Abstract
The PCR has been used to amplify a 2,181-bp ORF from Streptomyces coelicolor A3(2), designated SC9B1.20 (= SCO6073), encoding a protein of 726 amino acids and showing significant sequence similarity at the deduced amino acid level in both the N-terminal and C-terminal halves to the known sesquiterpene synthase pentalenene synthase. The full-length recombinant protein was expressed at high levels in Escherichia coli and shown to catalyze the Mg2+-dependent conversion of farnesyl diphosphate to the sesquiterpene alcohol (4S, 7R)-germacra-1 (10)E, 5E-diene-11-ol. The enzymatic cyclization had a kcat of 6.2 ± 0.5 × 10−3 s−1 and a Km for farnesyl diphosphate of 62 ± 8 nM. Expression of the N-terminal (366 amino acids) domain of the SC9B1.20 protein also gave a fully functional cyclase which converted farnesyl diphosphate to the identical sesquiterpene alcohol with a slightly lower kcat of 3.2 ± 0.4 × 10−3 s−1 and a twofold greater km of 115 ± 14 nM. By contrast, the expressed C-terminal domain of SC9B1.20 had no farnesyl diphosphate cyclase activity. The formation of the germacradienol seems to be the committed step in the formation of geosmin, the characteristic odoriferous constituent of Streptomyces species.
Keywords: secondary metabolism‖sesquiterpene synthase‖Streptomyces gene
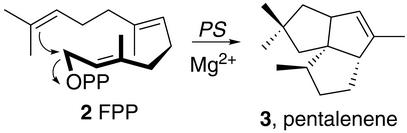
Terpenoid metabolites, the largest known group of natural products, include hormones, antibiotics, anti-tumor agents, flavor and odor constituents, and pigments, among a wealth of other physiologically or commercially important properties (1, 2). Of the >25,000 known terpenoids, the vast majority have been isolated from terrestrial or marine plants or from fungi, and only a relatively minor fraction from prokaryotes. No more than a dozen terpenoid metabolites have been obtained from Streptomyces species, even though these actinomycetes are a rich source of natural products of all structural classes, including over two-thirds of all naturally occurring antibiotics and a range of other medically useful metabolites, such as antitumor agents and immunosuppressants. In early studies of the odoriferous constituents of actinomycetes, Gerber (3) reported the isolation of several sesquiterpene alcohols, including epicubenol, as well as methyl isoborneol and geosmin 1, the latter two being modified monoterpene and sesquiterpene alcohols, respectively (4). The earthy odorant geosmin has itself been especially widely investigated because of its major contribution to the “off-flavor” of contaminated drinking water, wines, and other foodstuffs (5–8). Among Streptomyces-derived sesquiterpenes, the most thoroughly studied have been the pentalenolactone class of antibiotics, which have been isolated from several species of Streptomyces (9). Pentalenene synthase (EC 4.6.1.5) catalyzes the Mg2+-dependent cyclization of farnesyl diphosphate (FPP; Scheme S1 2) to pentalenene (Scheme S1 3), the parent hydrocarbon of the pentalenolactone family of metabolites (10). The pentalenene synthase gene from Streptomyces sp. UC5319 encodes a protein of MD 38 kDa, which has been cloned and expressed at high levels in Escherichia coli (11).
Scheme 1.
Cyclization of FPP 2 to pentalenene 3, catalyzed by pentalenene synthase (PS).
The amino acid sequence of pentalenene synthase has shown no significant similarity to any other known sesquiterpene synthase. Indeed, microbial sesquiterpene synthases in general show no overall sequence similarity either to one another (except for orthologs synthesizing the same sesquiterpene product) or to any other protein. This divergence in primary sequence has frustrated attempts to prospect for additional microbial synthases by techniques such as Southern hybridization or consensus PCR amplification (12). By contrast, the more than three dozen known plant monoterpene, sesquiterpene, and diterpene synthases have strongly conserved amino acid sequences, as well as similar intron organization and exon sizes, suggesting a common evolutionary origin (13). As a consequence, homology-based physical and bioinformatic screening methods have been very successful in identifying new plant terpene synthase genes. Interestingly, although plant and microbial sesquiterpene synthases differ significantly in primary structure, crystal structures of four different sesquiterpene cyclases of diverse biological origin have indicated a very high degree of conservation in the three-dimensional structure of all such synthases (14–18).
Streptomyces coelicolor A3(2) serves as the prototype of actinomycetes, a group of Gram-positive, soil-dwelling organisms, many of which exhibit a complex lifecycle involving mycelial growth and spore formation. The genetics of S. coelicolor is highly developed (19), and the genome sequence of the entire 8.7-Mb linear chromosome has recently been completed (20, 21). Among the predicted 7,825 proteins is a 2-kb ORF found by a TBLASTN search to have significant similarity to the pentalenene synthase gene. We now report the functional cloning of the full-length 2-kb gene, as well its 5′-terminal and 3′-halves, and the demonstration that it encodes a sesquiterpene synthase. A companion paper in this issue of PNAS by Gust et al. demonstrates the role of this same gene in geosmin production in S. coelicolor (22).
