Abstract
The mammalian soluble epoxide hydrolase (sEH) is an enzyme with multiple functions, being implicated in detoxification of xenobiotic epoxides as well as in regulation of physiological processes such as blood pressure. The enzyme is a homodimer, in which each subunit is composed of two domains. The 35-kDa C-terminal domain has an α/β hydrolase fold and harbors the catalytic center for the EH activity. The 25-kDa N-terminal domain has a different α/β fold and belongs to the haloacid dehalogenase superfamily of enzymes. The catalytic properties of the enzyme reported so far can all be explained by the action of the C-terminal domain alone. The function of the N-terminal domain, other than in structural stabilization of the dimer, has therefore remained unclear. By structural comparison of this domain to other haloacid dehalogenase family members, we identified a putative active site containing all necessary components for phosphatase activity. Subsequently, we found rat sEH hydrolyzed 4-nitrophenyl phosphate with a rate constant of 0.8 s−1 and a Km of 0.24 mM. Recombinant human sEH lacking the C-terminal domain also displayed phosphatase activity. Presence of a phosphatase substrate did not affect epoxide turnover nor did epoxides affect dephosphorylation by the intact enzyme, indicating both catalytic sites act independently. The enzyme was unable to hydrolyze 4-nitrophenyl sulfate, suggesting its role in xenobiotic metabolism does not extend beyond phosphates. Thus, we propose this domain participates instead in the regulation of the physiological functions associated with sEH.
Epoxide hydrolases (EHs) comprise a family of enzymes that convert oxiranes, i.e., heterocyclic compounds with a three-membered ring system containing one oxygen, to vicinal diols (1). Historically, this reaction has been regarded as a detoxification process, because epoxides often possess genotoxic potency due to their electrophilic reactivity (2). This function is attributable to the two major mammalian EHs, the microsomal EH and the soluble EH (sEH), which detoxify a large variety of xenobiotic-derived epoxides with complementary yet overlapping substrate specificity.
In recent years, evidence has accumulated that EHs also play a role in the processing of important signaling molecules. First, Hammock and coworkers (3) reported that sEH converts leukotoxin, the putative mediator of adult respiratory distress syndrome and multiple organ failure developing after large body burns, to leukotoxin diol. Further, the authors demonstrated that leukotoxin itself does not act as the inflammatory mediator but is actually the precursor of the biologically active leukotoxin diol. Second, it was recently reported that spontaneously hypertensive rats have increased renal expression of sEH and systemic administration of sEH inhibitors effectively lowered blood pressure in these animals (4). In line with this observation, mice lacking the sEH gene display a blood pressure phenotype (5). Male wild-type mice had a significantly higher systolic blood pressure than male sEH−/− mice; no such difference was evident among the female equivalents. These results are in perfect agreement with the large gender difference in mice with respect to sEH expression, with males having particularly high constitutive levels of sEH. Current thinking attributes the influence of sEH on blood pressure to its ability to hydrolyze vasoactive epoxyeicosatrienoic acids. These compounds have been described as a new class of endothelium-derived hyperpolarizing factors (6), and their increased breakdown is a likely cause of the sEH-dependent increase in blood pressure.
When the first mammalian sEHs were cloned (7–9), it became evident that the EH activity of the enzyme was attributable to a large C-terminal domain. This domain was suggested to adopt an α/β hydrolase fold with a catalytic triad, due to its sequence similarity to the bacterial haloalkane dehalogenase (10, 11). The identity of the amino acid residues constituting the catalytic triad of sEH and their essential role in the hydrolysis of epoxides were proven by biochemical analysis and site-directed mutagenesis (12–14). The C-terminal EH domain is connected via a proline-rich linker to a smaller N-terminal domain, showing sequence similarity to bacterial haloacid dehalogenase, an enzyme related by function but not by structure to haloalkane dehalogenase, and this N-terminal domain was thus expected to have a fold unrelated to that of the C-terminal domain (15). Its possible function remained unclear. Koonin and Tatusov (16) analyzed the relationship between haloacid dehalogenases, the N-terminal EH domain, and other enzymes and identified a large group of structural relatives called the haloacid dehalogenase (HAD) superfamily, which includes, among other enzymes, a number of phosphatases (16).
