plates of confluent cells were scraped, washed three times in PBS, and suspended in 2 ml ice cold buffer A (10 mM HEPES–KOH pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT and 0.2 mM PMSF) on ice for 10 min. An aliquot of 25 µl 10% NP-40 was added and the cells vortexed. Lysis of the plasma membrane was monitored by nuclear staining with 0.4% trypan blue. The nuclei were pelleted by centrifugation (in a Sorvall 6000D at 1100 r.p.m.), the supernatants discarded and the pellets resuspended in 100–200 µl ice cold buffer C (20 mM HEPES–KOH pH 7.9, 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT and 0.2 mM PMSF) on ice for 20 min. The samples were transferred to microfuge tubes, centrifuged at 13 000 r.p.m. in a microfuge at 4°C for 5 min and the supernatants removed and stored at –80°C. For EMSAs, double-stranded probes were generated by annealing complementary single-stranded oligonucleotides and labeling with in-fill reactions using Klenow fragment and [α32P]dCTP. Single-stranded probes were end-labeled with [γ32P]ATP and T4 polynucleotide kinase. Typically, nuclear extracts were incubated with 50 000–100 000 c.p.m. of probe in binding buffer (25 mM HEPES pH 7.0, 4 mM Tris–HCl pH 8.0, 5 mM MgCl2, 1 mM CaCl2 and 1 mM DTT) and 3 µg poly(dI-dC). When competitor oligonucleotides or antibodies were included, they were added to the binding reaction and incubated on ice for 15 min before the addition of probe, followed by incubation at room temperature for 30 min. The anti-hnRNP K monoclonal was the kind gift of Dr Gideon Dreyfuss, University of Pennsylvania School of Medicine (29), while the anti-hnRNP K polyclonal was kindly provided by Dr Miau, Institute of Molecular Medicine, Dr H.L. Tsai Memorial Laboratory, College of Medicine, National Taiwan University (30). The Ets-1 antibody was purchased from Santa Cruz, Inc. The reactions were then resolved on 6% polyacrylamide gels in 1× Tris–glycine running buffer [50 mM Tris base, 380 mM glycine and 2 mM EDTA] at 150 V for ∼3 h at 4°C. Gels were dried and visualized either by autoradiography or by phosphorimage analysis using a Bio-Rad Molecular FX Imager.
Ultraviolet crosslinking analysis
An EMSA gel containing a shifted SPy-TC1 double-stranded radiolabeled oligonucleotide complex was exposed to UV radiation in a Stratagene Stratalinker (auto cross-link mode). The SPy-oligonucleotide complex was then excised from the EMSA gel and either physically inserted into the well of an SDS–PAGE gel (12%) and the complex resolved at 150 V for 1 h, or eluted from the gel fragment by overnight incubation at 37°C in elution buffer (20 mM Tris–HCl pH 8.5, 1 mM EDTA) followed by SDS–PAGE analysis as described above. When complete, the gel was dried and visualized by autoradiography using Kodak X-Omat film at –80°C overnight.
Western blotting
For western blot analysis, polypeptides from affinity fractions were separated by 12% SDS–PAGE and transferred to supported nitrocellulose using a wet transfer apparatus running at 50 V for 2 h. The membrane was probed with monoclonal anti-hnRNP K antibody 3C2 (29) and detected using PicoWest Enhanced Chemiluminescent substrate (Pierce) and Kodak X-Omat XB-1 film at room temperature.
Purification of SPy
Crude SW620 nuclear extracts were dialyzed in buffer Z (10 mM Tris–HCl pH 7.7, 1 mM EDTA, 0.01% NP-40, 10% glycerol, 1 mM DTT) and applied to an SP-Sepharose column (1.5 × 4.0 cm). After loading, the column was washed extensively with buffer Z, and SPy activity subsequently eluted with a step gradient of 0.4 and 0.8 M NaCl in buffer Z. Fractions of 2 ml were collected and assayed for SPy binding activity by EMSA. Active fractions were pooled, dialyzed in buffer Z and applied to a second SP-Sepharose column (1.5 × 4.0 cm). The column was washed with buffer Z and eluted using a linear gradient of 0–0.4 M NaCl in buffer Z. Fractions containing SPy activity (determined by EMSA) were pooled, dialyzed and applied to a DNA affinity column (1.5 × 4.0 cm) constructed by coupling concatenated double-stranded oligonucleotides containing SPy binding sites to CNBr activated Sepharose according to the method of Kadonaga and Tjian (26). The affinity column was washed and eluted using a linear gradient of 0–1 M KCl in buffer Z. Fractions were assayed for SPy activity by EMSA and the active SPy fraction concentrated by lyophilization, analyzed by SDS–PAGE, and stained with SYPRO Ruby-Red protein gel stain (Bio-Rad). The gel was visualized on a Bio-Rad Molecular FX Imager.
Trypsin digestion of gel-separated polypeptides
The 60 kDa polypeptide present in the SPy-active affinity fraction, along with background spots from the gel, were excised from SYPRO Ruby-Red stained SDS–PAGE gel, cut into 1 mm squares and placed into a 1.5 ml tube. Gel manipulation was performed in a laminar flow hood to minimize keratin contamination. A 50 µl aliquot of 25 mM ammonium bicarbonate in 1:1 (v/v) water:acetonitrile was added and the samples vortexed for 10 min. The solution was discarded and the previous step repeated twice. Samples were then dried for 30 min in a vacuum centrifuge followed by incubation with 50 µl freshly prepared 10 mM DTT in H2O for 60 min at 56°C in the dark. The DTT was removed and 50 µl of 55 mM iodoacetamide (IAA) in H2O was added followed by incubation for 45 min at 22°C in the dark with occasional vortexing. The IAA solution was removed and samples vortexed in 50 µl of 25 mM ammonium bicarbonate in H2O for 10 min. The IAA solution was removed and the previous two steps repeated twice. Gel pieces were dried for 30 min in a vacuum centrifuge and resuspended in 100 µl of 25 mM ammonium bicarbonate/5 mM calcium chloride containing 25 ng/µl modified porcine trypsin (sequencing grade; Promega). Samples were incubated at 4°C for 30 min, the trypsin solution removed and 100 µl of 25 mM ammonium bicarbonate added. The tubes were sealed with parafilm and incubated overnight at 37°C. The supernatant was removed and placed into a new tube (tube B). The gel pieces were resuspended in 100 µl of H2O and vortexed for 5 min, followed by sonication for 10 min. The supernatant was transferred to tube B and the gel fragments re-suspended in 50 µl of 5% trifluoroacetic acid (TFA) in 1:1 (v/v) water: acetonitrile for 15 min. The supernatant was transferred to tube B and the previous step repeated once more. The pooled supernatants in tube B were evaporated in a vacuum centrifuge and the tryptic peptides re-suspended in 10 µl of 0.1% TFA in H2O.