Materials and Methods
Materials.
S. coelicolor A3(2) cosmid St9B1 (SC9B1, EMBL accession no. AL049727; ref. 20) was a gift from David A. Hopwood (John Innes Centre, Norwich, U.K.). S. coelicolor CH999, a derivative of S. coelicolor A3(2) in which the actinorhodin biosynthetic gene cluster has been deleted and the biosynthesis of prodigiosin has been blocked, was a gift from Chaitan Khosla (Stanford University, Stanford, CA). Oligonucleotide primers were purchased from Integrated DNA Technologies (Coralville, IA). E. coli XL1-Blue-, BL21(DE3)pLysS-, and BL21(DE3-RP)-competent cells, NcoI and XhoI restriction endonucleases, Pfu turbo DNA polymerase, and agarose (low EEO) were purchased from Stratagene. T4 DNA ligase, 1-kbp and 100-bp DNA ladders, nucleotide triphosphates, isopropyl β-d-thiogalactoside (IPTG), and prestained protein ladders were purchased from GIBCO/BRL. The pET21d(+) expression vector was obtained from Novagen. [1-3H]FPP [20 Ci/mmol (1 Ci = 37 GBq)] was purchased from Dupont/NEN, diluted with synthetic FPP to a specific activity of 80 Ci/mol, and preextracted with pentane to remove any contaminating alcohols. Qiaprep Spin miniprep kits and Qiaquick PCR purification kits were obtained from Qiagen (Chatsworth, CA). Superdex 200 and Q-Sepharose fast-flow resins were purchased from Amersham Pharmacia Biotech (Uppsala, Sweden). Chemicals and other buffer components were purchased from Sigma and were of the highest grade available.
Methods.
DNA sequencing of plasmid constructs was performed by the Howard Hughes Medical Institute Biopolymer/Keck Foundation Biotechnology Resource Laboratory (Yale University School of Medicine, New Haven, CT) on an ABI 377 DNA sequencing system (Applied Biosystems) using fluorescently labeled dideoxynucleotides (big-dye terminators.) N-terminal protein sequencing and electrospray-ionization MS (ESI-MS) were carried out at the Keck Laboratory. PCR was performed with a PTC-150 minicycler (MJ Research, Cambridge, MA). Radioactivity measurements used glass vials containing 7 ml of Optifluor scintillation mixture (Packard) using an LS5801 Liquid Scintillation Counter (Beckman Coulter). Optical rotations were measured on a Perkin–Elmer 241 polarimeter. NMR spectra were recorded on an AM 400 spectrometer (Bruker, Billerica, MA) operating at 400.134 MHz for 1H and 100 MHz for 13C. GC-MS was performed on a Hewlett–Packard 5898 quadrupole mass spectrometer in electron impact ionization mode, using a 30 m × 0.25 mm HP5MS capillary column. UV-visible absorption spectroscopy was performed on a Hewlett–Packard 8452A diode array spectrophotometer. Analysis of DNA and protein sequences used the COMPARE, BESTFIT, PILEUP, and other programs of the Wisconsin GCG Sequence Analysis Package, V.10.0 (Accelrys, San Diego, CA). The Sanger Centre Streptomyces coelicolor A3(2) Genomic Database is available at http://www.sanger.ac.uk/Projects/S_coelicolor/.
Ligations, restriction endonuclease digestions, transformations of competent cells, preparation of buffers and media, as well as other standard molecular biological procedures were carried out as described (23). Determination of protein concentrations was performed according to the method of Bradford (24) by using the Bio-Rad kit. Steady-state kinetic parameters were calculated by direct fitting to the Michaelis–Menten equation by nonlinear least squares regression using KALEIDAGRAPH V.3.05 (Adelbeck Software, Reading, PA).
PCR Amplification and Ligation.
Amplification from the St9B1 cosmid of the full-length SC9B1.20 gene and each of the corresponding 5′- and 3′-domains used 5 μM each of the following primer pairs: full-length SC9B1.20, rmwstartb, 5′-A TTTTATCCATGGTTATGACGCAACAGCCCTTCCAA-3′; rmwhalt4, 5′-TAATAACTCGAGTCAGTGCGTCAGTG CGGGACT-3′; N-terminal domain, rmwstartb, rmwhalt3, 5′-T AATATCTCGAGTCACGTGTAGGAGCGGGCCCGTT G-3′; C-terminal domain, rmwstart3, 5′-ATTTTATCCATGG TTATGCCCTACCCGCTGGAGCTG-3′, rmwhalt4. (Underlined bases indicate restriction sites for NcoI and XhoI, respectively.) Reaction mixtures used Pfu turbo polymerase and contained 7% DMSO. Each of the resultant PCR products was purified immediately by using a Qiaquick PCR purification kit (Qiagen), eluted with 50–60 μl of 10 mM Tris⋅HCl, pH 8.5, then double-digested with NcoI and XhoI at 37 °C for 2–2.5 h, and repurified with the Qiaquick purification kit.
