Abstract
Streptomycetes are high G+C Gram-positive, antibiotic-producing, mycelial soil bacteria. The 8.7-Mb Streptomyces coelicolor genome was previously sequenced by using an ordered library of Supercos-1 clones. Here, we describe an efficient procedure for creating precise gene replacements in the cosmid clones by using PCR targeting and λ-Red-mediated recombination. The cloned Streptomyces genes are replaced with a cassette containing a selectable antibiotic resistance and oriTRK2 for efficient transfer to Streptomyces by RP4-mediated intergeneric conjugation. Supercos-1 does not replicate in Streptomyces, but the clones readily undergo double-crossover recombination, thus creating gene replacements. The antibiotic resistance cassettes are flanked by yeast FLP recombinase target sequences for removal of the antibiotic resistance and oriTRK2 to generate unmarked, nonpolar mutations. The technique has been used successfully by >20 researchers to mutate around 100 Streptomyces genes. As an example, we describe its application to the discovery of a gene involved in the production of geosmin, the ubiquitous odor of soil. The gene, Sco6073 (cyc2), codes for a protein with two sesquiterpene synthase domains, only one of which is required for geosmin biosynthesis, probably via a germacra-1 (10) E,5E-dien-11-ol intermediate generated by the sesquiterpene synthase from farnesyl pyrophosphate.
Keywords: FLP recombinase‖lambda Red recombinase‖oriT‖SCO5222‖SCO6073
The recently completed 8.7-Mb Streptomyces coelicolor A3(2) genome sequence (1) has revealed a wealth of information. Notably, about 400 kb (more than 20 gene clusters) are apparently concerned with secondary metabolites. Predicted gene products include type I modular and type I and type II iterative polyketide synthases, chalcone synthases, nonribosomal peptide synthetases, terpene synthases, and genes with functions that are less well understood. Mutational analysis can provide a significant insight into the functions of these genes.
Here, we describe the adaptation to S. coelicolor of a PCR targeting-based gene disruption protocol (2) that permits the deletion of entire gene clusters for secondary metabolism and allows relatively rapid generation of nonpolar, in-frame deletions, thus avoiding polar effects on genes transcriptionally downstream. This procedure is much more efficient than those previously used to disrupt Streptomyces genes (3). Our approach was based on the discovery that allelic exchanges on the Escherichia coli chromosome can be achieved by recombination with a PCR-generated selectable marker flanked at both ends by extensions of only a few tens of nucleotides homologous to the desired region of the chromosome, when the Redα (exo), Redβ (bet), and Redγ (gam) proteins of the phage λ are present in the targeted strain (4–9). We have used λ-Red to promote recombination in E. coli between a PCR-amplified antibiotic resistance cassette selectable in E. coli and Streptomyces, and S. coelicolor DNA on a cosmid from the set used to sequence the S. coelicolor genome (10). The inclusion of an origin of transfer (oriT; RK2) in the disruption cassette allows the conjugal transfer of the PCR-targeted cosmid into S. coelicolor, readily yielding exconjugants with the desired gene replacement. The strategy also allows the elimination of the disruption cassette by FLP-recombinase-mediated site-specific recombination (11).
In a demonstration of the utility of this PCR-targeting method for the rapid construction of S. coelicolor knockout mutants, we set out to identify genes involved in the biosynthesis of the secondary metabolite geosmin. Many actinomycetes (12–14), certain cyanobacteria (15), myxobacteria, liverworts (16), and higher fungi (17) that inhabit aquatic or soil environments produce geosmin, which can impart an undesirable earthy or musty odor to food and water supplies (18, 19). The very ubiquitousness of geosmin, especially among streptomycetes, suggests that it has adaptive importance, but this remains unidentified at present. To the best of our knowledge, no genes that direct geosmin biosynthesis in any of these organisms have been identified. In streptomycetes, geosmin has been postulated to be synthesized from farnesyl pyrophosphate by means of a eudesmanoid or germacranoid sesquiterpene precursor (20). If so, an early step in geosmin biosynthesis should involve a sesquiterpene synthase. Two genes, here designated cyc1 and cyc2, encoding enzymes showing similarity to the sesquiterpene synthase that carries out a cyclization reaction involved in pentalenolactone synthesis in Streptomyces UC5319 (21), were found in the S. coelicolor genome sequence. Here, we use PCR targeting and transposon mutagenesis to show that one of these two genes and only one of the two protein domains encoded by that gene are essential for geosmin biosynthesis. The accompanying paper by Cane and Watt (22) in this issue of PNAS describes biochemical analysis of the protein encoded by this gene.
Materials and Methods
Strains and Culture Conditions.
Media and culture conditions for E. coli and S. coelicolor were the same as those described (2, 3). Strains are listed in Table 1. E. coli BW25113 (2) was used to propagate the recombination plasmid pIJ790 and S. coelicolor cosmids (10). E. coli DH5α carrying pCP20 (designated BT340) was used for FLP-mediated site-specific recombination (2). Both strains (CGSC#7636 and CGSC#7629, respectively) were obtained from the Escherichia coli Genetic Stock Center (Yale University, New Haven, CN). BW25113 and BT340 were grown in SOB or LB, respectively. E. coli DH5α (Stratagene) was used as a host for plasmid constructions. E. coli ET12567/pUZ8002 (23) was the nonmethylating plasmid donor strain for intergeneric conjugation with S. coelicolor strain M145 (1). Carbenicillin (Carb, 100 μg/ml), apramycin (Apra, 50 μg/ml), chloramphenicol (Cm, 25 μg/ml), kanamycin (Kan, 50 μg/ml) or viomycin (Vio, 30 μg/ml), all from Sigma, were added to growth media when required. Vio is currently available only from Research Diagnostics (Flanders, NJ). l-arabinose (10 mM final concentration; Sigma) was added as indicated to SOB medium from a 1 M sterile filtered stock solution to induce genes under control of the pBAD promoter (2).