MALDI-TOF mass spectrometry
Tryptic peptides re-suspended in 0.1% TFA were desalted by solid-phase extraction using disposable pipette tips (ZipTipC18, Millipore) according to the manufacturer’s instructions. Peptides were eluted in 1–2 µl of 0.1% TFA/75% acetonitrile directly onto a MALDI target plate. An equal volume of matrix solution, consisting of 5 mg/ml α-cyano-4-hydroxycinnamic acid (Aldrich) in 0.1% TFA/50% acetonitrile, was applied to the sample and allowed to dry. A 1:1 mixture (31) of matrix and mass calibrant containing des-Arg Bradykinin and ACTH clip 18-39 was also placed beside the sample for close external calibration. Mass spectra of tryptic peptides were acquired on a Voyager-DE STR MALDI-TOF mass spectrometer (Applied Biosystems, Framingham, MA) operating in the positive ion, delayed extraction and reflectron modes. Experimental conditions were as follows: accelerating voltage 20 000 V; grid voltage 72.5%; guide wire voltage 0.001%; delay 100 ns; laser power 2386. Spectra were obtained by combining 400 scans and applying mass calibration, baseline correction, noise filtering and de-isotoping procedures using the spectrometer software (Data Explorer v. 4.0, Applied Biosystems). Monoisotopic peptide masses were submitted to Protein Prospector (http://prospector.ucsf.edu/) for comparison with entries in the National Center for Biotechnology Information (NCBI) sequence database, using the MS Fit program.
RESULTS
UV cross-linking analysis suggests that SPy is a single polypeptide of ∼50–58 kDa
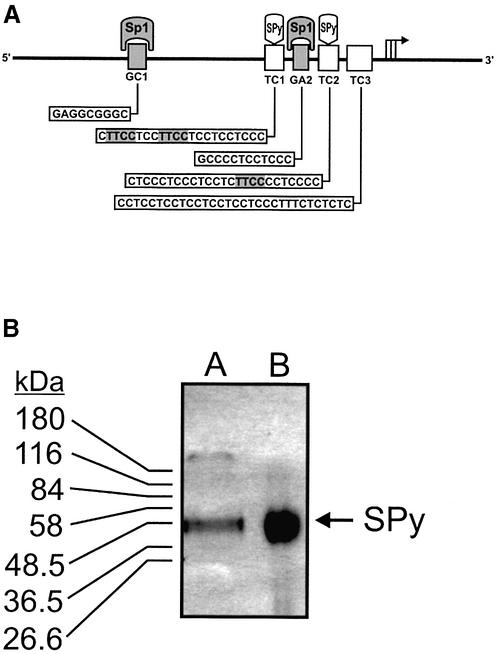
SRC1A expression is dependent upon several cis elements including the GC1 and GA2 sites, which interact with the Sp1 family; and three polypurine:polypyrimidine sites, TC1-3, which interact with SPy (Fig. 1A). SPy had been previously shown to produce a characteristically broad and fairly rapidly migrating complex with radiolabeled double-stranded TC1 and TC2 oligonucleotides in EMSAs (16). To further characterize the nature of this SPy-DNA complex we carried out ultra-violet cross-linking of an EMSA gel containing a SPy-double-stranded 32P-labeled TC1 complex. A gel fragment encompassing the radioactive cross-linked complex was then excised from the EMSA gel and either physically inserted into the well of an SDS–PAGE gel, or eluted out of the gel fragment and resolved by SDS–PAGE. As shown in Figure 1B, both elution (lane A) and excision (lane B) of the cross-linked complex from the EMSA gel generated a single polypeptide species migrating between 50 and 58 kDa on SDS–PAGE.
Figure 1.
Ultraviolet cross-linking analysis of a SPy-TC1 double-stranded oligonucleotide complex. (A) The SRC1A promoter region contains two critical Sp-family member DNA-binding sites, GC1 and GA2, as well as three TC tracts, TC1, TC2 and TC3. SPy double-stranded binding sites located within TC1 and TC2 are shaded gray. The major transcription start sites are located just downstream of TC3 and are marked by an arrow. (B) An EMSA was first performed by incubating wild-type TC1 double-stranded radiolabeled oligonucleotides with SW620 nuclear extract followed by exposure of the gel to UV light. The shifted species was isolated by excision, and the radiolabeled SPy-DNA complex either eluted out of the excised gel slice and subjected to analysis by SDS–PAGE (lane A), or electrophoresed out of the gel slice following insertion into the well of an SDS–polyacrylamide gel (lane B). The gels were then visualized by autoradiography.