The digested PCR products were ligated into NcoI/XhoI-digested pET21d by using a 3:1 molar ratio of insert:vector and T4 DNA ligase; the ligation mixture was used to transform competent cells of E. coli XL1-Blue under standard conditions. The resulting transformants were grown overnight at 37°C on LB-agar plates containing ampicillin (100 μg/ml). Plasmid DNA was isolated from 10 individual colonies after overnight growth at 37°C in LB-ampicillin (100 μg/ml) medium and purified by using Qiagen spin miniprep kits. Plasmids were screened by appropriate restriction digests and sequenced to confirm the integrity of the inserted DNA sequences. Three plasmids were selected for expression: pRW31 (full-length SC9B1.20), pRW22 (N-terminal domain), and pRW19 (C-terminal domain).
Expression of Recombinant Terpene Cyclase.
In a typical procedure, pRW31 was used to transform competent cells of E. coli BL21(DE3)pLysS. Transformants were screened for the presence of the correct inserts by restriction digestion of isolated plasmid. For expression of SC9B1.20 protein, a 50-ml culture of E. coli BL21(DE3)pLysS/pRW31 was grown at 37°C to an OD600 of 0.7. The culture was cooled to 28°C and induced with 0.4 mM IPTG, then incubated for 6 h at 28°C. The cells were harvested by centrifugation (7,500 × g) and resuspended in 7.5 ml of cell lysis buffer [50 mM Tris⋅HCl/20% (vol/vol) glycerol/1 mM EDTA/pH 8.2]. 2-Mercaptoethanol (10 mM), benzamidine (0.2 mM), PMSF (0.2 mM), and lysozyme (1.5 mg/ml) were added, and the suspended cells were incubated at 32–35°C, with periodic inversion, for 7–10 min until cell lysis was complete. DNase I (10–20 μg/ml), MgCl2 (10 mM), and Triton X-100 (0.1%) were added, and then the incubation was continued at the same temperature for a further 4–7 min, before cooling to 0–4°C.
All subsequent manipulations were carried out at 4°C. The insoluble fraction containing the protein inclusion bodies was collected by centrifugation (15,000 × g for 15 min), washed with 10 ml of lysis buffer, and recentrifuged (15,000 × g for 15 min). The supernatant fractions were combined and purified by ion-exchange chromatography on Q-Sepharose (40 ml, fast flow) by using a linear gradient of 100–450 mM NaCl in refolding buffer (lysis buffer plus 10 mM 2-mercaptoethanol). Fractions containing the purest samples of the desired protein, as judged by SDS/PAGE, were combined, diluted eightfold with refolding buffer, and subjected to a second round of Q-Sepharose ion-exchange chromatography under the same conditions. The purest fractions from the second purification were again recombined, diluted eight-fold with refolding buffer, and purified on 1.5 ml of Q-Sepharose resin, eluting with refolding buffer containing 500 mM NaCl. The most concentrated fractions were combined and subjected to a final purification by gel-filtration chromatography on Superdex 200 (preparative grade) using 5 mM ammonium bicarbonate as the elution buffer. By using this method, 2–3 mg of soluble full-length cyclase was isolated, with a purity of >97%, based on SDS/PAGE.
The insoluble inclusion bodies were resolubilized, refolded, and purified by using a modification of the reported method of Hill et al. (25). The washed inclusion bodies (60–70 mg wet mass) were suspended in 8 ml of lysis buffer and added dropwise to a stirring solution of 120 ml of 18 mM NaOH containing 5 mM 2-mercaptoethanol and 0.02% Triton X-100. Stirring was maintained until all of the cloudiness had disappeared. A few ml of 100 mM NaOH were added to aid protein solubilization if necessary. Insoluble material was removed by centrifugation (15,000 × g for 20 min), and then the supernatant was added at 6–8 ml/min by means of a peristaltic pump to a slowly stirred flask containing 2,400 ml of refolding buffer. Stirring was halted after completion of the addition, and the protein was allowed to equilibrate for at least 2.5 h. Q-Sepharose resin (fast-flow, 25 ml, preequilibrated with refolding buffer) was added, and the mixture was stirred for 2–3 h to adsorb the protein to the resin. After allowing the mixture to settle, most of the supernatant was decanted (2,000 ml), and the remainder of the resin was poured into a column equipped with a sintered glass filter. The resin was washed with 75 ml of refolding buffer containing 90 mM NaCl, then eluted with a gradient of 100–450 mM NaCl in 125 ml of refolding buffer. Fractions were analyzed and further purified as described above for the soluble protein fraction. Protein derived from pRW22 and pRW19 was expressed and purified in the same way from the appropriate expression hosts.
Terpene Synthase Assay.