Table 1.
Strains and plasmids used in this study
| Strain/plasmid | Relevant genotype/comments | Source/ref. |
|---|---|---|
| Plasmids | ||
| pKD20 | λ-RED (gam, bet, exo), bla, araC, rep101ts | 2 |
| pCP20 | FLP-recombination plasmid: flp, bla, cat, rep101ts | 11 |
| pIJ666 | Source of cat gene for pIJ790 | 24 |
| pOJ436 | Source of ApraRoriT sequence for pIJ773 | 25 |
| pIJ790 | λ-RED (gam, bet, exo), cat, araC, rep101ts | This study |
| pIJ773 | aac(3)IV (ApraR) + oriT | This study |
| pIJ778 | aadA from Ω-fragment (SpecR, StrepR) + oriT | This study |
| pIJ779 | aadA from Ω-fragment (SpecR, StrepR) | This study |
| pIJ780 | vph from Ω-fragment (VioR) + oriT | This study |
| pIJ781 | vph from Ω-fragment (VioR) | This study |
| Supercos-1 | bla, neo, cos | Stratagene |
| pUZ8002 | tra, neo, RP4 | 23 |
| pUB307 | neo, RP4 | 30 |
| pSETΩ | aadA, oriT, attP, int | 31 |
| E. coli | ||
| BW25113 | K12 derivative: ΔaraBAD, ΔrhaBAD | 2 |
| ET12567 | dam, dcm, hsdS, cat, tet | 29 |
| BT340 | DH5α/pCP20 | 11 |
| S. coelicolor | ||
| M145 | SCP1−, SCP2− | 1 |
| KF62 | cyc1∷Tn4560 insertion at aa position 75 | This study |
| J3001 | Δcyc2: aa 2–725 replaced with pIJ773 cassette | This study |
| J3002 | cyc1∷Tn4560,Δcyc2 | This study |
| J3003 | Δcyc2scar: aa 2–725 replaced with scar sequence | This study |
| J3004 | cyc2ΔN: aa 6–340 replaced with scar sequence | This study |
| J3005 | cyc2ΔC: aa 380–721 replaced with scar sequence | This study |
| J3006 | cyc1∷Tn4560,cyc2ΔN | This study |
| J3007 | cyc1∷Tn4560,cyc2ΔC | This study |
Plasmids.
The ampicillin resistance marker bla on pKD20 (in strain CGSC#7636) was replaced by cat (CmR) amplified from pIJ666 (24) with primers catforw and catrev (Table 2), thus generating pIJ790 (Table 1). The PCR product contained cat, with its own promoter, flanked by 40 bp of homology to regions adjacent to bla. CmR colonies were tested for ampicillin or Carb sensitivity. The relevant plasmid structure was confirmed by restriction analysis.
Table 2.
PCR primers used in this study (from Invitrogen)
| Primer name | Generation of | Sequence (5′–3′) |
|---|---|---|
| catforw | pIJ790 | ATGAGTATTCAACATTTCCGTGTCGCCCTTATTCCCTTTtcgggcacgtaagaggttcc |
| catrev | pIJ790 | TTACCAATGCTTAATCAGTGAGGCACCTATCTCAGCGAttaagggcaccaataactgc |
| apraforw | pIJ773 | tcatgagctcagccaatc |
| oriTrev | pIJ773 | cgccagcctcgcagagcag |
| Sc9B1.20forw | cyc2 deletion | CGGCGGATGCGGTCGAAGAGCCCTGGGTAGGGCCGGGCCattccggggatccgtcgacc |
| Sc9B1.20rev | cyc2 deletion | CGAGCCACGAAAGAGTGAGACTGAACGTCCGTCAGCGCGtgtaggctggagctgcttc |
| Sc9B1.20ΔNforw | cyc2ΔN deletion | AGCCCTGGGTAGGGCCGGGCCATGACGCAACAGCCCTTCattccggggatccgtcgacc |
| Sc9B1.20ΔNrev | cyc2ΔN deletion | GAGCAGTGCTCCCACGTCCGCCGCGGAGGTGCCGGGCCCtgtaggctggagctgcttc |
| Sc9B1.20ΔCforw | cyc2ΔC deletion | GTGCCCTTCCAGAAGGTCGGCCCGTCCGTCATCCCCGACattccggggatccgtcgacc |
| Sc9B1.20ΔCrev | cyc2ΔC deletion | CAGTGAACGTCCGTCAGCGCGTCAGTGCGTCAGTGCGGGtgtaggctggagctgcttc |
The 3′ homology of primers catforw and catrev to the cat gene of pIJ666 and the unique priming sites P1 and P2 of the disruption cassettes are underlined. The oligonucleotides have 39 nt of homology (uppercase) to the relevant gene. Start and stop codons within the 39-nt primer extension are bold.