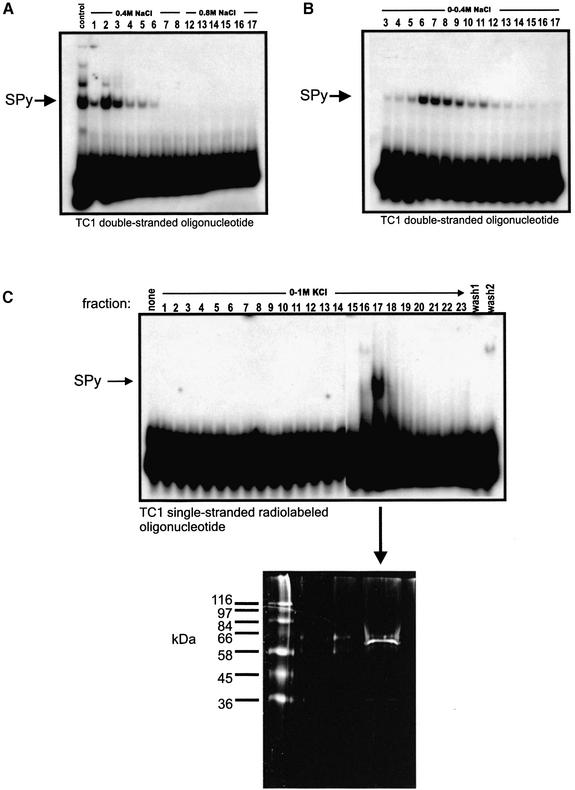
Purification of SPy by ion-exchange and DNA affinity chromatography
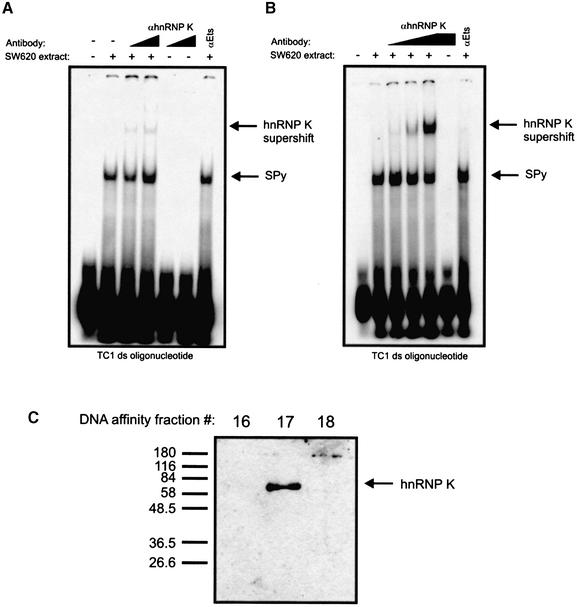
The purification of SPy comprised a combination of ion-exchange and DNA affinity chromatography. Initially, SW620 nuclear extracts were prepared and purified by ion-exchange on an SP-Sepharose column (see Materials and Methods), which was eluted in a step-wise manner with 0.4 and 0.8 M NaCl. Fractions containing SPy protein were identified by EMSAs using radiolabeled double-stranded TC1 oligonucleotides. The purpose of the Sepharose chromatography stage was to obtain SPy protein devoid of factors such as nucleases that could interfere later with the DNA affinity column. As shown in Figure 2A, SPy-binding activity was present within fractions of the 0.4 M eluate. Fractions 1–5 were pooled, dialyzed and reapplied to another SP-Sepharose column that was eluted with a linear NaCl gradient from 0–0.4 M NaCl. ‘Active’ SPy fractions (Fig. 2B) were identified by EMSAs as above, pooled and dialyzed in preparation for the DNA affinity column. Fractions from the second ion-exchange column were also analyzed by EMSA using a radiolabeled TC1-based double-stranded mutant oligonucleotide (see Fig. 6 for sequence), which failed to show any shifted species (data not shown). This was consistent with our previous observations that the TC1 double-stranded mutant probe was unable to bind SPy (16), suggesting that the complex observed in Figure 2B was indeed SPy. The DNA affinity column was constructed as described in Materials and Methods, and the pooled SPy-containing fractions from the second ion-exchange column loaded. The DNA affinity column was washed extensively, eluted with a linear gradient of 0–1.0 M KCl and fractions assayed for SPy activity by EMSA (Fig. 2C). Since the affinity column was constructed with double-stranded TC1 oligonucleotides, assaying the eluted fractions with a radiolabeled double-stranded TC1 oligonucleotide would likely have been unsuccessful, particularly with fractions containing high salt concentrations. Therefore, to circumvent these potential detection problems, the fractions were assayed using radiolabeled TC1 pyrimidine single-stranded oligonucleotides, which resulted in a single fraction (no. 17) showing SPy-binding (Fig. 2C). Fraction 17 was then concentrated by lyophilization and analyzed by SDS–PAGE, which revealed the presence of one prominent polypeptide with an apparent molecular weight of ∼60 kDa (Fig. 2C, bottom panel).
Figure 2.
EMSA analysis of ion-exchange and DNA-affinity chromatography fractions. (A) EMSA analysis of fractions from first round step-wise elution of ion-exchange column using a TC1 double-stranded radiolabeled oligonucleotide. (B) EMSA of fractions from second round linear elution ion-exchange column using a TC1 double-stranded radiolabeled oligonucleotide. Fraction numbers from each column are shown along the tops of each panel, including the salt concentration gradients used for each elution. (C) EMSA analysis of fractions eluted from the SPy DNA affinity column using a radiolabeled TC1 single-stranded pyrimidine oligonucleotide. DNA affinity fraction 17 was resolved by SDS–PAGE and fluorescently stained with SYPRO Ruby-Red.
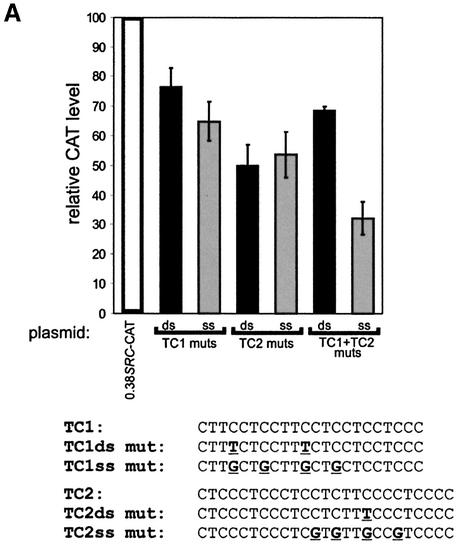
Figure 6.
