Abstract
Many therapeutic targets are intracellular proteins and molecules designed to interact with them must effectively bind to their target inside the cell. Intracellular antibodies (intrabodies) recognise and bind to proteins in cells and various methods have been developed to produce such molecules. Intracellular antibody capture (IAC) is based on a genetic screening approach and is a facile methodology with which effective intracellular antibodies can be obtained. During the development of the IAC technology, consensus immunoglobulin variable frameworks were identified which can form the basis of intrabody libraries for direct screening. In this paper, we describe the de novo synthesis of intrabody libraries based on the IAC consensus sequence. The procedure comprises in vitro production of a single antibody gene fragment from oligonucleotides and diversification of CDRs of the immunoglobulin variable domain by mutagenic PCR. Completely de novo intrabody libraries can be rapidly generated in vitro by these approaches. As an example, a single immunoglobulin VH domain intrabody library was screened directly in yeast with an oncogenic BCR-ABL antigen bait and distinct antigen binders were isolated illustrating the functional utility of the library. This second generation IAC approach (IAC2) has many practical advantages, in particular the ability to isolate intrabodies by direct genetic selection, which obviates the need for in vitro production of antigen for pre-selection of antibody fragments.
INTRODUCTION
Intracellular antibodies or intrabodies are antibody fragments that are used inside cells for interaction with target antigens and either for interference with function (1–3) or in some cases to mediate cell killing following antigen binding (4). Intrabodies have particular promise in the area of functional genomics where genome sequence projects are generating a plethora of open reading frames for which no functional data are available. Intrabodies have a role in defining these functions, especially where protein interactions can be defined. In therapeutics, the use of intracellular antibodies for functional ablation has been described and should be an invaluable format for disease-specific reagents.
Intracellular antibodies are typically formulated as single chain Fv (scFv) fragments which comprise immunoglobulin variable (V) domains of heavy (H) and light (L) chains held together by a short linker (5,6). Often, antigen-specific hybridomas have been used as a source of antibody genes from which scFv have been made for in-cell expression as intrabodies, and successes have been reported in which cellular phenotypes have been obtained due to scFv–antigen binding (7–9). Conversion of hybridoma antibodies into intracellular antibody fragments is laborious as this strategy requires an antigen-specific hybridoma from which the scFv derivative must be active in the cellular milieu (which is a reducing environment). Several different methods have been used to directly develop intrabodies without the need of hybridomas. These include genetic screening for intrabody– antigen interaction (10–12) based on two-hybrid screening (13) and use of fixed scFv frameworks for intrabodies (14–16). In the former approach, the intracellular antibody capture (IAC) technology (11,12) facilitated the identification of consensus frameworks comprising residues from VH and VL which are most commonly found in selected intracellular antibodies. When intracellular antibodies based on these scaffolds were expressed in mammalian cells, they were found to be soluble, well expressed and functionally efficient (17). In addition, recent studies have confirmed that the IAC consensus frameworks can be used to convert poor intracellular antibodies into efficient ones (17) by mutating framework residues to the IAC consensus whilst leaving the complementarity determining regions (CDRs) intact, which is the part most important for antigen binding.
It should be possible to build an intrabody library with only the knowledge of the intracellular antibody consensus sequence, without resorting to any pre-existing antibody gene clones. In this paper, we describe procedures to achieve this goal. Firstly, de novo antibody gene synthesis was carried out in which consensus scFv sequences (11) were used to generate oligonucleotides for gene synthesis and, secondly, cloned intracellular antibody genes were used as templates for CDR diversification, using a PCR method (18), which allows de novo intrabody libraries to be made in vitro with diversity at each CDR.
MATERIALS AND METHODS
Mammalian transactivation domain vector pEF-VP16
The vector pEF-VP16 was constructed for expression of scFv prey in mammalian two-hybrid assays. In this vector, scFv sequences may be cloned into SfiI and NotI sites in-frame with the VP16 transcriptional transactivator domain (AD) to make a fusion gene controlled by the promoter of the polypeptide elongation factor 1α (EF-1α), which allows high protein expression in mammalian cells (19). The VP16 AD fragment, including the nuclear localisation signal (NLS), was amplified by PCR using pNLVP16 (20) as template and the VP16 AD fragment was sub-cloned into the NotI site of pEF/myc/cyto (Invitrogen). To change the SfiI cloning site of pEF/myc/cyto for the SfiI site compatible for most scFv fragments, the SfiI region of this vector was mutagenised using two oligonucleotides 5′-CGTGAACACGTGGCCCAGCCGGCCCAGGTG CAGC and 5′-GCTGCACCTGGGCCGGCTGGGCCACG TGTTCACG using a QuikChange Site-directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions. The final clone has the EF-1α promoter, a multi-cloning site including SfiI and NotI sites compatible for scFv fragment insertions, an NLS and the VP16 AD (Fig. 1A).
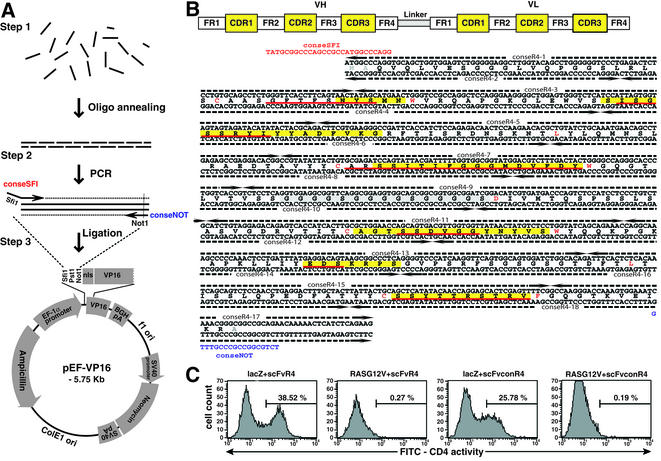
Figure 1.