The gene disruption cassette aac (3)IV (ApraR) and the oriT from RK2 were jointly amplified from pOJ436 (25) with primers apraforw and oriTrev (Table 2). FLP recognition target (FRT) sites and priming sites were then added to the resulting 1,246-bp fragment by amplification of left and right ends of the disruption cassette separately by PCR before joining them by XhoI digestion and ligation. The final disruption cassette was further cloned into the EcoRV site of pBluescript II SK(+) (Stratagene) to generate pIJ773 (Table 1). aadA (spectinomycin-resistance and streptomycin-resistance: SpecR, StrepR) and vph (VioR) were amplified from the omega fragments (accession nos. M60473 and X99314), and aac (3)IV (ApraR) of pIJ773 was replaced with the PCR products by λ-Red-mediated recombination, generating pIJ778 and pIJ780, respectively (Table 1). Disruption cassettes without oriT were constructed directly by PCR by using the omega fragments as templates and inserted into the EcoRV site of pBluescript II SK(+), generating pIJ779 (SpecR, StrepR) and pIJ781 (VioR), respectively (Table 1). All template plasmids contain the same FRT and priming sites, which were verified by sequencing.
Gene Replacement on Cosmids.
Electro-competent cells of an E. coli BW25113 transformant carrying pIJ790 (CmR) and the desired S. coelicolor cosmid (CarbR, KanR) containing the gene to be targeted were prepared from a 10-ml SOB culture containing 10 mM l-arabinose to induce the λ-red genes. The culture (30°C, OD600 ≈ 0.4) was immediately washed twice with ice-cold 10% glycerol and resuspended in the remaining drop of 10% glycerol after centrifugation, resulting in a 100-fold concentration of cells. Electro-competent cells (50 μl) were transformed with 100 ng purified PCR product in a 0.2-cm ice-cold electroporation chamber by using a Bio-Rad GenePulser II set to 200 Ω, 25 μF, and 2.5 kV. Shocked cells were incubated in 1 ml of LB at 37°C for 1 h and plated on LB containing Kan and Carb (to select the cosmid) and Apra (to select the integrated PCR product). Transformants were characterized by restriction analysis and PCR with a set of oligonucleotides priming outside the region of recombination.
PCR products were obtained by amplification of the twice gel-purified 1,383-bp EcoRI/HindIII pIJ773 disruption cassette with the appropriate primer pairs containing 39-nt extensions for λ-Red-mediated recombination (Table 2). Amplification was performed in a 50-μl reaction with 100 ng of template DNA, 200 μM dNTPs, 50 pmol each primer, 5% DMSO, according to the manufacturer's instructions (Expand High Fidelity Polymerase, Roche Molecular Biochemicals): denaturation at 94°C for 2 min, then 10 cycles with denaturation at 94°C for 45 s, annealing at 50°C for 45 s, and extension at 72°C for 1 min 30 s, were followed by 15 cycles with the annealing temperature increased to 55°C. A last elongation step was done at 72°C for 5 min. The PCR product was analyzed by gel electrophoresis and purified by using a PCR Purification kit (Qiagen, Chatsworth, CA).
Allelic Exchange in S. coelicolor.
The KanR, CarbR, and ApraR mutagenized cosmids were introduced into E. coli ET12567/pUZ8002 by electroporation as above and then transferred to Streptomyces by conjugation (3). ApraR exconjugants were screened for KanS, indicating a double-crossover allelic exchange in S. coelicolor, and were confirmed by PCR and Southern blot analysis.
In-Frame Deletion Mediated by FLP Recombinase.
pCP20 (CarbR, CmR) shows temperature-sensitive replication and thermoinducible expression of the FLP recombinase, which acts on FRT sites to remove the central part of the disruption cassette, leaving behind an 81-bp “scar” sequence (2). Electro-competent E. coli DH5α/pCP20 cells were transformed with the targeted cosmid DNA; CmR and ApraR transformants were selected on LB at 30°C. A few colonies were single colony-purified nonselectively at 42°C. Twenty to fifty percent of the colonies lost ApraR and pCP20 (CmR) simultaneously. Cosmid DNA containing the in-frame deletion was then introduced into the S. coelicolor mutant carrying the marked deletion in the gene of interest by polyethylene glycol (PEG)-mediated transformation (3). Selecting for single cross-over events with Kan (the neo gene of SuperCos1 confers KanR in Streptomyces) and subsequent screening for the loss of both KanR and the resistance derived from the marked deletion in the S. coelicolor mutant indicated the successful replacement of the resistance marker with an unmarked, in-frame deletion. Alternatively to the PEG-mediated transformation procedure, the cosmid containing the in-frame deletion can be made conjugation-proficient by replacing its neo gene with the VioRoriT (pIJ780) or SpecR,StrepRoriT (pIJ778) cassette by λ-Red-mediated recombination.
Gas Chromatography–Mass Spectrometry (GC-MS) Analysis.
Qualitative analysis was performed by coupled GC-MS (Hewlett-Packard gas chromatograph HP 5890 series II coupled with a Hewlett-Packard mass spectrometer HP 5989 A) (26). Volatiles released from S. coelicolor during 2 weeks' growth on MS plates at 30°C were adsorbed onto 100 mg of activated charcoal placed in the lid of each petri dish. The charcoal was extracted with 1 ml of chloroform (Sigma), which was then filtered through cotton wool. Five microliters of each extract was analyzed by GC-MS as follows: capillary column, fused silica (30 m; 0.25-mm inside diameter; 0.25-μm film thickness); carrier gas, He (0.82 kPa on column injection); temperature program: isothermal for 1 min at 40°C, change from 40 to 210°C at a rate of 10°C per min, and isothermal for 25 min at 210°C; energy of ionization, 70 eV. The presence of geosmin in the samples was confirmed by comparison with synthetic (±)-geosmin (Sigma).