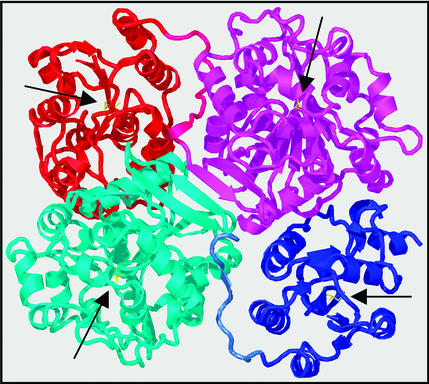
Very recently, the 3D structure of mouse sEH was elucidated (17). As expected, the enzyme was observed as a homodimer. Dimer formation is the result of interactions between the two C-terminal domains, as well as the association between the N-terminal domain of one subunit and the C-terminal domain of its dimerization partner; no interactions between the two N-terminal domains were observed (Fig. 1). The authors speculated that the structural role of the N-terminal domain in dimerization might be its most important function and, thus lacking any evidence for a catalytic activity, dubbed it the vestigial domain.
Figure 1.
3D structure of mammalian sEH. The sEH structure is displayed as a cartoon prepared by using the program O (21). Coordinates were taken from the Protein Data Bank (ID code 1cqz) (17). The N- and C-terminal domains of one subunit are colored in blue and cyan, respectively, whereas they are red and magenta in the other subunit. The side chains of the catalytic nucleophiles of all four active sites (see text) are shown in yellow. Their respective positions are indicated by black arrows.
Because the sEH orthologues in plants have EH activity similar to the mammalian enzymes yet lack the N-terminal domain and apparently exist as monomers (15), it seemed likely to us that the function of this domain was not exclusively structural. Inspection of the mouse sEH crystallographic structure showed a cavity in the N-terminal domain, which corresponds to the active site in all known HAD family members. In fact, this cavity was lined with conserved residues. Comparison with other sequences and structures suggested that the pattern of conservation was most consistent with the action of a phosphatase. It is now clear from our biochemical results that sEH can indeed act as a phosphatase and that the dephosphorylation activity is associated with the N-terminal domain. The possible physiological role of the two enzymatic functions in sEH is discussed.
Methods
In Silico Analyses.
The first 221 amino acid residues of rat, mouse, and human sEH were analyzed by using blast (18) and hidden Markov models (19). The coordinates of the same amino acid residues from the A chain of 1cqz, a Protein Data Bank entry for the mouse sEH structure (17), were analyzed by using dali (20). Structural alignments and interpretation made use of dali and the graphics program o (21). Fig. 1 was prepared with o and molray (22).
Purification of Native Rat sEH.
Six 6-week-old male Fischer rats were fed a standard lab chow (Altromin, Lage, Germany) fortified with 0.5% tiadenol to induce hepatic expression of sEH. After 8 days on the diet, animals were fasted for 1 day and killed by cervical dislocation, and the livers were excised and homogenized in ice-cold 100 mM potassium phosphate, pH 7.4. All subsequent steps were performed at 4°C. Cytosol was obtained as the supernatant after ultracentrifugation of the homogenate for 1 h at 100,000 × g. Purification of sEH was carried out as a single-step affinity purification on benzyl thiosepharose with 4-fluorochalcone oxide as the eluting agent as described by Wixtrom et al. (23). The enzyme eluted from the column at a concentration of 0.5 mg/ml and could be stored at −70°C until further use without loss of activity.
Enzyme Assays.
EH activity of sEH using trans-stilbene oxide, styrene oxide, and epoxystearic acid as substrates was measured as described (24). Phosphatase activity using 4-nitrophenyl phosphate as substrate was performed as a continuous assay after the formation of 4-nitrophenol at 405 nm in a Spectramax 250 microplate reader (Molecular Devices). Standard buffer conditions were 2 mM 4-nitrophenyl phosphate in 50 mM Tris⋅HCl/50 mM NaCl/10 mM MgCl2, pH 7.8, in a total volume of 250 μl. The reaction was started by addition of enzyme, typically corresponding to 0.5–20 μg of pure sEH, and allowed to proceed at 37°C for up to 60 min. Under these conditions, the product formation rate remained constant over time. Hydrolysis of 4-methylumbelliferyl phosphate was measured in the same buffer containing varying amounts of the substrate. Formation of 4-methylumbelliferone was monitored at excitation and emission wavelengths of 350 and 455 nm, respectively, in a Spectramax Gemini XS microplate fluorimeter. Due to the high absorbance of the substrate at the excitation wavelength, the change in fluorescence could not be monitored continuously when the substrate concentration exceeded 1 mM. In such a case, 10 μl of the assay mixture was transferred to a well containing 190 μl of EDTA, 100 mM, pH 8.0, to stop the reaction, and fluorescence was subsequently measured. This was done at three separate time points, usually 10, 20, and 30 min after the start of the reaction.