De novo antibody gene synthesis. (A) Flow diagram of de novo antibody gene synthesis. (Step 1) Oligonucleotides corresponding to both strands of the desired antibody fragment (in this case an scFv but could be VH or VL alone) are mixed, annealed and ligated. (Step 2) PCR amplification of whole scFv is achieved using flanking primers (conseSFI + conseNOT) carrying SfiI or NotI sites. (Step 3) The PCR product is cleaved with SfiI and NotI and cloned into a compatible vector, in this case pEF-VP16. This vector was constructed from pEF/myc/cyto (Invitrogen) by addition of the VP16 AD and mutation of the SfiI site for compatibility with most scFv cloned sequences (see Materials and Methods). (B) Sequence of hybrid scFv and location of oligonucleotides used for gene synthesis. The design of scFvconR4 used the IAC consensus framework sequence (11) with the VH and VLλ CDR sequences from anti-β-galactosidase scFvR4 (21). CDRs are in yellow (22) or underlined in red (23,24). There is little homology between ScFvR4 and the IAC framework consensus as the former comprises VH3-VLλ (21) whereas the latter is VH3-VLκ (11). Landmark framework residues are highlighted in red [according to IMGT, C23, W41, L89 and C104 for both VH and VL; also W118 (for VH), D1 and F118 (for VL)]. (C) Mammalian cell reporter assay for scFv intrabody activity. CHO-CD4 cells (26) were transfected as indicated and stimulation of CD4 surface expression was measured using FACS with anti-human CD4 antibody and FITC-anti-mouse antibody. Cells were transfected with combinations of plasmids encoding DBD-βgal (lacZ), scFvR4-VP16 (scFvR4), DBD-RASG12V (RASG12V) and/or scFvconR4-VP16 (scFvconR4) as indicated. Approximately 10 000 cells were counted for each panel. VP16, VP16 AD.
De novo antibody gene synthesis
For antibody gene synthesis, oligonucleotides were designed from the scFv coding sequence comprising the VH and VL framework of the intrabody consensus (11) and the CDRs of an anti-β-galactosidase scFvR4 (21) (Fig. 1B). The double strands of DNA were divided into 18 oligonucleotides, of which 16 are 90 bases long and the two oligonucleotides flanking the ends of the scFv are, respectively, 100 bases on the 5′ end and 60 bases on the 3′ end. Each opposite strand oligonucleotide overlaps by 40–50 bases to ensure good annealing. All crude oligonucleotides were purified on 8% polyacrylamide gels containing 7 M urea and visualised by UV shadowing, using fluorescent thin layer chromatographic plates. Oligonucleotides were eluted by soaking the gel slice in 0.3 M sodium acetate overnight at room temperature (∼20°C). The supernatant was collected by centrifugation and the oligonucleotides were precipitated with ethanol. The concentrations of the purified oligonucleotides were calculated from the absorption spectrum. One microgram of each of the purified oligonucleotides was phosphorylated in a final volume of 100 µl in the presence of 2 µl of T4 polynucleotide kinase (10 U/µl) and 1 mM rATP. The volume was increased to 100 µl using NTE (100 mM NaCl, 10 mM Tris, 1 mM EDTA) and phosphorylation carried out by incubation at 37°C for 30 min. The reaction was stopped by incubation at 70°C for 10 min. The oligonucleotides were annealed after boiling the reaction for 30 s and allowing to cool to room temperature (∼20°C) over 40 min. Ligation of the annealed oligonucleotides was carried out using 17 µl of the annealed mixture, 2 µl of 10× T4 ligase buffer and 1 µl of T4 DNA ligase (400 U/µl) in a final volume of 20 µl. The mixture was incubated at 15°C overnight. The assembled oligonucleotides were finally PCR amplified with conseSFI and conseNOT primers (see Fig. 1B), which include a SfiI site at the 5′ end and a NotI site at the 3′ end for sub-cloning into pEF-VP16. A master mix for five PCRs (final volume 30 µl) was prepared containing 500 ng of each primer (i.e. conseSFI and conseNOT), 2.5 U Pfu DNA polymerase, 0.2 mM dNTPs, 1× PCR buffer and 1 µl of the ligated oligo mixture. PCR conditions were denaturation at 94°C for 5 min, followed by 30 cycles of 94°C for 1 min, 55°C for 1 min and 72°C for 1 min, with a final extension at 72°C for 5 min. The PCR product was separated on a 1% agarose gel and purified using a QIAexII gel purification kit (Qiagen). The purified 757 bp product (eluted in 40 µl of elution buffer) and the expression vector were digested with 1 µl of SfiI (20 U/µl) in a volume of 30 µl at 50°C for 5–6 h and vector linearisation was checked on an aliquot before proceeding to the NotI digestion. If the SfiI digestions appeared complete, digestion with 1 µl of NotI (10 U/µl) was carried out at 37°C for 16 h. The digested PCR products were purified on agarose gels, ligated with vector using T4 ligase at 15°C overnight and transformed into Escherichia coli TG-1. The constructs were verified by restriction enzyme digestion using SfiI and NotI and by DNA sequence analysis.