Results
Adaptation of PCR Targeting for S. coelicolor.
The λ-Red expression plasmid pKD20 (2) could not be used with the S. coelicolor Supercos-1 clones because both contain the bla gene, preventing selection and providing sequence identity for undesirable cointegrate formation by homologous recombination. Thus, the bla gene was replaced by cat (CmlR) to give pIJ790. Like pKD20, pIJ790 expressed λ-Red when induced by arabinose and was compatible with the ColE1-derived Supercos-1.
For the disruption of genes cloned in Supercos-1 and the efficient transfer of the mutant derivatives to S. coelicolor, a gene replacement cassette (ApraRoriT) containing aac (3)IV (ApraR) and oriTRK2 was constructed in pBluescript II SK(+) (Fig. 1A). ApraR and KanR (encoded by neo of Supercos-1) can be selected independently in both E. coli and S. coelicolor. The ApraRoriT cassette is flanked at both ends by FRT sequences to allow efficient removal of ApraR and oriT by yeast FLP recombinase (Fig. 1 F–H). The template for the cassette amplification was the 1,383-bp EcoRI/HindIII fragment of pIJ773, which was twice purified by agarose gel electrophoresis to eliminate all traces of the intact plasmid. Without this precaution, ApraR colonies without the desired gene replacement predominated among the transformants in the next step.
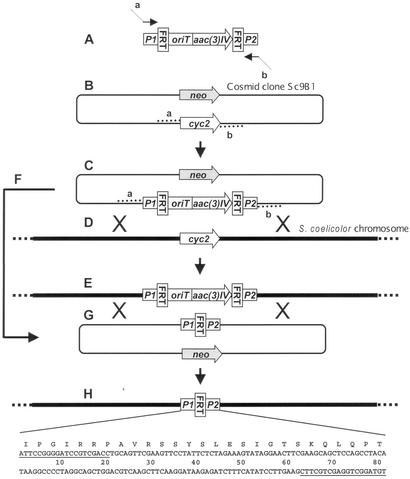
Figure 1.
Strategy for gene replacement in Streptomyces. (A) A gene replacement cassette containing aac (3)IV (ApraR), oriT of RK2, and two FRT sites was amplified by PCR by using primers containing 39-nt 5′ homology extensions corresponding to regions flanking the S. coelicolor target gene (dotted lines a and b) and 20-nt 3′ homology to the unique priming sites P1 and P2 of the disruption cassette. (B) The PCR product from A was used to transform E. coli BW25113/pIJ790 (expressing the λ-Red recombination functions), containing the Supercos-1 cosmid with the cloned target gene (cyc2). (C) ApraR transformants were selected, and the recombinant cosmid was identified by PCR and restriction analysis. (D) The potent methyl-specific restriction system of S. coelicolor was circumvented by introducing the recombinant cosmid into the methylation-deficient E. coli host ET12567/pUZ8002. The cosmid was mobilized in trans by pUZ8002 to Streptomyces by conjugation. (E) Selected exconjugants (ApraR) were screened for KanS (loss of the Kan resistance gene neo), and the double cross-over allelic exchange was confirmed by PCR and Southern blot analysis. (F) E. coli DH5α cells containing pCP20 were transformed with the mutagenized cosmid DNA, and FLP synthesis was induced. (G) The loss of the disruption resistance marker was tested, and the cosmid was introduced by polyethylene glycol (PEG)-mediated transformation into the S. coelicolor mutant carrying a marked deletion within the gene of interest. (H) KanR transformants arising from single cross-over events were restreaked nonselectively and screened for the loss of both KanR and ApraR, indicating a successful replacement by the in-frame deletion marked only by a “scar” sequence of 81 bp containing priming sites P1 and P2 (underlined), and one FRT site.
For generation of multiple gene disruptions on either a different or the same S. coelicolor cosmid, cassettes similar to the ApraRoriT cassette containing aadA (SpecR/StrepR) or vph (VioR) instead of aac (3)IV, both with and without oriT, were also constructed (Table 1). All of these cassettes contain the same priming sequences for PCR amplification and leave the same “scar” sequence after FLP recombinase-mediated excision. Other cassette constructs with different resistance genes (not shown) failed to give the desired gene replacements because they recombined preferentially with Supercos-1 sequences.
In designing primers for PCR amplification, we took into account that λ-Red-mediated recombination frequencies approach their maximum levels with a 40-bp targeting sequence (27). We used 39 bp rather than 40 bp because it comprises an integral number of codons, simplifying primer design. The 39-bp targeting sequence “tails” were supplied as the 5′ ends of long PCR primers, whose 3′ ends consisted of 19- or 20-bp priming sequences (Fig. 1A). The 39-bp 5′ target sequence tails were designed to generate “scar” sequences that were in a reading frame that lacked in-frame stop codons. A Perl program (JIC_GC_BMW) is available to assist in the primer design and in the analysis of the mutants generated (http://jic-bioinfo.bbsrc.ac.uk/S.coelicolor/).
Disruptions of cyc2.