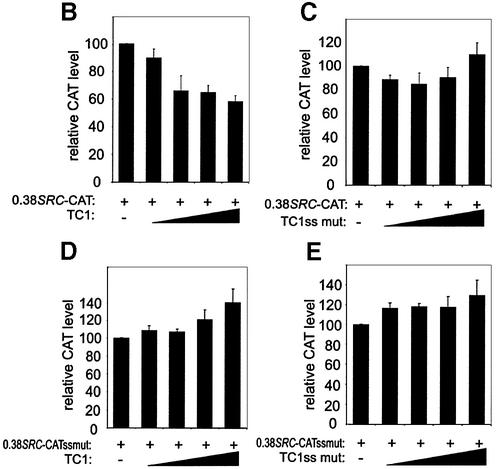
Effect of SPy/hnRNP K DNA-binding mutations and titration of SPy/hnRNP K protein on SRC1A promoter activity. (A) Mutations that abolish SPy single-stranded binding (i.e. four incorporated purines within either the pyrimidine strand of TC1, TC2 or both together) were introduced into 0.38SRC-CAT and reporter levels assayed by transient transfection in SW480 cells. Solid black bars represent SPy double-stranded mutations; gray bars represent SPy single-stranded mutations. CAT levels are expressed relative to wild-type 0.38SRC-CAT (open bar). Sequences of the mutations are shown below. (B) Single-stranded oligonucleotides capable of SPy/hnRNP K binding (TC1 CT) as well as (C), the mutant form, incapable of SPy/hnRNP K binding (TC1ss mut CT) were co-transfected with 0.38SRC-CAT into SW480 cells. 0.38SRC-CAT harboring TC1+TC2ss mutations was cotransfected with both TC1 CT (D) and TC1ss mut CT (E), single-stranded oligonucleotides into SW480 cells. The black triangle represents increasing concentrations (see Materials and Methods) of the competitor oligonucleotide in the presence of consistent quantities of reporter plasmid. CAT levels were standardized to total protein and β-galactosidase expression, and are the average of three duplicate experiments.
Identification of SPy by MALDI-TOF mass spectrometry
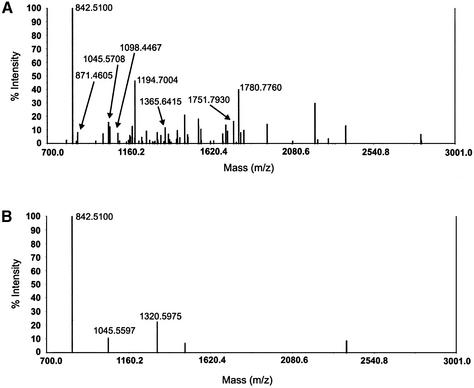
To reveal the identity of SPy, in-gel tryptic digestion was performed followed by analysis of the tryptic peptides by MALDI-TOF mass spectrometry. The 60 kDa polypeptide was excised from the Ruby-Red-stained SDS–PAGE gel (Fig. 2C) along with several protein-free gel pieces as background controls. The gel fragments were then digested with trypsin and their masses determined by MALDI-TOF mass spectrometry. As shown in Figure 3A, the gel slice containing the 60 kDa SPy polypeptide had a spectrum containing many more peptides than the background spectrum, which showed peaks for the trypsin enzyme only (Fig. 3B). The resulting experimental peptide masses generated from the gel slice containing the 60 kDa polypeptide were then searched against predicted tryptic peptide masses for all the proteins contained within the NCBI protein database, using the MS-Fit sequence database search program available at the ProteinProspector website (http://prospector.ucsf.edu). As shown in Figure 4A and B, 14 (out of 32 submitted) masses matched precisely to the Homo sapiens pyrimidine binding protein hnRNP K. The correlation of the matches was significant, with a maximum difference in mass of –21.2 p.p.m. between the submitted and matched peptides, with a majority of the differences within 10 p.p.m. Submitted peptide masses that fall within 50 p.p.m. of the matched masses are considered significant, and protein identification unambiguous, if >15% of the matched protein sequence is covered by these peptides (32). The locations of the experimentally derived tryptic peptides according to the amino acid sequence of hnRNP K are shown in Figure 4B.
Figure 3.
MALDI-TOF mass spectrometric analysis of SPy tryptic peptides. The polypeptide present in affinity fraction 17 was resolved by SDS–PAGE, stained with Ruby-Red, excised from the gel and subject to tryptic digestion. MALDI-TOF mass spectrometric analysis of the subsequent tryptic peptides produced the spectrum shown in (A). The spectrum in (B) represents the masses of tryptic peptides present in a background piece from the SDS–PAGE.
Figure 4.
ProteinProspector search results of SPy tryptic peptide masses. The peptide masses resulting from MALDI-TOF analysis of SPy tryptic peptides were searched using the ProteinProspector website against the masses of virtually derived tryptic peptides present in the NCBI protein database. Fourteen of 32 submitted peptide masses (A) were found to match precisely with virtually derived tryptic peptide masses of hnRNP K. The table shows submitted and matched masses of the corresponding peptides, including the difference in mass between the two (delta p.p.m.). Matches to within 50 p.p.m. are considered significant (32). (B) Amino acid sequence of hnRNP K showing the regions that matched to SPy tryptic peptides (bold and underlined). Numbered regions correspond to the locations within hnRNP K of identified tryptic peptides from (A). These peptides cover 32% of the matched protein sequence, providing unambiguous identification.