Recombinant Expression and Purification of sEH.
Human sEH cDNA was obtained via RT-PCR from human liver RNA isolated by the method of Chomczynski and Sacchi (25) and verified by sequencing. The coding sequence corresponding to the N-terminal 221 amino acids of the enzyme was equipped with a 3′-extension coding for a histidine hexamer (His-tag) and cloned into pGEF II (26), a T7 RNA polymerase-dependent bacterial expression vector. Transformation of this construct into Escherichia coli BL21-AI and induction with arabinose in LB led to recombinant expression of the His-tagged N-terminal domain of human sEH in the bacteria. A mutant coding for an Asp-9→Ala substitution was prepared from the above expression construct by using the QuikChange Site-Directed Mutagenesis kit (Stratagene), with the mutation primers 5′-GCCGTCTTCGCCCTTGACGGG-GTGCTGGCG-3′ and 5′-CCCCGTCAAGGGCGAAGACGGCCGCGCGCA-3′. For enzyme production, bacteria were grown at 30°C in a Fernbach flask in LB containing 100 μg of ampicillin per milliliter. At an OD600 of 0.6, cultures were induced with 1–100 μM arabinose and allowed to grow further overnight at 30°C or room temperature. After harvesting by centrifugation, the pellet was resuspended in ammonium hydrogen carbonate buffer, pH 7.4, 20 mM. Cells were subsequently broken by one pass through a FrenchPress pressure cell (Aminco) at 30,000 psi. The resulting lysates were clarified by centrifugation at 4,000 × g for 10 min. Purification of the recombinant His-tagged sEH fragment was performed on a HiTrap chelating HP column (AP Biotech, Freiburg, Germany) according to the instructions of the manufacturer.
For the production and purification of the full-length human sEH, the complete coding sequence was cloned into pASK-IBA7, a tetracylin-inducible bacterial expression vector. After transformation into E. coli XL1blue and induction of recombinant enzyme synthesis with anhydrotetracyclin, this construct directs the synthesis of sEH carrying an N-terminal strep-tag fusion. This tag is essentially an 8-aa oligopeptide that mimics biotin, and it could thus be used as an affinity tag for the single-step isolation of the protein with a streptavidin column, according to the instructions of the manufacturer (IBA, Goettingen, Germany). All expression constructs were sequenced before use to confirm the correctness of the encoded amino acid sequence.
The full-length rat sEH was expressed in bacteria by using the expression construct pRSET B-sEH as described (7, 14), with the exception that E. coli BL21-AI was used as the expression host.
Results
Structural Similarity of the sEH N Terminus to Haloacid Dehalogenases and Phosphatases.
First, we reinvestigated the phylogenetic relationship between the sEH N-terminal domain and other enzymes available in the public databases. A blast search using the human sEH N-terminal domain as the probe unearthed a large number of significantly related sequences. Due to the efficacy of the different genome projects, most of these identified hits were deduced from nucleic acid sequences and represented proteins of as-yet-unproven function. The closest relative found with this strategy that has a documented, non-sEH catalytic activity was the haloacid dehalogenase from Pseudomonas sp. (27). A number of other enzymes, including several phosphatases and mutases, received somewhat poorer scores.
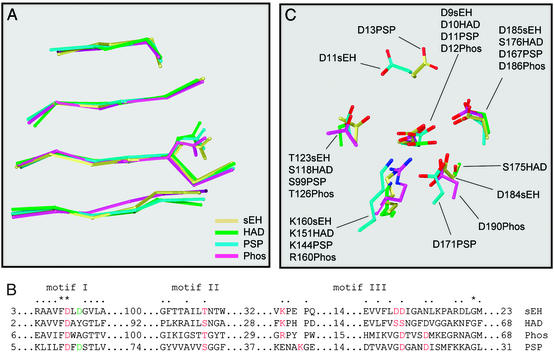
A second search, based on structural similarity as assessed by the program dali, came up with a slightly different result. As judged from the Z score, an indicator of the statistical significance of the observed relation, three proteins were identified as being particularly similar to the N-terminal domain (Z scores in the range of 9–12, each having ≈135 residues matching with a rms distance of 3 Å). These were the phosphonoacetaldehyde hydrolase from Bacillus cereus (28), the haloacid dehalogenase from Xanthobacter autotrophicus (29), and the phosphoserine phosphatase from Methanococcus jannaschii (30), in order of decreasing similarity. The relationship was limited in each case to the α/β fold that bears most of the catalytic residues; the structures and sequences of the active site lids vary widely.