Assays were performed in 1 ml of assay buffer (50 mM Tris⋅HCl/20% (vol/vol) glycerol/10 mM MgCl2, pH 8.2) with FPP (80 Ci/mol) at concentrations of 8, 16, 40, 100, 500, 750, and 1,000 nM. Reactions in 10-ml test tubes were initiated by the addition of 10 μl (1.2 pmol, 100 ng) of purified enzyme, followed by the immediate overlay of 1.2 ml of hexane. After 16 min of incubation at 30°C, the reaction was terminated by the addition of 75 μl of 500 mM EDTA (pH 8.0). Ether (100 μl) was added, and the mixture was vortexed for 30 s. The organic layer was loaded onto a silica gel column (7 mm) in a Pasteur pipette and expelled with an N2 stream into a scintillation vial containing 7 ml of Opti-Fluor. The samples were extracted with a further two 750-μl portions of 11:1 hexane/ether. After passage of the extracts through the same minicolumn, the silica gel was subjected to a final wash with 2 × 750 μl of ether. The steady-state kinetic parameters, kcat and Km, were calculated by fitting the liquid scintillation data to the Michaelis–Menten equation.
GC-MS Analysis.
For GC-MS analysis of 4, recombinant purified cyclase obtained from E. coli BL21(DE3)pLysS/pRW31 or pRW22 (1 nmol) was incubated with 60 nmol of FPP in 1.0 ml of 50 mM Tris⋅Cl, pH 8.2, containing 20% (vol/vol) glycerol, 5 mM MgCl2, and 0.2 mM 2-mercaptoethanol overlaid with 1.5 ml of HPLC-grade pentane in a 4-ml Teflon-lined screw-capped vial for 2 h at 30°C. The reaction mixture was extracted with 2 × 1.5 ml pentane/CH2Cl2 (5:1), and the combined organic layers were dried by passage through a Pasteur pipette containing 1 g of MgSO4 before concentration under reduced pressure at 0°C to 100 μl. A 2-μl portion of the concentrated extract was analyzed by capillary GC-MS using a temperature program of 50–280°C, with a gradient of 20°C/min and a solvent delay of 3 min. Enzymatically generated 4, which constituted >98% of the total product, eluted at 10.35 min, with an [M]+ of m/z = 222 and a base peak of 59.
Preparative-Scale Incubation.
In a typical preparative-scale incubation, FPP (15 mg, 30 μmol) was incubated with 0.6 μmol of purified recombinant S. coelicolor cyclase in 500 ml of kinetics buffer with the MgCl2 concentration reduced to 5 mM to avoid precipitation of the Mg2+⋅FPP or Mg2+⋅PPi complex. The level of 2-mercaptoethanol, which came from the stock enzyme solution, was no more than 0.1–0.2 mM, because higher concentrations led to accumulation of an unidentified sulfur-containing by-product. The incubation mixture was overlaid with 300 ml of HPLC-grade pentane, and the incubation was carried out for 18 h at 30°C, with the enzyme being added in three equal portions every 6 h. The product was extracted as above, the extract was concentrated to 1 ml and then purified over 4 g of SiO2 by using a linear pentane/CH2Cl2 gradient. The recovered 4 [1.5 mg, mp 65–67° (ref. 26, mp 50–53°; ref. 27, 72–73°) after recrystallization from pentane] had an Rf of 0.35 on SiO2 TLC plates (hexane/ether 1:1, visualized with anisaldehyde). [α]24 (nm) (CHCl3, c = 0.16) −142.5° (589), −153.8° (578), −181.3° (546), −375.6° (436), −762.5° (365), −1,738.1° (313) (ref. 26). [α]D20 − 82° (CHCl3, c = 0.23); (ref. 27). [α]D20 − 153.3° (CHCl3, c = 1.5). 1H NMR (CDCl3) δ 5.67 (dd, J5–6 = 15.99, J4–5 = 3.55 Hz, 1 H, H-5), 5.06 (br d, J1–2β = 11.5, H-1), 4.99 (ddd, J5–6 = 15.99, J6–7 = 9.82, J = 1.88 Hz, 1 H, H-6), 2.43 (m, J2α−2β = 14.1 Hz, 1 H, H-2β), 2.42 (m, J4–5 = 3.55, J4–15 = 6.92 Hz, 1 H, H-4), 2.24 (m, J6–7 = 9.82, 1 H, H-7), 2.22 (m, 2 H, H-9), 1.91 (br d, J2α−2β = 14.1, 1 H, H-2α), 1.71 (m, 1 H, H-3α), 1.57 (br s, 3 H, H-14), 1.55 (m, 1 H, H-3β), 1.49 (m, 1 H, H-8α), 1.28 (m, 1 H, H-8β), 1.18 (s, 3 H, H-13), 1.13 (d, J4–15 = 6.92 Hz, 3 H, H-15), 1.10 (s, 3 H, H-12). 13C NMR (CDCl3) ppm 143.59 (CH, C-5), 131.55 (C, C-10), 130.99 (CH, C-1), 124.12 (CH, C-6), 72.16 (C, C-11), 59.33 (CH, C-7), 41.71 (CH2, C-9), 34.27 (CH, C-4), 33.19 (CH2, C-3), 27.29 (CH3, C-13), 26.66 (CH3, C-12), 22.48 (CH2, C-8), 22.14 (CH2, C-2), 17.16 (CH3, C-14), 15.14 (CH3, C-15).