Replacement of cyc2 in cosmid Sc9B1 (see below) was achieved by introducing a PCR-amplified, tailed gene disruption cassette ApraRoriT by electroporation into E. coli BW25113/pIJ790 containing Sc9B1 (Fig. 1B), prepropagated at 30°C in the presence of 10 mM arabinose to induce the λ-Red genes on pIJ790. About 250 ApraR, CarbR, KanR colonies were obtained (Fig. 1C). These colonies were CmS because the temperature-sensitive pIJ790 was lost during cultivation at 37°C. No ApraR transformants were obtained when arabinose was omitted or when a different cosmid without cyc2 was substituted for Sc9B1. Agarose gel analysis of digested plasmid DNA from these transformants revealed in most cases mixtures of the original cosmid clone and the expected mutant cosmids. The mutant cosmid constituted between 10% and 90% of the plasmid DNA. [Note that even ApraR, CarbR, KanR clones containing only a minority of the desired mutant cosmids were still useful for introducing the mutation into Streptomyces (see below).]
The high precision of the recombination occurring between 39-bp sequences was more extensively documented in experiments with another gene, whiI (28). Cosmid DNA samples from 50 transformants were all shown to contain the desired gene replacement both by restriction analysis and by PCR, using primers annealing 100 bp outside the 39-bp tail sequences. Gene replacement was maximal with 100 ng of the tailed PCR product.
The ApraRoriT cassette which replaced cyc2 in cosmid Sc9B1 was flanked by 28,085 bp and 3735 bp of S. coelicolor DNA. [Note that the EMBL entry for Sc9B1, accession no. AL049727, does not show the entire cosmid sequence; complete (real) cosmid sequences are available from ftp://ftp.sanger.ac.uk/pub/S_coelicolor/cosmid_inserts.]
The next step was to introduce the mutant allele into Streptomyces. Because S. coelicolor A3(2), like some other streptomycetes, strongly restrict methylated DNA from wild-type E. coli K12, the plasmid DNA had to be passed through a nonmethylating host. We used ET12567/pUZ8002 for this purpose, because the plasmid supplies transacting functions for the mobilization of oriT-containing cosmids (3, 23, 29). Cosmid DNA from the above ApraR, CarbR, KanR transformants was introduced by electroporation into ET12567/pUZ8002. By using 100 ng CCC DNA, ≈100 ApraR, CmR, KanR transformants were recovered after 24 h of incubation at 37°C. The low transformation frequency and the long time required for visible colonies to develop were typical for this strain (note that the KanR selective marker of pUZ8002 becomes ineffectual in the presence of the similarly marked Supercos-1).
As an alternative to electroporation, mutant cosmids containing oriT could also be introduced into ET12567 by RP4 tra-mediated mobilization. BW25113 containing the self-transmissible pUB307 instead of the transfer-deficient pUZ8002 was used for this purpose (30). ET12567 contains chromosomal tetracycline resistance and CmR markers to counterselect E. coli BW25113.
Once in Streptomyces, Supercos-1 does not replicate autonomously, but the long regions of sequence identity in the inserts promote efficient integration by homologous recombination. Thus, intergeneric conjugation between ET12567/pUZ8002/Sc9B1∷ApraRoriT and S. coelicolor M145 produced ≈104–105 ApraR colonies per conjugation. Most of these colonies were also KanR and presumably contained the entire plasmid integrated into the chromosome by a single crossover event. However, a sizable minority (>10%) were KanS, indicating double-crossover gene replacement and loss of the Supercos-1 vector (Fig. 1 D and E). In the case of the cyc2 replacement, this finding was verified by PCR and Southern blot analysis for three independent ApraR KanS S. coelicolor exconjugants.
The cyc2 Gene Is Essential for Geosmin Production.
One of the best known characteristics of streptomycetes is their production of the earthy odor geosmin, which is thought to be derived by the action of a sesquiterpene synthase on farnesyl pyrophosphate, which in turn is synthesized via the mevalonate pathway. S. coelicolor contains homologues of the genes needed for farnesyl pyrophosphate synthesis, as well as two homologues, cyc1 (SCO5222) and cyc2 (SCO6073), of the gene for a known sesquiterpene synthase (pentalenene synthase, which is involved in pentalenolactone synthesis) from Streptomyces UC5319 (21). The cyc1 product (Cyc1) contains one conserved sesquiterpene synthase domain, and the cyc2 product (Cyc2) contains two such domains (Fig. 2). cyc2 was replaced by ApraRoriT in both S. coelicolor M145 (generating J3001, Table 1) and an M145 derivative, KF62, containing a Tn4560 insertion in cyc1 (generating J3002, Table 1). The wild type and the cyc1 mutant produced amounts of geosmin easily detectable by GC-MS (GC-retention time and MS-fragmentation pattern were identical to those of authentic geosmin). In contrast, no geosmin was detectable in the cyc2 mutant (Table 3).
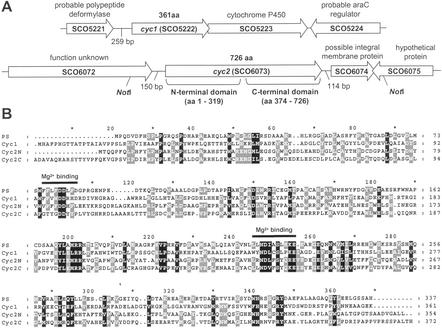
Figure 2.