PCR mutagenesis
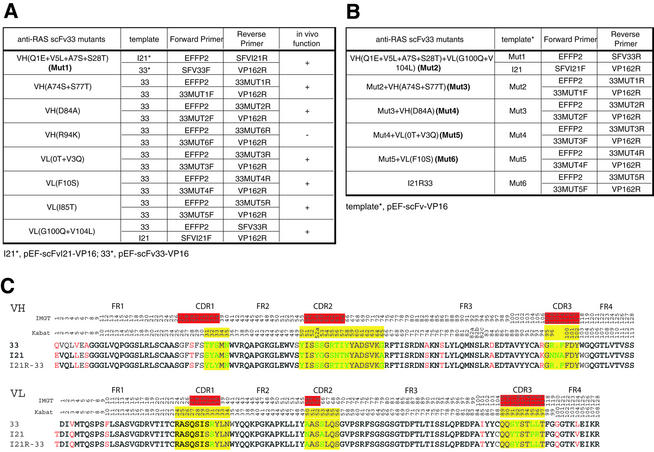
Specific mutations of the framework regions (FRs) of anti-RAS scFv33 (sequence shown in Table 1C) (17) into those of anti-RAS scFvI21 (Table 1C) was achieved by PCR-based mutagenesis based on the method of Hoogenboom and Winter (18) (herein called footprint mutagenesis). This was done firstly to investigate whether specific amino acid substitutions would affect in vivo function of the anti-RAS intrabodies (i.e. antigen-binding ability) (detailed functional data are described elsewhere, 17). Anti-RAS scFv33 mutants are listed in Table 1A and were constructed following the flow chart of the footprint mutagenesis method shown in Figure 2A. The locations of the mutant primer sequences relative to the scFv33 sequence are shown in Figure 2B. Two initial templates were used, either pEF-scFv33-VP16 or pEF-scFvI21-VP16 [respectively scFv33 and scFvI21 (17) cloned in pEF-VP16], for PCR as listed in Table 1. Each mutagenesis comprised synthesis of two overlapping PCR products using mutant oligonucleotides (Fig. 2A, step 1) followed by complete assembly (Fig. 2A, step 2) and cloning into the pEF-VP16 vector (Fig. 2A, step 3) to generate the mutated template for the next round of PCR mutagenesis (repeat). At each step the functional validity of the changes was estimated (17). Step 1 PCRs (final volume 20 µl) contained 0.5 µM each primer pair, 2.5 U Pfu DNA polymerase, 0.2 mM dNTPs, 1× PCR buffer and 50 ng pEF-scFv-VP16 template. PCRs were carried out by denaturation at 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 60°C for 30 s and 75°C for 45 s, with a final extension at 75°C for 10 min. Following PCR amplification, the amplified DNA fragments were electrophoresed on 2% agarose, extracted and purified using a QIAquick Gel Extraction Kit (Qiagen). Purified PCR fragments were assembled and amplified by PCR in step 2 with pEF-VP16 vector primers EFFP (5′-TCTCAAGCCTCA GACAGTGGTTC-3′) and VP162R (5′-CAACATGTCCAG ATCGAA-3′) by denaturation at 95°C for 5 min, followed by a gradient annealing at 60 to 30°C (0.1°C/s reduction in temperature) and gradient extension at 30 to 75°C (0.1°C/s increase in temperature), followed by 29 cycles with denaturation at 95°C for 45 s, annealing at 60°C for 45 s and extension at 75°C for 90 s. The amplified DNA fragment was digested with SfiI and NotI, purified by electrophoresis and gel extraction and re-cloned into the SfiI and NotI sites of pEF-VP16 in step 3. The constructs were verified by restriction enzyme digestion using SfiI and NotI and confirmed by DNA sequencing and tested for antigen binding in vivo (17).
Table 1. Templates and primers for stepwise PCR mutagenesis to convert scFv33 to scFvI21R33.
The primers were used in mutagenesis, illustrated in Figure 2, of the framework of the scFv33 sequence, to convert it to the I21R33 sequence. (A) At the first round, both pEF-scFv33-VP16 and pEF-scFvI21-VP16 were used as templates: individual mutations were incorporated with the primers, as indicated. (B) At subsequent rounds, the PCR template used was the previously mutated version, except round 2, in which either Mut1 or pEF-scFvI21R-VP16 was used. (C) An alignment of the derived protein sequences of scFv33 (33), scFvI21 (I21) and scFvI21R33 (I21R-33) in the single letter code. Residue numbering and CDRs according to the IMGT (top row) (23,24) or Kabat databases (second row) (22) are shown. The framework residues which differ between the three are in red and within the CDRs in green. CDRs highlighted in red (underlined) and yellow are from IMGT and Kabat, respectively.
Figure 2.
PCR-based mutagenesis of scFv frameworks. Specific framework residues of the intrabody scFv33 (17) were mutated to those of scFvI21 to yield scFvI21R33 (17) by stepwise mutagenesis. (A) At each step of mutagenesis, the scFv template, cloned in pEF-VP16, was PCR amplified using a fixed primer (EFFP2 or VP162R) together with a mutant primer (indicated by broken arrows) at a specific position; this yields two PCR fragments which are assembled with EFFP + VP162R primers and cloned into pEF-VP16 for the next round of mutagenesis. (B) Nucleotide and derived protein sequences of scFv33, indicating the amino acid residues mutated in the stepwise mutagenesis to scFvI21R33. The PCR primers are shown above the template sequence (red, forward primers; blue, reverse primers). The CDRs are highlighted in yellow (22) or underlined in red (23,24) and the linker between VH and VL in grey. Landmark framework residues are highlighted in red [according to IMGT, C23, W41, L89 and C104 for both VH and VL; also W118 (for VH), D1 and F118 (for VL)]. VP16, VP16 AD.
The construction of pEF-scFvI21R33-VP16 (17) [i.e. scFvI21R33 having the CDRs of anti-RAS scFv33 and the framework of anti-RAS scFvI21 except for the lysine at VH position 94 (22) or 106 (23,24), which was changed to arginine] was performed by repeating the PCR– assembly–cloning procedures described above according to Table 1B, starting with Mut1 as a template. Each round of mutation gave the mutated template for the next round of the stepwise mutagenesis using the conditions described above.
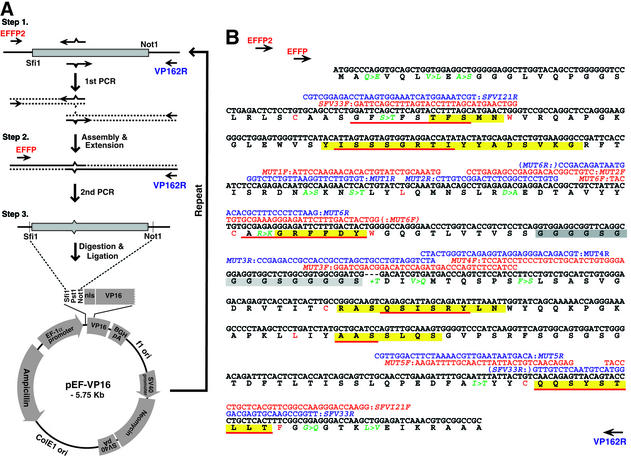
Diversification of VH CDR3 by mutagenesis for intrabody library construction
A flow chart outlining the construction of a scFv library based on VH CDR3 randomisation (18) is shown in Figure 3A. The primers used to randomise CDR3 of the VH domain in anti-ABL scFvA25 (11) and the partial nucleotide and protein sequences of anti-ABL scFvA25 are shown in Figure 3B. The template encoding anti-ABL scFvA25 was subcloned into the pEF-VP16 vector. In step 1, two PCR products were made using the pEF-scFvA25-VP16 template, namely the VH domain plus FR4 using the PCR primers EFFP2 plus A25C3Bn and the VL domain using the PCR primers A25CDR3F plus VP162R. The two PCRs yield overlapping products (Fig. 3A). A25C3Bn comprised three distinct oligonucleotides, each with a homologous sequence footprint around mutagenic regions of 3, 6 and 10 codons to generate mutations within VH CDR3. Amplified PCR fragments (for the VH and VL domains) were individually electrophoresed on agarose and purified. The two PCR products were assembled in a second PCR using oligonucleotides EFFP and VP162R, which encompass the whole scFv (i.e. VH and VL). The final PCR product was digested with SfiI and NotI and subcloned into the SfiI and NotI sites of pEF-VP16. Ligated DNA was electroporated in E.coli strain DH10B (Invitrogen). Clones were randomly picked from each final ligation (i.e. from A25C3B3, A25C3B6 and A25C3B10) and sequenced to verify the insert and the correct integration of CDRs. Primer sequences (M = A or C; N = any nucleotide; n = 3, 6 or 10 to randomise amino acid residues in CDR3 of the VH domain): EFFP2, 5′-GGAGGGGTTTTATGCGATGG-3′; EFFP, 5′-TCTCAAGCCTCAGACAGTGGTTC-3′; A25C3B, 5′-GACGGTGACCAGGGTTCCCTGGCCCC(MNN)nTCTC GCACAGTATATTAC-3′; A25CDR3F; 5′-GGGGCCAGG GAACCCTGGTCACCGTC-3′; VP162R, 5′-CAACATGTC CAGATCGAA-3′.