EMSA and western analysis confirm SPy identity as hnRNP K
The precise match between SPy tryptic peptides and those of hnRNP K suggested with a high probability that SPy was hnRNP K; however, we proceeded to confirm our identification using hnRNP K antibodies in EMSAs and western blot analyses. EMSAs using both polyclonal and monoclonal anti-hnRNP K antibodies produced supershifted SPy complexes (Fig. 5A, polyclonal and B, monoclonal). The EMSA in Figure 5A was performed with an hnRNP K polyclonal antibody (30) and produced a faint supershifted species, while the hnRNP K monoclonal antibody 3C2 (29), as shown in Figure 5B, produced a robust supershifted species and an associated reduction in intensity of the SPy complex. Furthermore, western blot analysis of fractions 16–18 of the DNA affinity purification using the anti-hnRNP K monoclonal antibody (3C2) showed immunoreactivity with a 60 kDa species in fraction 17 only. This finding was consistent with the identification of SPy as hnRNP K as revealed by the mass spectrometry data, and strongly suggested that the factor previously referred to as SPy was indeed hnRNP K.
Figure 5.
Antibodies specific for hnRNP K recognize SPy. EMSA analysis of a SPy-TC1 double-stranded DNA complex in the presence of increasing amounts of anti-hnRNP K (A) polyclonal or (B) monoclonal 3C2 antibody. Addition of Ets antibody to the EMSA reactions in both (A) and (B) is shown as a control. (C) Western analysis of DNA affinity fractions 16–18 (of Fig. 2C) using anti-hnRNP K monoclonal antibody 3C2.
SRC1A promoter activity is dependent on hnRNP K binding
Previously we reported that SPy/hnRNP K bound specifically to double-stranded polypurine:polypyrimidine sequences located within the SRC1A promoter region (Fig. 1A) (16). Point mutations in two of these tracts, TC1 and TC2, abolished the ability of SPy/hnRNP K to bind its double-stranded target and resulted in reduced promoter activity, especially in concert with mutations in critical Sp1 binding sites (16). However, our work also showed that even these mutated pyrimidine sequences, when in single-stranded form, were still able to bind SPy/hnRNP K with a high affinity. Thus, some of the observed residual promoter activity might have been the result of such single-stranded binding. Because of this relaxed SPy/hnRNP K single-stranded binding specificity, we hypothesized that more drastic mutations disrupting the pyrimidine content of the single-stranded targets would be required to completely prevent SPy/hnRNP K single-stranded binding. Single-stranded pyrimidine oligonucleotides, based on the TC1 and TC2 sequences and harboring four pyrimidine to purine substitutions (Fig. 6A), were subsequently designed and shown by EMSA to completely abolish SPy/hnRNP K’s single- and double-stranded binding ability (data not shown). These TC1 and TC2 single-stranded SPy/hnRNP K-binding mutations were then introduced both individually and collectively into the 0.38SRC-CAT reporter construct by site-directed mutagenesis. Surprisingly, when assayed by transient transfection into SW480 cells, the constructs containing individual mutations abolishing SPy/hnRNP K single-stranded binding at either TC1 or TC2 resulted in CAT levels similar to those observed with the previously reported individual double-stranded mutations (Fig. 6A). However, there was a significant difference observed between the single-stranded SPy/hnRNP K mutations in TC1 and TC2 together compared to the TC1 and TC2 double-stranded mutations together. The TC1-TC2 single-stranded mutations together reduced CAT levels to 30% of wild-type while the TC1-TC2 double-stranded mutant reduced CAT levels to just <70% (Fig. 6A). These findings suggest that ultimately it may be the ability of SPy/hnRNP K to bind these TC tracts in single-stranded form that is critical for maximum SRC1A promoter activity.
To further examine the effects of SPy/hnRNP K’s interaction with the SRC1A promoter, we co-transfected the wild-type 0.38SRC-CAT vector with increasing concentrations of wild-type TC1 single-stranded oligonucleotides. The objective here was to titrate out endogenous SPy/hnRNP K, which is present at relatively high levels in SW480 cells. As shown in Figure 6B, addition of wild-type TC1 oligonucleotides reduced CAT levels in a dose-dependent manner, by ∼50%. However, the addition of TC1ss mut oligonucleotide, which is unable to bind SPy/hnRNP K in vitro (sequence shown in Fig. 6A), showed no effect on 0.38SRC-CAT reporter levels, even at the highest concentration (Fig. 6C). Lastly, we repeated these experiments using the 0.38SRC-CAT vector containing both TC1 and TC2 single-stranded mutations. In this case, co-transfection of either wild-type or mutated versions of the TC1 oligonucleotide had no effect (Fig. 6D and E, respectively). These results demonstrate that the in vitro binding properties of SPy/hnRNP K correlate strongly with the activity of the SRC1A promoter in vivo. In conjunction with the mutational analysis we conclude that SPy/hnRNP K is integrally involved in regulating SRC1A promoter activity.
DISCUSSION
In this paper we further dissect the DNA binding characteristics of SPy and its effect on SRC1A promoter activity. Using a combination of DNA affinity chromatography and MALDI-TOF mass spectrometry we also report the identification of this factor as hnRNP K. The relatively rapid migration of the SPy complex on EMSA gels, coupled with our UV-crosslinking experiments and subsequent purification of SPy as a single 60 kDa protein, were entirely consistent with the hypothesis that the SPy complex observed on EMSA gels represented a single protein species. This notion was further strengthened by the robust supershift generated in EMSA gels with the hnRNP K monoclonal antibody, which also confirmed the identity of SPy as hnRNP K. Interestingly, while the calculated mass of hnRNP K is 50–51 kDa, it is known to migrate as a 60–65 kDa species on SDS–polyacrylamide gels (17).
hnRNP K was first identified as one of more than 20 proteins found in ribonucleoprotein particles where it binds RNA (19). Its highly modular structure allows a number of diverse interactions with an unusually large range of molecules. This includes single-, double- and triple-stranded DNA as well as various proteins including transcriptional regulators such as TATA binding protein (TBP) (23) and Zik-1 (33), inducible serine threonine kinases (34), the oncoprotein Vav (20) and members of the c-Src family of tyrosine kinases (35). This has led to proposals that hnRNP K acts as some form of platform or scaffold to facilitate diverse molecular interactions (35,36). Mirroring this diversity, hnRNP K to date has been directly implicated in numerous cellular processes including transcription, mRNA transport, splicing and translational regulation (22–25). In addition to its many partners and roles, hnRNP K is also modified by a number of kinases, including casein kinase II (37), PKC (38) and tyrosine kinases such as c-Src and Lck (21,39). While the effect of such phosphorylations is not well understood, they have been shown to modulate DNA and RNA binding ability (36).