Results
A TBLASTN search of the S. coelicolor A3(2) genome sequence in the Sanger Centre database using the 337-amino acid sequence of pentalenene synthase from Streptomyces sp. UC5319 as a query yielded a high probability match to a single 2,181-bp ORF from S. coelicolor cosmid SC9B1 designated SC9B1.20 (= SCO6073 in the reannotated sequence; ref. 21), encoding an apparent protein of 726 amino acids. Interestingly, pentalenene synthase showed significant similarities to both the N-terminal and C-terminal halves of the deduced amino acid sequence of the SC9B1.20p protein, with 28% identity and 41% similarity over the N-terminal 319 amino acids and 29% identity and 46% similarity to the C-terminal 339 amino acids, with a gap of ≈55 amino acids separating the two regions of homology (see Fig. 1, which is published as supporting information on the PNAS web site, www.pnas.org). The upstream 319 amino acids contained slight variants of the two universally conserved terpenoid synthase signature motifs, both of which have been associated with binding of catalytically essential Mg2+ ions (14–18, 28). The first is an acid-rich domain with a high proportion of aromatic amino acids, 83FFFDDHFLE, comparable to the aspartate-rich domain of pentalenene synthase, 76FFLFDDLFD, but with three amino acids instead of four between the second and third acidic amino acid residues. The second conserved motif found in SC9B1.20, 265LVNDVLTSRLHQ, corresponds to the 217LLNDIASLEKE sequence of pentalenene synthase, in which the triad of residues in bold has also been implicated in the binding of Mg2+ (15, 16, 18). In the downstream C-terminal domain, there is a weaker analog of the aspartate-rich domain consisting of 453YGDDYP and a characteristic 596LLNDYFSYQKE motif that is found at the usual ≈140-amino acids downstream of the aspartate-rich motif. To determine whether the SC9B1.20p protein was indeed a functional terpenoid cyclase and whether this synthase activity was resident in the N-terminal and/or C-terminal domains, we separately expressed the full-length protein as well as the upstream and downstream halves in E. coli and tested each protein for its ability to catalyze the Mg2+-dependent cyclization of FPP.
PCR was used to amplify both the full-length SC9B1.20 ORF as well as the N-terminal and C-terminal domains. For the full-length ORF, the start primer carried an NcoI restriction site just upstream of the deduced start codon, thus appending two additional amino acids, Met-Val- to the N terminus of the derived protein. The reverse primer was designed to introduce an XhoI restriction site just downstream of the normal stop codon. The N-terminal half of SC9B1.20 was amplified by using the same start primer in combination with a reverse primer designed to insert a stop codon and XhoI site immediately after Thr-366. Finally, the C-terminal pentalenene synthase homology region was amplified by using a start primer that placed an NcoI site and the codons for Met-Val immediately upstream of the natural Met-383 and the same halt primer that was used for amplification of the full-length gene. Each of the resulting PCR amplicons was digested with NcoI and XhoI and ligated into the corresponding sites of the expression vector pET21d to give plasmids harboring the full-length (pRW31), N-terminal (pRW22), and C-terminal (pRW19) gene fragments.
Both pRW31 and pRW22 were used to transform the expression host E. coli BL21(DE3)pLysS. The resultant transformants were induced with 0.4 mM IPTG and incubated at 28–30°C for 6 h before cell harvest and lysozyme-catalyzed cell-disruption. The desired proteins, which migrated with Mr 85,000 and 41,000, respectively, on SDS/PAGE (predicted MD 81,704 and 41,140 Da), usually constituted 30–45% of total protein, with ≈50% or more being obtained as insoluble inclusion bodies. Use of higher concentrations of IPTG (2–10 mM) or higher induction temperatures (30–37°C) increased the fraction of material in inclusion bodies to 90–95% or higher. The soluble protein fractions could each be purified by adsorption onto Q-Sepharose followed by two rounds of ion exchange chromatography to give 2–3 mg of soluble protein from a 50-ml culture with a purity of 97–98%. A final FPLC cleanup by gel-filtration on Superdex 200 gave protein >99% pure by SDS/PAGE. The desired proteins could also be purified from the corresponding inclusion bodies by resolubilization in 18 mM NaOH, followed by simultaneous dilution and neutralization by slow addition to a 20-fold excess (vol/vol) of 50 mM Tris buffer, pH 8.2. Bulk adsorption onto Q-Sepharose and ion-exchange purification gave protein that was indistinguishable in purity, gel mobility, and catalytic properties from the native soluble protein. ESI-MS analysis of the full-length pRW33 protein revealed the presence of a major protein species with calculated mass of 81,631 Da, consistent with retention of the N-terminal methionine (predicted 81,703.6 Da), whereas N-terminal sequencing gave (X)-X-X-Thr-Gln-Gln-Pro-Phe-Gln-Leu-Pro-His, corresponding to amino acids 4–12 after the (Met)-Val-Met of the full-length protein. Similarly, ESI-MS analysis of the N-terminal domain protein encoded by pRW22 revealed the presence of a 40:60 mixture of two species with molecular masses of 41,136 (predicted mass of 41,140) and 41,004 (predicted mass of 41,008) Da, corresponding to retention and loss, respectively, of the N-terminal methionine residue. N-terminal sequencing of the pRW22 protein gave results identical to those obtained for the full-length pRW31 protein. To express the C-terminal domain protein, pRW19 was used to transform a variety of expression hosts, including E. coli BL21(DE3)-RP and BL21(DE3)pLysS. In both cases, recombinant protein migrating on SDS/PAGE with Mr 40,000 (predicted mass of 39,015 Da) was obtained as 30–35% of total protein, almost exclusively as insoluble inclusion bodies. The inclusion bodies could be solubilized with 18 mM NaOH and refolded by dilution into excess buffer, then purified by Q-Sepharose ion exchange chromatography as for the full-length protein.