Sesquiterpene synthase homologues cyc1 and cyc2 of S. coelicolor. (A) Flanking genes and their proposed functions. The stop codon of cyc1 overlaps the start of SCO5223, suggesting the possibility of translational coupling of the two genes. SCO5223 encodes a cytochrome P450 enzyme that catalyzes sterol 14α-demethylation (35). cyc2 is flanked by SCO6072, encoding a 666-aa protein of unknown function, and SCO6074, encoding a 203-aa protein annotated as a possible integral membrane protein. (B) clustalx alignment of Cyc1 and the N- and C-terminal domains of Cyc2 (Cyc2N, Cyc2C) to the 337-aa sequence of the pentalenene synthase (PS) from Streptomyces UC5319 reveals two conserved terpenoid synthase motifs. The residues Asp-80, Asp-81, and Asp-84 within the sequence 76FFLFDDLFD of pentalenene synthase are believed to form an Mg2+-binding site (36). Mg2+ is required to facilitate pyrophosphate departure in the first step of the cyclization reaction. Cyc1 contains all three conserved aspartate residues, whereas both the N- and the C-terminal domains of Cyc2 contain only the first two aspartate residues. Residues within a second sequence of pentalenene synthase (217LLNDIASLEKE) are also thought to be associated with binding of Mg2+ ions and are conserved in Cyc1 and both domains of Cyc2.
Table 3.
Geosmin production by S. coelicolor mutants
| Mutant strain | Genotype | Geosmin production | Complemented by | Geosmin production |
|---|---|---|---|---|
| KF62 | cyc1∷Tn4560 | + | ||
| J3001 | Δcyc2 | − | cyc2 | + |
| J3002 | cyc1∷Tn4560, Δcyc2 | − | cyc2 | + |
| J3003 | Δcyc2scar | − | cyc2 | + |
| J3004 | cyc2ΔN | − | cyc2 or cyc2 N terminus | + |
| J3005 | cyc2ΔC | + | ||
| J3006 | cyc1∷Tn4560, cyc2ΔN | − | cyc2 or cyc2 N terminus | + |
| J3007 | cyc1∷Tn4560, cyc2ΔC | + |
Production (+) was indicated by a peak corresponding to the geosmin standard within the total ion current chromatogram (TIC), and a mass spectrum (MS) with the expected Mr = 182 and a characteristic fragmentation pattern identical to that observed for the authentic standard. Where a geosmin peak was observed (+), the typical signal to noise ratio was ≈100, although this varied from sample to sample. Where no geosmin peak was observed (−), no ions were detected above background with a mass spectrum similar to authentic geosmin.
It was possible that the putative integral membrane protein gene SCO6074, downstream of cyc2 (Fig. 2), might have been inactivated by polar effects from the insertion of the ApraRoriT cassette. To eliminate such polar effects, the ApraRoriT sequence was removed by FLP-mediated recombination from the mutant Sc9B1 cosmid. PCR amplification and sequencing confirmed that the expected “scar” sequence without any in-frame stop codons had been generated.
Sc9B1 containing the scar-marked cyc2 deletion had only the KanR encoded by Supercos-1 for selection in Streptomyces and lacked oriT. After passage through E. coli ET12567, the plasmid was introduced into S. coelicolor cyc2∷ApraRoriT (strain J3001) by polyethylene glycol (PEG)-mediated protoplast transformation. About 30 KanR transformants were obtained with 100 ng CCC DNA. Nonselective propagation and screening for ApraS transformants readily yielded an unmarked deletion of cyc2 (strain J3003) containing a “scar” sequence of 81 bp (27 codons), which was confirmed by PCR and Southern blot analysis.
To confirm that the loss of geosmin production in the constructed cyc2 mutants was directly caused by the cyc2 deletions, we were able to restore geosmin production by reintroducing cyc2 as a 3,574-bp NotI fragment in the φC31 attP integrating vector pSETΩ (ref. 31; Table 3).
Only the Amino-Terminal Sesquiterpene Synthase Domain of Cyc2 Is Required for Geosmin Biosynthesis.
cyc2 contains two sesquiterpene synthase-like domains. To find out whether both were needed for geosmin synthesis, each domain was deleted separately in S. coelicolor M145 by using the FLP system to generate precisely tailored in-frame mutations, as described above. The amino-terminal deletion, cyc2ΔN (strain J3004), removed codons 6–340 and retained the ATG start codon, and the carboxyl-terminal deletion, cyc2ΔC (strain J3005), removed codons 380–721 and retained the stop codon. Geosmin production was abolished in J3004 (cyc2ΔN) but not in J3005 (cyc2ΔC), which produced similar amounts to wild-type S. coelicolor (Table 3). This result demonstrates that the C-terminal domain was not required for geosmin biosynthesis. The possibility that Cyc1 might complement the loss of the C-terminal domain of Cyc2 was eliminated because the constructed cyc1,cyc2ΔC double-mutant J3007 still produced geosmin. To verify that the N terminus of Cyc2 without the C terminus of Cyc2 was sufficient for normal geosmin production, the C-terminally deleted gene was cloned into pSETΩ (31) and introduced into the N-terminally deleted cyc2 mutant. Geosmin production was restored (Table 3).