hnRNP K has been implicated in the transcriptional regulation of several genes (23,24,40). The work described here shows that SPy/hnRNP K interacts in vitro with functionally important SRC1A promoter elements and therefore likely regulates SRC expression in vivo too. Our reasons for this conclusion can be summarized as follows. (i) Single point mutations within either TC1 or TC2, which disrupted the ability of hnRNP K to bind double-stranded targets in vitro, affected promoter activity in vivo. (ii) More drastic mutations that destroyed the ability of SPy/hnRNP K to bind single-stranded targets in vitro had an even more pronounced effect in vivo. (iii) Titration of endogenous hnRNP K with single-stranded oligonucleotides shown to bind SPy/hnRNP K strongly in vitro reduced SRC1A promoter activity in vivo, while mutated versions of these oligonucleotides had no effect.
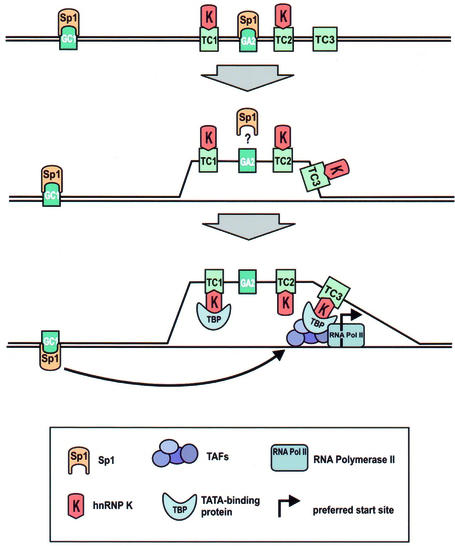
What then might be the role of hnRNP K in SRC1A regulation? Previous studies have shown that hnRNP K can activate the c-myc promoter through interactions with a similar polypurine:polypyrimidine tract (29). Michelotti et al. (23) reported that hnRNP K can interact with a CT element in vitro and induce a single-stranded conformation when present in a supercoiled plasmid. They proposed a model whereby hnRNP K could induce a single-stranded ‘bubble’ in the c-myc promoter by virtue of its high affinity for polypyrimidine sequences, thereby facilitating the entry of other components of the basal transcriptional machinery. Our previous characterization of SPy/hnRNP K DNA-binding specificity leads us to propose a similar model for SRC (Fig. 7). However, we have shown for the first time that SPy/hnRNP K actually has a distinct double-stranded DNA sequence binding requirement, namely the CTTCC motif present in TC1 and TC2. We therefore propose that this could be an important factor determining promoter selectivity by SPy/hnRNP K, which would be dependent on the presence of this motif. Interestingly, the c-myc CT element previously mentioned contains exactly such a motif (TCCTCCCCACCTTCC CCACCCTCCCCACCCTCCCCA). In addition, hnRNP K has been shown to interact with a similar sequence (ACC CTCCCCTCCCCTGTAACTCCACCCCTTCCCCA) found in the neuronal nicotinic acetylcholine receptor promoter, which also contains the same motif (41).
Figure 7.
Possible model for SRC1A transcriptional regulation by Sp1 and hnRNP K. Our data suggest a model by which hnRNP K could recognize and bind specifically to double-stranded sequences within TC1 and TC2 followed by strand separation that would be facilitated by the increased affinity of hnRNP K for single-stranded DNA. The resulting single-stranded ‘bubble’ could encompass the entire TC-tract region or more discrete regions therein. For example, the ability of hnRNP K to bind TBP could recruit TFIID to the TC3 region and aid in the assembly of a pre-initiation complex. Transactivation would be accomplished by Sp1, most notably in this model, from binding to the GC1 site. Such a structure would be in agreement with the location of the preferred transcription start sites, which are located ∼20 bp downstream of TC3 and would also account for other minor sites, which are located throughout the TC-tract region.
Thus, we propose that following the binding of SPy/ hnRNP K to its double-stranded target, melting of the SRC1A promoter region would be facilitated by the higher affinity of SPy/hnRNP K for the single-stranded pyrimidine strands. SPy/hnRNP K could then recruit other elements of the basal transcriptional machinery, namely TFIID, through the known interaction between hnRNP K and TBP (23). Our mutation and transfection experiments (Fig. 6) also support such a model, since to significantly disrupt SRC1A promoter activity, the ability of SPy/hnRNP K to bind to both TC1 and TC2 in single-stranded form must be destroyed. However, it should be noted that these constructs still contain an intact TC3 tract, which is also capable of binding SPy/hnRNP K with high affinity in single-stranded form. We are currently assessing the contribution of TC3 to the remaining promoter activity.
It is interesting to note that despite the long-term observation that hnRNP K has remarkable nucleic acid binding abilities there has not been a systematic study to examine DNA-binding specificity. This is especially true of the double-stranded DNA-binding ability of hnRNP K. Indeed, where binding studies have been carried out the source of hnRNP K tends to be recombinant hnRNP K isolated from prokaryotic sources. Given the results of our studies and the likelihood that phosphorylation could influence binding ability, we feel such binding studies are a priority and should help identify other promoters potentially regulated by hnRNP K. Polypurine:polypyrimidine tracts are not uncommon in eukaryotic promoter regions and the models described here could represent a method by which hnRNP K could target the basal transcriptional machinery to specific promoters, especially ones with high GC content which often lack TATA boxes.