To test the predicted terpene synthase activity of the SC9B1.20 protein, each of the above-prepared recombinant proteins was incubated with 60 μM FPP in 1 ml of 50 mM Tris⋅HCl, pH 8.2, in the presence of 5 mM MgCl2 for 2 h at 30 °C. The organic-soluble product was extracted with 5:1 pentane/CH2Cl2 and the concentrated extract was analyzed by capillary GC-MS. Both the full-length protein obtained from pRW31 and the N-terminal domain encoded by pRW22 gave a single major product 4 of identical retention time and mass, m/z = 222, consistent with formation of a sesquiterpene alcohol of molecular formula C15H26O. The same alcohol, which constituted >97% of the total product formed, was obtained from both native soluble protein and resolubilized inclusion bodies. The steady-state kinetic parameters were determined for each protein construct by using [1-3H]FPP: full-length synthase from pRW31 had a kcat of 6.2 ± 0.5 × 10−3 s−1 and a Km for FPP of 62 ± 8 nM, whereas the N-terminal synthase domain encoded by pRW22 displayed an only slightly lower kcat of 3.2 ± 0.4 × 10−3 s−1 and a twofold greater Km of 115 ± 14 nM. The higher homolog, geranylgeranyl diphosphate (GGPP), did not function as a substrate. By contrast, when the refolded pRW19 protein, corresponding to the C-terminal half of the parent synthase, was incubated with FPP, only small quantities of a mixture of farnesol and nerolidol, identified by capillary GC-MS, were formed, at a rate no greater than five to six times greater than background solvolysis, over a range of pH from 5.5 to 9.5 in the presence of a wide variety of metal cations, including Mg2+, Mn2+, Co2+, Fe2+, Fe3+, Cu2+, Ni2+, and Zn2+. GGPP also was not a substrate for the C-terminal domain of the cyclase.
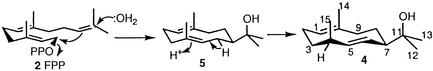
Having demonstrated the terpene synthase activity of both the full-length SC9B1.20 protein and its N-terminal domain, we next determined the structure of the cyclization product 4. Chromatographic purification of the crude pentane/CH2Cl2 extract of a preparative-scale (500 ml) incubation of 55–65 μM FPP 2 with either the pRW31 or pRW22 proteins typically yielded 1–2 mg of 4 as a colorless solid found to be >98% pure by GC-MS. Besides the parent M+ peak, m/z = 222, the electron impact mass spectrum of 2 showed ions at m/z = 204 (M+ − H2O), 189 (M+ − [H2O+CH3]), 164 (M+ − [(CH3)2CO]), 149 (M+ − [(CH3)2CO+CH3]) and a base peak at 59 [(CH3)2COH+]. The 400 MHz 1H NMR spectrum of 4 revealed the presence of two methyl groups attached to a quaternary carbon (δ 1.10 and 1.18), a secondary methyl attached to a C-H methine (δ 1.13, d, J = 6.92 Hz), and an allylic methyl attached to a fully substituted olefinic carbon (δ 1.57, br s). Of the three olefinic protons, two (δ 4.99, 1 H, ddd; 5.67, 1 H, dd) were coupled to one another (J = 15.99 Hz), indicating the presence of an E-disubstituted alkene, and the third (δ 5.06, br d, J = 11.5 Hz) was located on a trisubstituted double bond. The 106.1 MHz 13C NMR spectrum displayed 15 unique signals, with 4 clearly identifiable olefinic carbons [143.59 (CH), 131.55 (CH), 130.99 (C), and 124.12 (CH) ppm], confirming the presence of one trisubstituted and one disubstituted double bond. Distortionless enhancement by polarization transfer experiments established that altogether there were four methyl, four methylene, five methine, and two quaternary carbons. The signal at 72.16 ppm for the non-olefinic quaternary carbon was consistent with the presence of a tertiary alcohol. Together with the mass spectrometric data, it was clear, therefore, that 4 was a monocyclic alcohol containing two double bonds. Detailed analysis of the 1H-1H COSY, heteronuclear multiple quantum correlation (HMQC), and heteronuclear multiple bond correlation spectra led to the assignment of 4 as germacra-1(10)E, 5E-dien-11-ol (Scheme S2), completely consistent with the reported 1H and 13C NMR data for the (4S,7R)-sesquiterpene alcohol 4 previously isolated from cultures of Streptomyces citreus CBS 109.60 and from the liverwort Dumortiera hirsuta (26, 27).‡ The absolute configuration of 4 was readily assigned by comparison of the observed optical rotation {[α]D24 − 142.5° (CHCl3, c = 0.16)} with the literature (ref. 27) value {[α]D − 153.3° (CHCl3, c = 1.5)}.