Discussion
Murphy et al. (6) showed that induction of the λ-Red genes (gam, bet, and exo) can significantly increase homologous recombination in E. coli. Datsenko and Wanner (2) developed this strategy into a rapid and efficient protocol, which they used to disrupt 13 genes on the E. coli chromosome. In other work, genes in a cosmid library have been targeted with PCR products, and the mutagenized cosmids were used subsequently to transform the filamentous fungus Aspergillus nidulans (32). Higher recombination frequencies resulted from increasing the length of homologous DNA flanking the transformation marker.
The inclusion of oriT from an IncP-group plasmid in the disruption cassette allowed conjugation to be used for intergeneric transfer of cosmid DNA from E. coli to Streptomyces. In comparison to transformation protocols described for Streptomyces (3), conjugation is much more efficient, and it is successful with many actinomycetes (33). It also avoids the processes of protoplasting and regeneration used during transformation procedures, which are often difficult to develop for other strains and which can cause genetic changes.
Typically, tens to hundreds of E. coli BW25113 transformants were obtained when using a PCR product with 39 bp of flanking DNA homologous to the targeted S. coelicolor sequence; this result reinforces previously published results (6, 27), which showed that 40–50 bp of sequence identity is sufficient to sustain recombination when Redα and Redβ are produced in combination with Redγ (which inhibits the RecBCD exonuclease V).
So far, more than 100 segments of the S. coelicolor genome ranging in size from 4 bp to over 7 kb have been replaced by PCR targeting by us alone or in collaboration with others (data not shown). On average, 10–30% of S. coelicolor exconjugants showed the desired allelic replacement. Interestingly, the frequency of double crossovers in Streptomyces remained higher than 5%, even when only 2–3 kb homology was present on one side and >30 kb on the other side of the gene replaced within the cosmid (results not shown).
In this study, this methodology has been applied both to replace the entire cyc2 gene and to generate independent in-frame deletions in the regions of cyc2 coding for the N- and C-terminal sesquiterpene synthase-like domains. In conjunction with a recently established transposon library (34), these mutations have enabled us to identify, for the first time, a gene involved in the biosynthesis of the well known “earthy odor” metabolite geosmin. We have also shown that only the N-terminal domain of the Cyc2 protein is required for geosmin biosynthesis. Consistent with our findings, Cane and Watt report in the accompanying paper (22) that both the full-length Cyc2 protein and its N-terminal domain catalyze the cyclization of farnesyl pyrophosphate to germacra-1 (10)E,5E-dien-11-ol, whereas the recombinant C-terminal domain has no detectable sesquiterpene synthase activity in vitro.
These findings reinforce and extend incorporation experiments in Streptomyces antibioticus, which first indicated that geosmin is synthesized from farnesyl pyrophosphate via a sesquiterpene precursor (20), and the finding that a geosmin over-producing strain of Streptomyces citreus also produced considerable quantities of the sesquiterpene alcohol germacra-1 (10)E,5E-dien-11-ol, suggesting that this compound is the sesquiterpene precursor of geosmin (26). Taken together, these results suggest plausible mechanisms for geosmin biosynthesis in Streptomyces spp., which are explored in detail in the accompanying paper (22). The Cyc2 synthase is predicted to carry out cyclization of farnesyl pyrophosphate, to give germacra-1 (10)E,5E-dien-11-ol after capture of a tertiary carbocation intermediate by water and allylic transposition of the 2,3-double bond. Several further reactions, involving at least three additional enzymes (a cyclase, a reductase, and a hydroxylase) would be required to convert germacra-1 (10)E,5E-dien-11-ol into geosmin. Genes for the biosynthesis of Streptomyces secondary metabolites are usually clustered on the chromosome. Interestingly, the genes flanking cyc2 (Fig. 2A) do not appear to code for enzymes likely to be involved in the conversion of germacradienol into geosmin, suggesting that genes for the biosynthesis of the secondary metabolite geosmin may, in fact, be scattered on the S. coelicolor chromosome.
In preliminary experiments, we have observed that certain cyc2 mutations cause reduced sporulation. We anticipate that the further use of PCR-targeted mutagenesis will greatly facilitate further investigations of the role of geosmin in the life of streptomycetes.
Acknowledgments
We thank David Cane for prepublication information about the biochemical attributes of the SCO6073 (Sc9B1.20) gene product, Nick Le Brun at the University of East Anglia, United Kingdom, for help with GC-MS analysis, and Charlotte E. G. Schoeller at the Technical University of Denmark for access to her Ph.D. thesis, which contained much valuable background information about geosmin. This work was funded by Biotechnological and Biological Research Council (BBSRC) Grant 208/IGF12432 (to K.F.C.), Wellcome Trust Research Fellowship Grant 053086 (to G.L.C.), a BBSRC Research Studentship (to T.K.), a Competitive Strategic Grant from the BBSRC to the John Innes Centre, and the John Innes Foundation.