In summary, we have identified the SRC pyrimidine-binding protein, SPy, as hnRNP K and suggest that it plays an important role in the regulation of SRC transcription. Our previous studies showing that SPy/hnRNP K has distinct double-stranded binding requirements also suggest a mechanism by which hnRNP K could selectively regulate specific promoters. Lastly, we are particularly intrigued by the frequent observations that hnRNP K and pp60c–src interact, including a recent report showing that such interactions can lead to pp60c–src activation and hnRNP K tyrosine phosphorylation (39). While no evidence yet exists, it is tempting to speculate that these interactions may be a sign that some positive or negative feedback regulation of SRC transcription exists.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by operating grants from the Canadian Institutes of Health Research (CIHR) as well as the Saskatchewan Regional Partnership Program.
REFERENCES
- 1.Abram C.L. and Courtneidge,S.A. (2000) Src family tyrosine kinases and growth factor signaling. Exp. Cell Res., 254, 1–13. [DOI] [PubMed] [Google Scholar]
- 2.Biscardi J.S., Tice,D.A. and Parsons,S.J. (1999) c-Src, receptor tyrosine kinases and human cancer. Adv. Cancer Res., 76, 61–119. [DOI] [PubMed] [Google Scholar]
- 3.Brown M.T. and Cooper,J.A. (1996) Regulation, substrates and functions of src. Biochim. Biophys. Acta, 1287, 121–149. [DOI] [PubMed] [Google Scholar]
- 4.Thomas S.M. and Brugge,J.S. (1997) Cellular functions regulated by Src family kinases. Annu. Rev. Cell Dev. Biol., 13, 513–609. [DOI] [PubMed] [Google Scholar]
- 5.Bolen J.B., Veillette,A., Schwartz,A.M., DeSeau,V. and Rosen,N. (1987) Activation of pp60c-src protein kinase activity in human colon carcinoma. Proc. Natl Acad. Sci. USA, 84, 2251–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cartwright C.A., Coad,C.A. and Egbert,B.M. (1994) Elevated c-Src tyrosine kinase activity in premalignant epithelia of ulcerative colitis. J. Clin. Invest., 93, 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cartwright C.A., Kamps,M.P., Meisler,A.I., Pipas,J.M. and Eckhart,W. (1989) pp60c-src activation in human colon carcinoma. J. Clin. Invest., 83, 2025–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Talamonti M.S., Roh,M.S., Curley,S.A. and Gallick,G.E. (1993) Increase in activity and level of pp60c-src in progressive stages of human colorectal cancer. J. Clin. Invest., 91, 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobs C. and Rubsamen,H. (1983) Expression of pp60c-src protein kinase in adult and fetal human tissue: high activities in some sarcomas and mammary carcinomas. Cancer Res., 43, 1696–1702. [PubMed] [Google Scholar]
- 10.Luttrell D.K., Lee,A., Lansing,T.J., Crosby,R.M., Jung,K.D., Willard,D., Luther,M., Rodriguez,M., Berman,J. and Gilmer,T.M. (1994) Involvement of pp60c-src with two major signaling pathways in human breast cancer. Proc. Natl Acad. Sci. USA, 91, 83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ottenhoff-Kalff A.E., Rijksen,G., van Beurden,E.A., Hennipman,A., Michels,A.A. and Staal,G.E. (1992) Characterization of protein tyrosine kinases from human breast cancer: involvement of the c-src oncogene product. Cancer Res., 52, 4773–4778. [PubMed] [Google Scholar]
- 12.Verbeek B.S., Vroom,T.M., Adriaansen-Slot,S.S., Ottenhoff-Kalff,A.E., Geertzema,J.G., Hennipman,A. and Rijksen,G. (1996) c-Src protein expression is increased in human breast cancer. An immunohistochemical and biochemical analysis. J. Pathol., 180, 383–388. [DOI] [PubMed] [Google Scholar]
- 13.Irby R.B., Mao,W., Coppola,D., Kang,J., Loubeau,J.M., Trudeau,W., Karl,R., Fujita,D.J., Jove,R. and Yeatman,T.J. (1999) Activating SRC mutation in a subset of advanced human colon cancers. Nature Genet., 21, 187–190. [DOI] [PubMed] [Google Scholar]
- 14.Dehm S., Senger,M.A. and Bonham,K. (2001) SRC transcriptional activation in a subset of human colon cancer cell lines. FEBS Lett., 487, 367–371. [DOI] [PubMed] [Google Scholar]
- 15.Bonham K., Ritchie,S.A., Dehm,S.M., Snyder,K. and Boyd,F.M. (2000) An alternative, human SRC promoter and its regulation by hepatic nuclear factor-1α. J. Biol. Chem., 275, 37604–37611. [DOI] [PubMed] [Google Scholar]
- 16.Ritchie S., Boyd,F.M., Wong,J. and Bonham,K. (2000) Transcription of the human c-Src promoter is dependent on Sp1, a novel pyrimidine binding factor SPy, and can be inhibited by triplex-forming oligonucleotides. J. Biol. Chem., 275, 847–854. [DOI] [PubMed] [Google Scholar]
- 17.Bomsztyk K., Van Seuningen,I., Suzuki,H., Denisenko,O. and Ostrowski,J. (1997) Diverse molecular interactions of the hnRNP K protein. FEBS Lett., 403, 113–115. [DOI] [PubMed] [Google Scholar]
- 18.Guillonneau F., Guieysse,A.L., Le Caer,J.P., Rossier,J. and Praseuth,D. (2001) Selection and identification of proteins bound to DNA triple-helical structures by combination of 2D-electrophoresis and MALDI-TOF mass spectrometry. Nucleic Acids Res., 29, 2427–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krecic A.M. and Swanson,M.S. (1999) hnRNP complexes: composition, structure and function. Curr. Opin. Cell Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- 20.Hobert O., Jallal,B., Schlessinger,J. and Ullrich,A. (1994) Novel signaling pathway suggested by SH3 domain-mediated p95vav/heterogeneous ribonucleoprotein K interaction. J. Biol. Chem., 269, 20225–20228. [PubMed] [Google Scholar]