Scheme 2.
Cyclization of FPP 2 to (4S, 7R)-germacra-1(10)E, 5E-diene-11-ol 4 via hedycaryol 5, catalyzed by the sesquiterpene synthase encoded by SC9B1.20.
Discussion
The protein encoded by SC9B1.20 of S. coelicolor, which shows significant levels of amino acid sequence similarity to pentalenene synthase, has thus been shown to catalyze the cyclization of FPP to (4S,7R)-germacra-1(10)E, 5E-dien-11-ol 4. This transformation can be readily explained by the mechanism illustrated in Scheme S2, in which ionization and cyclization of FPP is followed by capture of water by the 2-propyl cation side chain to give the hypothetical intermediate, (+)-hedycaryol 5, a known compound which has been isolated from the leaves of Hedycarya augustifolia (29). Protonation of 5 at C-5 and the loss of one of the H-7 protons will give 4. The N-terminal domain of SC9B1.20 is also fully competent catalytically, converting FPP to 4 with only minor differences in steady-state kinetic parameters compared with the wild-type, full-length cyclase. By contrast, the C-terminal half of SC9B1.20 did not process either FPP or its diterpene homolog, GGPP. It is possible that this latter domain is catalytically silent, despite its significant sequence similarity to pentalenene synthase and other terpene synthases. Alternatively, the C-terminal domain may catalyze another terpene synthase-like step that requires one or more substrates other than FPP.
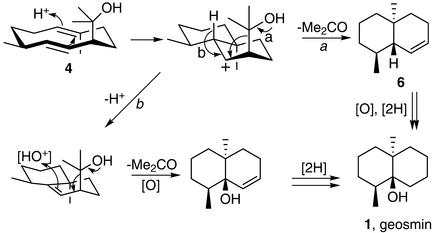
Attempts to isolate 4 from cultures of S. coelicolor CH999 were unsuccessful using the protocols described for the isolation of the same metabolite from S. citreus (26). Therefore, it is unlikely that 4 accumulates during normal, submerged culture of S. coelicolor, at least under the conditions examined. Upon completion of this study, we learned that Challis, Chater, Gust, and coworkers (22) have independently investigated the role of the SC9B1.20 gene in S. coelicolor. As described in the companion PNAS paper (22), targeted deletion of the entire SC9B1.20 gene or the corresponding N-terminal domain abolished production of geosmin 1, the characteristic odoriferous constituent of Streptomyces species (5). Geosmin 1 has previously been proposed to be a degraded sesquiterpene, but little experimental information has been available concerning its biosynthesis, other than the fact that it was apparently labeled when either [1-14C]- or [2-14C]acetate was administered to S. antibioticus (4). After cyclization of FPP 2 to (4S,7R)-germacra-1(10)E, 5E-dien-11-ol 4 catalyzed by the gene product of SC9B1.20, the formation of geosmin 1 could take place by a short sequence of biochemically well precedented steps, illustrated in Scheme S3. Proton-initiated cyclization of 4 and fragmentation of the side chain alcohol, with loss of acetone, would generate the bicyclic diene 6. Oxidation of 6 at the bridgehead and reduction of the disubstituted double bond would yield geosmin 1. It is not yet known whether the requisite genes encoding the enzyme activities needed to mediate the conversion of 4 to geosmin 1 are proximal to the SC9B1.20 ORF or located elsewhere in the S. coelicolor genome. Because deletion of the C terminus of SC9B1.20 does not abolish geosmin production, however, (22) this portion of the protein plays no role in the formation of geosmin.
Scheme 3.
Proposed mechanism of conversion of (4S, 7R)-germacra-1 (10)E, 5E-diene-11-ol 4 to the odoriferous compound geosmin 1.
Supplementary Material
Acknowledgments
We thank Prof. D. A. Hopwood of the John Innes Centre (Norwich, U.K.) for a gift of S. coelicolor cosmid St9B1 and for bringing to our attention the related work of Challis, Chater, Gust, and coworkers (22) reported in this issue of PNAS. This work was supported by National Institutes of Health Merit Award GM30301 (to D.E.C.).