Abbreviations
- FRT
FLP-recombinase recognition target
- Carb
carbenicillin
- Apra
apramycin
- Cm
chloramphenicol
- Kan
kanamycin
- Vio
viomycin
- R
resistance
- S
sensitivity
- Strep
streptomycin
- Spec
spectinomycin
- oriT
origin of transfer
- GC-MS
gas chromatography–MS
References
- 1.Bentley S D, Chater K F, Cerdeno-Tarraga A M, Challis G L, Thomson N R, James K D, Harris D E, Quail M A, Kieser H, Harper D, et al. Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 2.Datsenko K A, Wanner B L. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kieser T, Bibb M J, Buttner M J, Chater K F, Hopwood D A. Practical Streptomyces Genetics. Norwich, U.K.: John Innes Foundation; 2000. [Google Scholar]
- 4.Zhang Y, Muyrers J P, Testa G, Stewart A F. Nat Biotechnol. 2000;18:1314–1317. doi: 10.1038/82449. [DOI] [PubMed] [Google Scholar]
- 5.Muyrers J P, Zhang Y, Stewart A F. Genet Eng. 2000;22:77–98. doi: 10.1007/978-1-4615-4199-8_6. [DOI] [PubMed] [Google Scholar]
- 6.Murphy K C, Campellone K G, Poteete A R. Gene. 2000;246:321–330. doi: 10.1016/s0378-1119(00)00071-8. [DOI] [PubMed] [Google Scholar]
- 7.Muyrers J P, Zhang Y, Testa G, Stewart A F. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Buchholz F, Muyrers J P, Stewart A F. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 9.Murphy K C. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 11.Cherepanov P P, Wackernagel W. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 12.Gerber N N. CRC Crit Rev Microbiol. 1979;7:191–214. doi: 10.3109/10408417909082014. [DOI] [PubMed] [Google Scholar]
- 13.Gerber N N, Lechevalier H A. Appl Microbiol. 1965;13:935–938. doi: 10.1128/am.13.6.935-938.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scholler C E, Gurtler H, Pedersen R, Molin S, Wilkins K. J Agric Food Chem. 2002;50:2615–2621. doi: 10.1021/jf0116754. [DOI] [PubMed] [Google Scholar]
- 15.Saadoun I, Schrader K K, Blevins W T. J Basic Microbiol. 2001;41:51–55. doi: 10.1002/1521-4028(200103)41:1<51::AID-JOBM51>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 16.Sauerwein M, Becker H. Planta Med. 1990;56:364–367. doi: 10.1055/s-2006-960983. [DOI] [PubMed] [Google Scholar]
- 17.Kiviranta H, Tuomainen A, Reiman M, Laitinen S, Liesivuori J, Nevalainen A. Cent Eur J Public Health. 1998;6:296–299. [PubMed] [Google Scholar]
- 18. Dionigi, C. P. (1993) Food Flavour Safety, 322–377.
- 19.Wood S, Williams S T, White W R. Int Biodeterior Bull. 1983;19:83–97. [Google Scholar]
- 20.Bentley R, Meganathan R. FEBS Lett. 1981;125:220–222. doi: 10.1016/0014-5793(81)80723-5. [DOI] [PubMed] [Google Scholar]
- 21.Cane D E, Sohng J K, Lamberson C R, Rudnicki S M, Wu Z, Lloyd M D, Oliver J S, Hubbard B R. Biochemistry. 1994;33:5846–5857. doi: 10.1021/bi00185a024. [DOI] [PubMed] [Google Scholar]
- 22.Cane D E, Watt R M. Proc Natl Acad Sci USA. 2003;100:1547–1551. doi: 10.1073/pnas.0337625100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paget M S, Chamberlin L, Atrih A, Foster S J, Buttner M J. J Bacteriol. 1999;181:204–211. doi: 10.1128/jb.181.1.204-211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kieser T, Melton R E. Gene. 1988;65:83–91. doi: 10.1016/0378-1119(88)90419-2. [DOI] [PubMed] [Google Scholar]
- 25.Bierman M, Logan R, O'Brien K, Seno E T, Rao R N, Schoner B E. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 26.Pollak F C, Berger G B. Appl Environ Microbiol. 1996;62:1295–1299. doi: 10.1128/aem.62.4.1295-1299.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu D, Ellis H M, Lee E C, Jenkins N A, Copeland N G, Court D L. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ainsa J A, Parry H D, Chater K F. Mol Microbiol. 1999;34:607–619. doi: 10.1046/j.1365-2958.1999.01630.x. [DOI] [PubMed] [Google Scholar]
- 29.MacNeil D J, Gewain K M, Ruby C L, Dezeny G, Gibbons P H, MacNeil T. Gene. 1992;111:61–68. doi: 10.1016/0378-1119(92)90603-m. [DOI] [PubMed] [Google Scholar]
- 30.Flett F, Mersinias V, Smith C P. FEMS Microbiol Lett. 1997;155:223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x. [DOI] [PubMed] [Google Scholar]
- 31.O'Connor T J, Kanellis P, Nodwell J R. Mol Microbiol. 2002;45:45–57. doi: 10.1046/j.1365-2958.2002.03004.x. [DOI] [PubMed] [Google Scholar]
- 32.Chaveroche M K, Ghigo J M, d'Enfert C. Nucleic Acids Res. 2000;28:e97. doi: 10.1093/nar/28.22.e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsushima P, Broughton C G, Turner J R, Baltz R H. Gene. 1994;146:39–45. doi: 10.1016/0378-1119(94)90831-1. [DOI] [PubMed] [Google Scholar]
- 34.Fowler K. Ph.D. thesis. Norwich, U.K.: John Innes Centre; 2002. [Google Scholar]
- 35.Lamb D C, Fowler K, Kieser T, Manning N, Podust L M, Waterman M R, Kelly D E, Kelly S L. Biochem J. 2002;364:555–562. doi: 10.1042/BJ20011380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lesburg C A, Zhai G, Cane D E, Christianson D W. Science. 1997;277:1820–1824. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]