- 21.Van Seuningen I., Ostrowski,J., Bustelo,X.R., Sleath,P.R. and Bomsztyk,K. (1995) The K protein domain that recruits the interleukin 1-responsive K protein kinase lies adjacent to a cluster of c-Src and Vav SH3-binding sites. Implications that K protein acts as a docking platform. J. Biol. Chem., 270, 26976–26985. [DOI] [PubMed] [Google Scholar]
- 22.Gorlach M., Wittekind,M., Beckman,R.A., Mueller,L. and Dreyfuss,G. (1992) Interaction of the RNA-binding domain of the hnRNP C proteins with RNA. EMBO J., 11, 3289–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michelotti E.F., Michelotti,G.A., Aronsohn,A.I. and Levens,D. (1996) Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell. Biol., 16, 2350–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomonaga T. and Levens,D. (1995) Heterogeneous nuclear ribonucleoprotein K is a DNA-binding transactivator. J. Biol. Chem., 270, 4875–4881. [DOI] [PubMed] [Google Scholar]
- 25.Ostareck D.H., Ostareck-Lederer,A., Wilm,M., Thiele,B.J., Mann,M. and Hentze,M.W. (1997) mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell, 89, 597–606. [DOI] [PubMed] [Google Scholar]
- 26.Kadonaga J.T. and Tjian,R. (1986) Affinity purification of sequence-specific DNA binding proteins. Proc. Natl Acad. Sci. USA, 83, 5889–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall C.V., Jacob,P.E., Ringold,G.M. and Lee,F. (1983) Expression and regulation of Escherichia coli lacZ gene fusions in mammalian cells. J. Mol. Appl. Genet., 2, 101–109. [PubMed] [Google Scholar]
- 28.Andrews N.C. and Faller,D.V. (1991) A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res., 19, 2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takimoto M., Tomonaga,T., Matunis,M., Avigan,M., Krutzsch,H., Dreyfuss,G. and Levens,D. (1993) Specific binding of heterogeneous ribonucleoprotein particle protein K to the human c-myc promoter, in vitro. J. Biol. Chem., 268, 18249–18258. [PubMed] [Google Scholar]
- 30.Miau L.H., Chang,C.J., Shen,B.J., Tsai,W.H. and Lee,S.C. (1998) Identification of heterogeneous nuclear ribonucleoprotein K (hnRNP K) as a repressor of C/EBPβ-mediated gene activation. J. Biol. Chem., 273, 10784–10791. [DOI] [PubMed] [Google Scholar]
- 31.Bouziane M., Cherny,D.I., Mouscadet,J.F. and Auclair,C. (1996) Alternate strand DNA triple helix-mediated inhibition of HIV-1 U5 long terminal repeat integration in vitro. J. Biol. Chem., 271, 10359–10364. [DOI] [PubMed] [Google Scholar]
- 32.Jensen O.N., Wilm,M., Shevchenko,A. and Mann,M. (1999) Sample preparation methods for mass spectrometric peptide mapping directly from 2-DE gels. Methods Mol. Biol., 112, 513–530. [DOI] [PubMed] [Google Scholar]
- 33.Denisenko O.N., O’Neill,B., Ostrowski,J., Van Seuningen,I. and Bomsztyk,K. (1996) Zik1, a transcriptional repressor that interacts with the heterogeneous nuclear ribonucleoprotein particle K protein. J. Biol. Chem., 271, 27701–27706. [DOI] [PubMed] [Google Scholar]
- 34.Van Seuningen I., Ostrowski,J. and Bomsztyk,K. (1995) Description of an IL-1-responsive kinase that phosphorylates the K protein. Enhancement of phosphorylation by selective DNA and RNA motifs. Biochemistry, 34, 5644–5650. [DOI] [PubMed] [Google Scholar]
- 35.Ostrowski J., Schullery,D.S., Denisenko,O.N., Higaki,Y., Watts,J., Aebersold,R., Stempka,L., Gschwendt,M. and Bomsztyk,K. (2000) Role of tyrosine phosphorylation in the regulation of the interaction of heterogenous nuclear ribonucleoprotein K protein with its protein and RNA partners. J. Biol. Chem., 275, 3619–3628. [DOI] [PubMed] [Google Scholar]
- 36.Ostrowski J., Kawata,Y., Schullery,D.S., Denisenko,O.N., Higaki,Y., Abrass,C.K. and Bomsztyk,K. (2001) Insulin alters heterogeneous nuclear ribonucleoprotein K protein binding to DNA and RNA. Proc. Natl Acad. Sci. USA, 98, 9044–9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wadd S., Bryant,H., Filhol,O., Scott,J.E., Hsieh,T.Y., Everett,R.D. and Clements,J.B. (1999) The multifunctional herpes simplex virus IE63 protein interacts with heterogeneous ribonucleoprotein K and with casein kinase 2. J. Biol. Chem., 274, 28991–28998. [DOI] [PubMed] [Google Scholar]
- 38.Schullery D.S., Ostrowski,J., Denisenko,O.N., Stempka,L., Shnyreva,M., Suzuki,H., Gschwendt,M. and Bomsztyk,K. (1999) Regulated interaction of protein kinase Cdelta with the heterogeneous nuclear ribonucleoprotein K protein. J. Biol. Chem., 274, 15101–15109. [DOI] [PubMed] [Google Scholar]
- 39.Ostareck-Lederer A., Ostareck,D.H., Cans,C., Neubauer,G., Bomsztyk,K., Superti-Furga,G. and Hentze,M.W. (2002) c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol. Cell. Biol., 22, 4535–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomonaga T. and Levens,D. (1996) Activating transcription from single stranded DNA. Proc. Natl Acad. Sci. USA, 93, 5830–5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du Q., Melnikova,I.N. and Gardner,P.D. (1998) Differential effects of heterogeneous nuclear ribonucleoprotein K on Sp1- and Sp3-mediated transcriptional activation of a neuronal nicotinic acetylcholine receptor promoter. J. Biol. Chem., 273, 19877–19883. [DOI] [PubMed] [Google Scholar]