Abbreviations
- ESI-MS
electrospray ionization–MS
- FPP
farnesyl diphosphate
Footnotes
References
- 1.Glasby J S. Encyclopedia of Terpenoids. Chichester: Wiley; 1982. [Google Scholar]
- 2.Sacchettini J C, Poulter C D. Science. 1997;277:1788–1789. doi: 10.1126/science.277.5333.1788. [DOI] [PubMed] [Google Scholar]
- 3.Gerber N N. Phytochemistry. 1971;10:185–189. [Google Scholar]
- 4.Bentley R, Meganathan R. FEBS Lett. 1981;125:220–222. doi: 10.1016/0014-5793(81)80723-5. [DOI] [PubMed] [Google Scholar]
- 5.Gerber N N. CRC Crit Rev Microbiol. 1979;7:191–214. doi: 10.3109/10408417909082014. [DOI] [PubMed] [Google Scholar]
- 6.Hassett A J, Rohwer E R. J Chromatogr. 1999;849:521–528. doi: 10.1016/s0021-9673(99)00621-4. [DOI] [PubMed] [Google Scholar]
- 7.Darriet P, Pons M, Lamy S, Dubourdieu D. J Agric Food Chem. 2000;48:4835–4838. doi: 10.1021/jf0007683. [DOI] [PubMed] [Google Scholar]
- 8.Heil T P, Lindsay R C. J Environ Sci Health. 1988;23:489–512. doi: 10.1080/03601238809372621. [DOI] [PubMed] [Google Scholar]
- 9.Cane D E, Oliver J S, Harrison P H M, Abell C, Hubbard B R, Kane C T, Lattman R. J Am Chem Soc. 1990;112:4513–4524. [Google Scholar]
- 10.Cane D E, Tillman A M. J Am Chem Soc. 1983;105:122–124. [Google Scholar]
- 11.Cane D E, Sohng J-K, Lamberson C R, Rudnicki S M, Wu Z, Lloyd M D, Oliver J S, Hubbard B R. Biochemistry. 1994;33:5846–5857. doi: 10.1021/bi00185a024. [DOI] [PubMed] [Google Scholar]
- 12.Hohn T M. In: Comprehensive Natural Products Chemistry: Isoprenoids Including Carotenoids and Steroids. Cane D E, editor. Oxford: Elsevier; 1999. pp. 201–215. [Google Scholar]
- 13.Trapp S C, Croteau R B. Genetics. 2001;158:811–832. doi: 10.1093/genetics/158.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lesburg C A, Zhai G, Cane D E, Christianson D W. Science. 1997;277:1820–1824. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
- 15.Seemann M, Zhai G, de Kraker J W, Paschall C M, Christianson D W, Cane D E. J Am Chem Soc. 2002;124:7681–7689. doi: 10.1021/ja026058q. [DOI] [PubMed] [Google Scholar]
- 16.Starks C M, Back K, Chappell J, Noel J P. Science. 1997;277:1815–1820. doi: 10.1126/science.277.5333.1815. [DOI] [PubMed] [Google Scholar]
- 17.Caruthers J M, Kang I, Rynkiewicz M J, Cane D E, Christianson D W. J Biol Chem. 2000;275:25533–25539. doi: 10.1074/jbc.M000433200. [DOI] [PubMed] [Google Scholar]
- 18.Rynkiewicz M J, Cane D E, Christianson D W. Proc Natl Acad Sci USA. 2001;98:13543–13548. doi: 10.1073/pnas.231313098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kieser T, Bibb M J, Buttner M J, Chater K F, Hopwood D A. Practical Streptomyces Genetics. Norwich, U.K.: John Innes Foundation; 2000. [Google Scholar]
- 20.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 21.Bentley S D, Chater K F, Cerdeno-Tarraga A M, Challis G L, Thomson N R, James K D, Harris D E, Quail M A, Kieser H, Harper D, et al. Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 22.Gust B, Challis G L, Fowler K, Kieser T, Chater K F. Proc Natl Acad Sci USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 24.Bradford M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Hill A M, Cane D E, Mau C J, West C A. Arch Biochem Biophys. 1996;336:283–289. doi: 10.1006/abbi.1996.0559. [DOI] [PubMed] [Google Scholar]
- 26.Gansser D, Pollak F C, Berger R G. J Nat Prod. 1995;58:1790–1793. [Google Scholar]
- 27.Toyota M, Yoshida T, Matsunami J, Asakawa Y. Phytochemistry. 1997;44:293–298. [Google Scholar]
- 28.Peters R J, Croteau R B. Proc Natl Acad Sci USA. 2002;99:580–584. doi: 10.1073/pnas.022627099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jones, R. V. H. & Sutherland, M. D. (1968) J. Chem. Soc. Chem. Commun., 1229–1230.
- 30.Valle M G, Appendino G, Nano G M, Picci V. Phytochemistry. 1987;26:253–256. [Google Scholar]
- 31.Appendino G, Jakupovic J, Alloatti S, Ballero M. Phytochemistry. 1997;45:1639–1643. [Google Scholar]
- 32.Zhabinskii V N, Minnaard A J, Wijnberg J B P A, de Groot A. J Org Chem. 1996;61:4022–4027. doi: 10.1021/jo9602534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.