Figure 3.
Preparation of the scFv intrabody library with randomised CDR3. The intracellular antibody scFvA25, recognising the BCR-ABL oncogenic protein (11), was used as a template for a diversified library with randomised mutations of the VH CDR3 region. (A) scFvA25 was cloned into the pEF-VP16 vector and two mutagenesis PCRs carried out with primers EFFP2 + A25C3Bn (in which the central region of the primer has n = 3, 6 or 10 to randomise amino acid residues in CDR3 of the VH domain) and A25CDR3F + VP162R. The two PCR products were mixed, assembled and cloned into the mammalian pEF-VP16 vector. CDRs are highlighted in yellow (22) or underlined in red (23,24). (B) The sequence of the A25 VH CDR3 region (highlighted in yellow) and PCR primers A25CDR3F + A25C3Bn. (C) The DNA sequences of randomly selected clones from each first PCR were obtained and the derived VH CDR3 protein sequences of these clones are shown [highlighted in yellow (22) or underlined in red (23,24)]. VP16, VP16 AD.
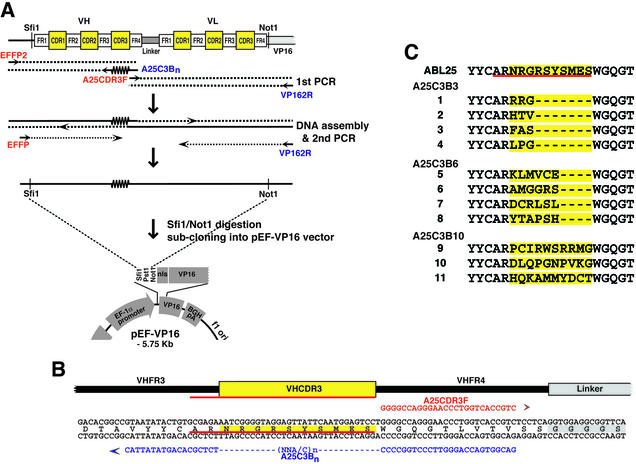
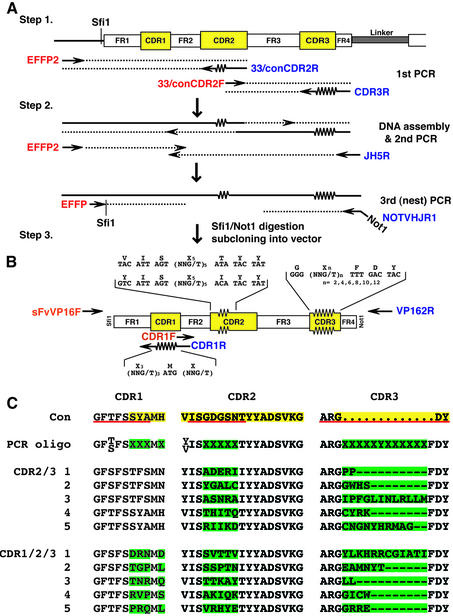
Diversification of VH CDR1, CDR2 and CDR3 for intrabody library construction
A flow chart outlining the randomisation of the VH CDRs is shown in Figure 4A. Two templates were used, one encoding the VH domain from anti-RAS scFvI21R33 (17) and the other from the canonical intrabody consensus sequence (11), each subcloned into the pEF-VP16 vector.
Figure 4.
Diversification of VH CDR1, CDR2 and CDR3 to make intrabody libraries. (A) PCR mutagenesis to generate a V-region segment with randomised CDRs. The template illustrated is a VH segment and the CDR2 and CDR3 regions are mutated as shown. (Step 1) Two PCRs were carried out with EFFP2 + CDR2R [either 33CDR2R or conCDR2R, which randomise CDR2 as shown in (B)] and CDR2F [either 33CDR2F or conCDR2F) + CDR3R, which randomises CDR3 as shown in (B)]. (Step 2) The two reaction products were assembled into a complete VH sequence using EFFP2 + JH5R flanking primers and this in turn amplified with partially nested primers EFFP + NOTVHJR1 to incorporate SfiI and NotI restriction sites. (Step 3) Cloning PCR products into yeast pVP16* (10). CDRs are highlighted in yellow (22) or underlined in red (23,24). Forward primers are shown in red and reverse primers in blue. (B) This illustrates a VH domain depicting FRs and CDRs (highlighted in yellow) with the PCR oligonucleotide sequences used for mutagenesis. For first round mutagenesis, CDR2 and CDR3 were simultaneously changed as indicated. For second round mutagenesis, CDR1 was changed as indicated. (C) Derived protein sequences of the CDRs of five selected clones made by mutation of CDR2 + CDR3 compared with the CDR1/2/3 sequences (top line) of the canonical consensus VH sequence (11) (CDR2/3) or five selected clones made by randomisation of CDR1 from a library of 3.4 × 106 clones with mutated CDR2 + CDR3 (CDR1/2/3); CDR residues are highlighted in yellow (22). The regions randomised in the PCR oligonucleotides are shown in the second line, with mutant residues highlighted in green. In CDR1, the mutated positions correspond to three positions of CDR-IMGT (positions 32–34) and two positions of FR2-IMGT (positions 39 and 40) (23,24); the approach described in this paper could easily be applied to these positions. For CDR2, the CDR2-IMGT limits corresond to eight amino acids, positions 56–63. The CDR3-IMGT limits are of 18 amino acids; there are five additional positions between 111 and 112, namely 111.1, 111.2, 112.3, 112.2, 112.1 (24).