Superposition of the core β sheets of the three structures on the β sheet in the N-terminal domain of mouse sEH confirmed strong similarity and allowed the alignment of residues implicated in the catalytic mechanism of the different (non-sEH) enzymes (Fig. 2). The equivalent of sEH Asp-9 has been identified as the catalytic nucleophile of the three other enzymes and is invariably an aspartic acid. The surrounding residues are also very well conserved (motif I, Fig. 2B), with some important exceptions. The equivalent of Asp-11 is only aspartate in the serine phosphatase sequence (Fig. 2B); its role in this is to bind to both the phosphate group and the divalent cation (presumably magnesium) that is required for catalysis. The cation facilitates the binding of a phosphate substrate near the catalytic aspartate, by buffering their electrostatic repulsion. In motif II, all enzymes have a serine or threonine in the same spatial position; this is also associated with binding the phosphate group in the phosphatases. In all of the enzymes, a basic group (from the side chain of arginine or lysine) is found at the same position in the active site, although it may be derived from a different position within the sequence and main-chain structure. The role of this residue is generally to stabilize a negatively charged intermediate. It is particularly interesting to see that the sEH, on the one hand, resembles the structure of haloacid dehalogenase in that the two potentially catalytic residues in motif III (Fig. 2B) are neighbors, whereas they are three residues apart from each other in the phosphatase sequences. However, the nature and placement of these side chains of sEH (Asp-184 and -185; Fig. 2C) are clearly more similar to their equivalents in the phosphatases; the haloacid dehalogenases have serine residues in these positions instead. In the phosphatases, the two aspartic acid residues are implicated in Mg2+ binding, either directly or indirectly through a water molecule. The above analyses led us to suspect that the N-terminal domain of sEH could act as a phosphatase.
Figure 2.
Structural/sequence alignment of mammalian sEH with haloacid dehalogenase, phosphonoacetaldehyde hydrolase, and phosphoserine phosphatase. Based on the DALI results (see text), four structures were superimposed. (A) The alignment of the four central strands of the β sheet, in the order β2, -1, -4, and -5 from top to bottom [strand designation according to Morais et al. (28)], including the catalytic nucleophile sidechain. (B) A structure-based sequence alignment in the vicinity of (potential) catalytic residues, which are marked in red. Designation of motifs is adopted from Wang et al. (30), with the numbers of residues in the linking segments given for each sequence. (C) The spatial arrangement of these residues. Phos, phosphonoacetaldehyde hydrolase; PSP, phosphoserine phosphatase. Protein Data Bank entries are 1cqz (sEH), 1jud (HAD), and 1fez (Phos). 1f5s (PSP). The spatial arrangement observed for PSP has also been seen more recently in the structure from another phosphatase from the HAD family (38).
Native Rat sEH Displays Phosphatase Activity.
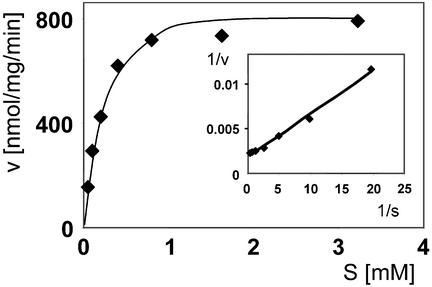
To determine whether sEH does catalyze dephosphorylation, we incubated purified rat sEH with a generic phosphatase substrate, 4-nitrophenyl phosphate, under standard conditions. The enzyme did indeed hydrolyze this substrate with a turnover number of 0.8 per sec and a Km of 0.235 mM (Fig. 3). This is at approximately the same rate as the reaction observed for sEH with its diagnostic epoxide, the substrate trans-stilbene oxide (turnover rate 0.5 s−1), thus indicating that the native holoenzyme is a bona fide phosphatase. In the absence of Mg2+ from the incubation buffer, the enzyme showed strongly reduced phosphatase activity that could be completely eliminated by the inclusion of 20 mM EDTA in the buffer. This is in good agreement with the expectation that a divalent cation would be a necessary cofactor in the catalytic site of the N-terminal domain.