Library 1 (CDR2/3). Randomisation of VH CDR2 and CDR3 in each case was done by footprint mutagenesis as described above. In the first round of PCR amplification (Fig. 4A, step 1), the two parts of the VH domain were separately amplified by PCR using two pairs of oligonucleotides: EFFP2 plus conCDR2R (to randomise CDR2) and conCDR2F plus CDR3R (to randomise CDR3) for template scFv625 (consensus VH); EFFP2 plus 33CDR2R (to randomise CDR2) and 33CDR2F plus CDR3R (to randomise CDR3) for template scFvI21R33 (I21 VH). Amplified PCR fragments were electrophoresed on agarose and purified. In the second round of PCR (Fig. 4A, step 2), the two PCR fragments were assembled using PCR oligonucleotides EFFP2 and JH5R. After purification of PCR product, a final PCR (Fig. 4A, step 3) was performed using EFFP and NOTVHJR1 to allow digestion with SfiI and NotI and ligation into yeast pVP16* vector (20) cut with SfiI + NotI. Ligated DNA was electroporated into competent E.coli strain DH10B. This facilitated the generation of two libraries (each called VH CDR2/3 library 1) with diversities of 2 × 106 (I21R33-derived library) and 1.4 × 106 (consensus library).
Library 2 (CDR1/2/3). For randomisation of VH CDR1, the two CDR2/3 libraries (library 1) were used as templates. Two PCRs were carried out with the pairs of oligonucleotides: sFvVP16F plus CDR1R (to randomise CDR1); CDR1F and VP162R (to copy the remaining part of the VH segment) (Fig. 4B). The two PCR fragments were assembled using sFvVP16F and VP162R, digested with SfiI and NotI and ligated into yeast pVP16* vector cut with SfiI and NotI. This facilitated the generation of two libraries with diversities of 3.04 × 107 (I21R33-derived library) and 2.215 × 107 (consensus library). Clones were randomly picked from each library and sequenced to verify the insert and the correct integration of CDRs (Fig. 4C). Primer sequences (M = A or C; N = any nucleotide; n = 1–6 to randomise amino acid residues in CDR3 of the VH domain): conCDR2R, 5′-CAGAGT CTGCATAGTATGT(MNN)5ACTAATGACTGAAACCCAC-3′; conCDR2F, 5′-ACATACTATGCAGACTCTGTG-3′; 33CDR2R, 5′-CAGAGTCTGCATAGTATAT(MNN)5ACT AATGTATGAAACCCAC-3′; 33CDR2F: 5′-ATATACTA TGCAGACTCTG-3′; CDR3R, 5′-TCCCTGGCCCCAGT AGTCAAA(MNNMNN)nCCCTCTCGCACAGTAATAG-3′; JH5R, 5′-GGTGACCAGGGTTCCCTGGCCCCAGTAGTC-3′; NOTVHJR1, 5′-ATAAGAATGCGGCCGCCGCTCGA GACGGTGACCAGGGTTCCCTG-3′; sFvVP16F, 5′-TGGG TCCGCCAGGCTCCAGG-3′; CDR1R, 5′-CCTGGAG CCTGGCGGACCCAMNNCAT(MNN)3CTGAAGCTGAATCCAGAGG-3′; CDR1F, 5′-TGGGTCCGCCAGGCTCC AGG-3′.
Library 1 (CDR2/3) was screened with a yeast pBTM116 bait encoding BCR-ABL p190 fused to the LexA DNA-binding domain (DBD) as described (11,17,25). Positive clones from the first round of yeast antibody–antigen interaction screening (growth by histidine prototropy and activation of β-galactosidase) were isolated. The selected 10 clones were re-transfected into yeast together with pBTM116 encoding LexA-DBD-BCR-ABL or LexA-DBD-HRAS (non-relevant bait) to confirm true positive clones by eliminating anti-LexA-DBD binders or other false positive clones.
Mammalian two-hybrid assay in CHO-CD4 using FACS analysis
Chinese hamster ovary (CHO) cells were grown in minimal essential medium α (α-MEM; Invitrogen) with 10% fetal calf serum, penicillin and streptomycin. FACS analysis using the CHO-CD4 reporter line (26) was performed as described previously (11) with small modifications. Aliquots of 3 × 105 CHO-CD4 cells were seeded in 6-well plates on the day before transfection. Samples of 0.5 µg pM1-HRASG12V (DBD-RAS) or pM1-βgal (DBD-βgal) and 1 µg pEF-scFvR4-VP16 or pEF-scFvconR4-VP16 were co-transfected into the cells using lipofectAMINE™ (Invitrogen) according to the manufacturer’s instructions. Forty-eight hours after transfection, cells were washed, dissociated using cell dissociation solution (Sigma) and resuspended in phosphate-buffered saline. The induction of cell surface CD4 expression was detected using anti-human CD4 antibody (Pharmingen) and FITC-conjugated anti-mouse IgG (Pharmingen). The relative fluorescence of the cells was measured with a FACSCalibur (Becton Dickinson) and the data were processed using the CELLQuest software.
RESULTS
De novo antibody gene synthesis
The production of antibody V genes from known protein sequence data was carried out by development of a set of overlapping oligonucleotides corresponding to the intracellular antibody scFv consensus framework (11) together with VH and VL CDRs from the anti-β-galactosidase scFvR4 (21) (Fig. 1B). Annealing and ligation of the mixture of oligonucleotides was followed by PCR of the assembled scFv and finally cloning into the mammalian expression vector pEF-VP16 vector after SfiI and NotI digestion (Fig. 1A, pEF-VP16). The synthetic scFv was cloned to derive pEF-scFvconR4-VP16 and this was sequenced to verify the scFv and its junction with the VP16 AD domain. The effectiveness of the hybrid scFvconR4 as an intrabody was assayed in a reporter assay, co-transfecting the pEF-scFvconR4-VP16 clone plus DBD-lacZ (4) into the CHO-CD4 line (this line carries a CD4 gene with a minimal promoter regulated by five repeated Gal4 DNA-binding sites and can be transcriptionally activated and monitored by expression of cell surface CD4) (26). When pEF-scFvconR4-VP16 was transfected into CHO-CD4 cells with a clone encoding a Gal4 DBD fused to β-gal, we detected around 25% activation of CD4 expression (Fig. 1C). This compares with analogous experiments using DBD-βgal with the original scFvR4 (as pEF-scFvR4-VP16). However, no activation of CD4 expression was observed with either pEF-scFvconR4-VP16 or pEF-scFvR4-VP16 co-transfected with a non-relevant bait, DBD-RAS. This shows that the de novo gene synthesis method is efficient for cloning antibody fragments which retain their specificity and verifies the consensus framework as an intrabody expression scaffold.
CDR mutagenesis to create diverse de novo intrabody libraries
The IAC method defined a scFv consensus sequence which proved particularly advantageous for intracellular use (11) because the method selects intrabodies based on in vivo screens (10–12). One specific antibody derived using this method was scFv33, which is an anti-RAS antibody able to bind RAS in mammalian cells (17). A second scFv, scFvI21, was derived from a RAS yeast screen but did not bind RAS in mammalian cells (17), although its expression level was superior to scFv33. We wished to assess the importance of specific IAC consensus framework residues and we have used a PCR-based mutagenesis procedure to make mutations in the scFv33 framework (Table 1A) for evaluation in a mammalian cell luciferase reporter assay (11). With a series of changes, the scFv33 framework was effectively converted to scFvI21 in a step-wise manner (Table 1), which exemplifies the consensus framework (11) and which retains the parental ability to bind to RAS antigen (summarised in Table 1A). The templates scFv33 and scFvI21 were cloned into pEF-VP16 to be used as PCR templates and sequential mutagenesis was carried out. Full conversion required seven rounds of PCR, assembly and cloning (Table 1B) (note the only change to the scFvI21 framework was the lysine residue at position 94, which was changed to an arginine, consistent with the canonical intrabody consensus; 11). At each step a new template was created and sequenced to verify the specificity of the PCR and each mutation was tested for function (17). For sequential mutation, each round provides the template for the next mutagenesis step (Table 1B and Fig. 2). With this approach, a new scFv could be created with the framework of scFvI21R and the CDRs of scFv33 (scFvI21R33). The PCR mutagenesis can be applied to create single or small changes in a specified region. We have used this method to diversify one CDR in an scFv as described (18) (VH CDR3 of the anti-ABL scFvA25) (11) to generate a library of different sequences. The PCR mutagenesis was achieved in this case using as the template scFvA25 cloned in pEF-VP16 and an internal mixture of PCR primers covering scFv VH CDR3 in which the oligonucleotides contain 3, 6 or 10 codon equivalence of randomised sequence (Fig. 3A and B, primer A25C3Bn). After the first PCR, the two PCR products were assembled (Fig. 3A, second PCR) and the final product was cloned into pEF-VP16 to create a library of individual clones. Randomly picked clones were sequenced in the CDR3 region (Fig. 3C), confirming that CDR3 had been changed by 3, 6 or 10 residues, respectively.
In the procedure illustrated in Figures 2 and 3, each step introduced a mutation(s) at only one position in the scFv. Mutations can be made at two, and potentially more, positions by using mutant oligonucleotides for each PCR step, prior to assembly. This is shown in Figure 4, which illustrates simultaneously randomising the CDR2 and CDR3 regions of a VH template and subsequent randomisation of CDR1 (the same strategy would apply to mutagenesis of VL and, in addition, simultaneous randomisation of the three CDRs should be possible). In the examples for CDR2/3 changes, two VH subregion PCRs were carried out, each using a fixed sequence primer together with a randomising primer (Fig. 4A, EFFP2 + CDR2R or CDR2F + CDR3R, where the two reverse primers are mutagenic for CDR2 and CDR3, respectively). The two PCR products overlap in the CDR2 region and PCR assembly was achieved using the flanking primers, followed by a final PCR with EFFP plus NOTVHJR1 for cloning into the SfiI and NotI sites of the yeast pVP16* vector. Sequences of a selection of clones showed diversity of the CDR2 and CDR3 regions (Fig. 4C). A mixed library of 3.4 × 106 clones from the above was used as a substrate for a second round of mutagenesis at CDR1 using the pairs of primers EFFP2 plus CDR1R (a primer for mutagenic CDR1) and CDR1F plus VP162R. Sequences of a selection of clones showed diversity of CDR1, as well as in the CDR2 and CDR3 regions (Fig. 4C). This two step procedure thus allows production of randomised CDR1, CDR2 and CDR3. It should be possible to devise a similar protocol for simultaneously mutating CDR1, CDR2 and CDR3.
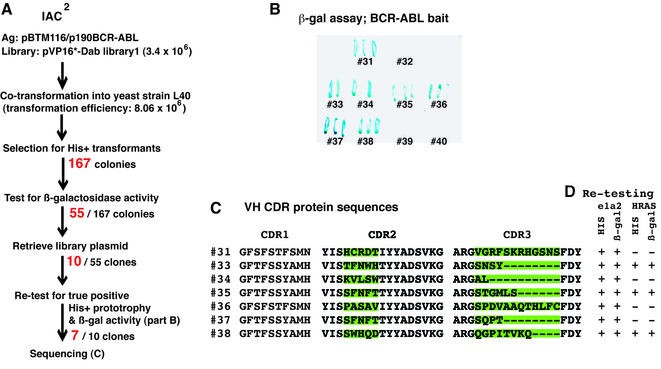
The intrabody CDR2/3 randomised library 1 was screened with a BCR-ABL bait (BCR-ABL is the fusion protein generated by the chromosomal translocations resulting in the Philadelphia chromosome; 27,28) directly in yeast, without the primary phage antibody enrichment step used in the first generation IAC (11,12). Fifty-five positive clones showed histidine-independent yeast growth and β-galactosidase activity (Fig. 5A). Ten of these clones were randomly selected and re-tested in a β-galactosidase assay using the original LexA-DBD-BCR-ABL fusion bait and seven allowed activation of the β-gal reporter gene (Fig. 5B). The DNA sequence of these seven revealed different CDR2 and CDR3 (Fig. 5C), showing that these were of different clonal origin. These seven clones were also tested in the yeast histidine prototrophy and β-galactosidase activation assays in response to LexA-BCR-ABL and a non-relevant bait, LexA-DBD-RAS. Three intrabody-expressing clones activated the his3 and lacZ genes in response to both baits, indicating that these antibody fragments recognise the common LexA-DBD domain of the baits (Fig. 5D), whilst four only activated the reporter in response to the BCR-ABL reporter, showing these to be recognising this antigen.
Figure 5.
Isolation of anti-BCR-ABL single domain intrabodies by direct yeast IAC2 library screening. (A) 8.06 × 106 clones from library 1 (CDR2/3) (diversity 3.4 × 106) were screened in yeast strain L40 co-transformed with LexA-BCR-ABL bait. 167 colonies grew on histidine– plates and 55 colonies of these were shown to have β-gal activation. Ten colonies were randomly selected and re-tested in yeast with LexA-BCR-ABL and non-relevant HRAS bait. Seven clones were isolated and DNA sequences obtained. (B) The β-galactosidase filter assay showing interaction between isolated clones and BCR-ABL antigen in yeast. (C) Alignment of CDR sequences of selected intracellular antibodies. The nucleotide sequences were obtained and the derived protein translations (shown in the single letter code) were aligned. The VH CDR2 and CDR3 regions corresponding to that randomised by PCR mutagenesis are highlighted in green. (D) Re-testing the antigen–antibody interaction in yeast using relevant BCR-ABL and non-relevant HRAS, to verify the specificity of antibodies with antigen. +, growth on his– plates or β-gal activation as indicated.
DISCUSSION
The methodology described here is a facile, transferable molecular tool kit for making antibody gene fragments, the acquisition of immunoglobulin mutants, for instance for affinity maturation which would involve CDR changes, and de novo formation of specific intrabody libraries based on the intracellular antibody consensus (11). We show that the de novo antibody gene synthesis method is a simple, oligonucleotide-based annealing, ligation and PCR procedure to make an antibody fragment suitable for cloning into a compatible vector. In our specific example of de novo intrabody production, a hybrid scFv was made in which the IAC consensus was the scaffold and an anti-β-galactosidase antibody (21) provided the CDR sequences. We chose the IAC consensus because it is advantageous for mammalian in-cell expression and anti-β-galactosidase antibody because it was specially developed in bacteria for soluble expression (21). Our hybrid scFvconR4 was able to bind to its target antigen in CHO cells the same as to the parental scFvR4 (Fig. 1C). The efficiency of β-gal binding, as judged by the intensity of CD4 reporter activation, was somewhat lower than the parental scFvR4. The apparent loss of binding of the chimeric intrabody (i.e. scFvconR4) may be attributable to several factors. It may reflect loss of in vivo affinity or lower expression in CHO cells. If the former, this could reflect on either the CDRs or the FRs. The slight loss of binding seen in the chimeric antibody may be attributable to the CDRs being present on the Vλ light chain in the parental scFv and Vκ light chain in the chimeric scFv. Nonetheless, it is an example that shows de novo production of functional intrabodies. Further, the result adds validation to the IAC consensus scaffold as a suitable intrabody scaffold (11). The IAC consensus derived in our work is representative of a well expressed, soluble scFv format suited to intrabody work but differs somewhat from another one similarly derived (11,12). However, our consensus was obtained from the most commonly occurring residue at any position (11) and variants do exist. This has been confirmed by our further studies, in which key residues of the FR and to a lesser extent the CDRs were studied in mutagenesis experiments (17). As a well expressed, soluble framework for library design, our IAC consensus seems ideal.
Diversification of the antibody fragments was carried out by mutagenesis allowing one or two step conversion of a V gene allowing generation of whole, diverse intrabody libraries. Thus intrabody libraries can be made starting simply with the consensus intrabody sequence (11). These libraries are ready for direct in vivo screening with any antigen that can be made as a bait in a suitable yeast two-hybrid vector. This facile methodology should make the application of intrabody-mediated interference with protein function a general procedure. These de novo intrabody libraries have proven useful in isolation of anti-BCR-ABL intrabodies by direct use of yeast antibody–antigen interaction screens (Fig. 5) and we have also shown that distinct functional antigen-specific intracellular antibodies can be isolated from these libraries (T.Tanaka, M.N.Lobato and T.H.Rabbitts, submitted for publication). Our results indicate that these libraries should be generally applicable to a second generation IAC approach (IAC2) which does not involve in pre-selecting potential intrabodies from antibody phage libraries in vitro. This advantage moves IAC2 into the realms of simple, generally available yeast genetic screening technology.
NOTE ADDED IN PROOF
A detailed protocol for intracellular antibody capture appears as a link within the Laboratory of Molecular Biology website http://www2.mrc-lmb.cam.ac.uk.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Pierre Martineau and Greg Winter for the scFvR4 clones and sequences and Dr Chi Van Dang for CHO-CD4 cells. This work was supported by the Medical Research Council. T.T. was the recipient of a grant from the National Foundation of Cancer Research and M.N.L. was the recipient of a grant from the Kay Kendall Leukaemia Fund.
REFERENCES
- 1.Biocca S., Pierandrei-Amaldi,P. and Cattaneo,A. (1993) Intracellular expression of anti-p21ras single chain Fv fragments inhibits meiotic maturation of Xenopus oocytes. Biochem. Biophys. Res. Commun., 197, 422–427. [DOI] [PubMed] [Google Scholar]
- 2.Tavladoraki P., Benvenuto,E., Trinca,S., De Martinis,D., Cattaneo,A. and Galeffi,P. (1993) Transgenic plants expressing a functional single-chain Fv antibody are specifically protected from virus attack. Nature, 366, 469–472. [DOI] [PubMed] [Google Scholar]
- 3.Rondon I.J. and Marasco,W.A. (1997) Intracellular antibodies (intrabodies) for gene therapy of infectious diseases. Annu. Rev. Microbiol., 51, 257–283. [DOI] [PubMed] [Google Scholar]
- 4.Tse E. and Rabbitts,T.H. (2000) Intracellular antibody-caspase mediated cell killing: a novel approach for application in cancer therapy. Proc. Natl Acad. Sci. USA, 97, 12266–12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bird R.E., Hardman,K.D., Jacobson,J.W., Johnson,S., Kaufman,B.M., Lee,S.-M., Lee,T., Pope,S.H., Riordan,G.S. and Whitlow,M. (1988) Single-chain antigen-binding proteins. Science, 242, 423–426. [DOI] [PubMed] [Google Scholar]
- 6.Huston J.S., Levinson,D., Mudgett-Hunter,M., Tai,M.-S., Novoty,J., Margolies,M.N., Ridge,R.J., Bruccoleri,R.E., Haber,E., Crea,R. and Oppermann,H. (1988) Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc. Natl Acad. Sci. USA, 85, 5879–5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marasco W.A., Haseltine,W.A. and Chen,S. (1993) Design, intracellular expression and activity of a human anti-human immunodeficient virus type 1 gp120 single-chain antibody. Proc. Natl Acad. Sci. USA, 90, 7899–7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan L., Bagasra,O., Laughlin,M.A., Oakes,J.W. and Pomerantz,R.J. (1994) Potent inhibition of human immunodeficiency virus type 1 replication by an intracellular anti-Rev single-chain antibody. Proc. Natl Acad. Sci. USA, 91, 5075–5079. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Cochet O., Kenigsberg,M., Delumeau,I., Virone-Oddos,A., Multon,M.C., Fridman,W.H., Schweighoffer,F., Teillaud,J.L. and Tocque,B. (1998) Intracellular expression of an antibody fragment-neutralizing p21 ras promotes tumor regression. Cancer Res., 58, 1170–1176. [PubMed] [Google Scholar]
- 10.Visintin M., Tse,E., Axelson,H., Rabbitts,T.H. and Cattaneo,A. (1999) Selection of antibodies for intracellular function using a two-hybrid in vivo system. Proc. Natl Acad. Sci. USA, 96, 11723–11728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tse E., Lobato,M.N., Forster,A., Tanaka,T., Chung,G.T.Y. and Rabbitts,T.H. (2002) Intracellular antibody capture technology: application to selection of single chain Fv recognising the BCR-ABL oncogenic protein. J. Mol. Biol., 317, 85–94. [DOI] [PubMed] [Google Scholar]
- 12.Visintin M., Settanni,G., Maritan,A., Graziosi,S., Marks,J.D. and Cattaneo,A. (2002) The intracellular antibody capture technology (IACT): towards a consensus sequence for intracellular antibodies. J. Mol. Biol., 317, 73–83. [DOI] [PubMed] [Google Scholar]
- 13.Fields S. and Song,O. (1989) A novel genetic system to detect protein-protein interactions. Nature, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- 14.Ohage E.C., Wirtz,P., Barikow,J. and Steipe,B. (1999) Intrabody construction and expression. II. A synthetic catalytic Fv fragment. J. Mol. Biol., 291, 1129–1124. [DOI] [PubMed] [Google Scholar]
- 15.Wirtz P. and Steipe,B. (1999) Intrabody construction and expression III: engineering hyperstable VH domains. Protein Sci., 8, 2245–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desiderio A., Franconi,R., Lopez,M., Villani,M.A., Viti,F., Chiaraluce,R., Consalvi,V., Neri,D. and Benvenuto,E. (2001) A semi-synthetic repertoire of intrinsically stable antibody fragments derived from a single-framework scaffold. J. Mol. Biol., 310, 603–615. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka T. and Rabbitts,T.H. (2003) Intrabodies based on intracellular capture frameworks that bind the RAS protein with high affinity and impair oncogenic transformation. EMBO J., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoogenboom H.R. and Winter,G. (1992) By-passing immunisation. Human antibodies from synthetic repertoires of germline VH gene segments rearranged in vitro. J. Mol. Biol., 227, 381–388. [DOI] [PubMed] [Google Scholar]
- 19.Mizushima S. and Nagata,S. (1990) pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res., 18, 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollenberg S.M., Sternglanz,R., Cheng,P.F. and Weintraub,H. (1995) Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol., 15, 3813–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martineau P., Jones,P. and Winter,G. (1998) Expression of an antibody fragment at high levels in the bacterial cytoplasm. J. Mol. Biol., 280, 117–127. [DOI] [PubMed] [Google Scholar]
- 22.Kabat E.A., Wu,T.T., Perry,H.M., Gottesman,K.S. and Foeller,C. (1991) Sequences of Proteins of Immunological Interest, 5th Edn. National Institutes of Health, Bethesda, MD.
- 23.Lefranc M.-P. and Lefranc,G. (2001) The Immunoglobulin FactsBook. Academic Press, London, UK.
- 24.Lefranc M.-P. (2002) IMGT, the international ImMunoGeneTics database: a high-quality information system for comparative immunogenetics and immunology. Dev. Comp. Immunol., 26, 697–705. [DOI] [PubMed] [Google Scholar]
- 25.Tse E., Chung,G. and Rabbitts,T.H. (2000) Isolation of antigen-specific intracellular antibody fragments as single chain Fv for use in mammalian cells. Methods Mol. Biol., 185, 433–446. [DOI] [PubMed] [Google Scholar]
- 26.Fearon E.R., Finkel,T., Gillison,M.L., Kennedy,S.P., Casella,J.F., Tomaselli,G.F., Morrow,J.S. and Dang,C.V. (1992) Karyoplasmic interaction selection strategy: a general strategy to detect protein-protein interaction in mammalian cells. Proc. Natl Acad. Sci. USA, 89, 7958–7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nowell P.C. and Hungerford,D.A. (1960) A minute chromosome in human granulocytic leukemia. Science, 132, 1497–1500. [Google Scholar]
- 28.Rowley J.D. (1973) A new consistent chromosomal abnormality in chronic myelogeneous leukemia identified by quinacrine fluorescence and giemsa staining. Nature, 243, 290–293. [DOI] [PubMed] [Google Scholar]