Figure 3.
4-Nitrophenyl phosphate hydrolysis by native rat sEH. Concentration-dependent 4-nitrophenol dephosphorylation is shown in the Michaelis–Menten representation. (Inset) The corresponding Lineweaver–Burk plot.
The Recombinant N-Terminal Domain of Human sEH Shows Phosphatase Activity.
Because we could not exclude the possibility that the phosphatase activity measured with the native sEH resides in the C-terminal (EH) domain of the enzyme, we expressed the N-terminal domain, corresponding to the first 221 amino acid residues of the human sEH, in E. coli. Recombinant bacteria expressed the truncated enzyme at appreciable levels as a soluble protein under permissive conditions, reaching ≈5% of the total cellular protein. The phosphatase activity in crude extracts from bacteria expressing the N-terminal sEH domain was enhanced by a factor of 3 as compared with the vector-only control, indicating that the recombinant protein contributed significantly to the enzymatic activity.
To purify the recombinant protein, we expressed the N-terminal domain fused to a His-tag. This allowed us to recover the soluble fusion protein by means of immobilized metal affinity chromatography. The recombinant expression product displayed a half life of 18 h when stored at +4°C. The freshly prepaired enzyme had a turnover rate of 0.013 s−1 and a Km of 2.3 mM with 4-nitrophenyl phosphate, thus being considerably less active with this substrate than the native rat sEH (see Table 1).
Table 1.
Kinetic constants of sEH with 4-nitrophenyl phosphate
| Km, mM | Vmax, nmol/mg/min | kcat, s−1 | kcat/Km, M−1⋅s−1 | |
|---|---|---|---|---|
| Native rat sEH | 0.24 | 810 | 0.84 | 3,500 |
| Recombinant rat sEH | 0.37 | 1450 | 1.61 | 4,400 |
| Human N-terminal domain, wild type | 2.3 | 31 | 0.013 | 5.7 |
| Human N-terminal domain, Asp-9→Ala | ND | ND | ND | ND |
| Human full-length sEH | 1.1 | 5.8 | 0.0061 | 5.5 |
ND, not detectable.
Because the structural alignment identified Asp-9 as the potential catalytic nucleophile of the N-terminal domain that should play an essential role in catalysis, we substituted this residue with alanine by site-directed mutagenesis. Indeed, the respective purified mutant enzyme lacked any detectable phosphatase activity (Table 1; detection limit for turnover number: 0.0005 s−1), in line with our deductions from the structural comparisons.
Recombinant Expression of Full-Length Rat sEH in E. coli Gives High 4-Nitrophenyl Phosphatase Activity.
The striking difference in activity between the native rat enzyme and the recombinant human N-terminal domain could be due to either a species difference or a technical artifact. To find out whether the heterologous expression compromises the phosphatase activity of sEH, we expressed the rat holoenzyme in E. coli as described earlier, with the exception that the strain BL21-AI was used as the expression host. This turned out to be a substantial improvement as compared with our previous work (14), because this time, we could recover most of the recombinant protein as soluble active enzyme under optimized conditions (Fig. 4). The purified recombinant rat sEH produced this way displayed a turnover rate rather similar to the native enzyme purified from rat livers (Table 1), indicating that expression in E. coli does not compromise the catalytic activity of sEH, in general. However, with the results obtained so far, it was not clear whether the difference in turnover rate between rat holoenzyme and human N-terminal domain was due to a species difference, a structural stabilization of the N-terminal domain by the C-terminal one, or a contribution of the C-terminal domain to phosphatase activity.
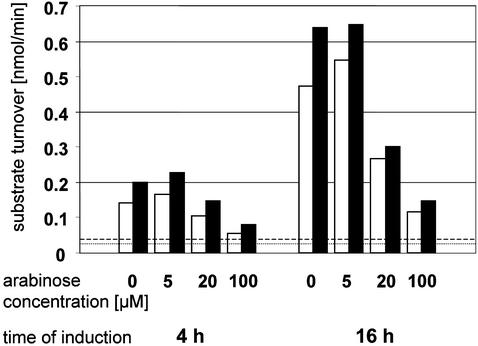
Figure 4.
Recombinant expression of the full-length rat sEH in E. coli BL21-AI. The bar graph shows the relationship between the concentration of the inducer arabinose and the yield of enzymatically active sEH. The open bars represent the hydrolysis of trans-stilbene oxide obtained with 60 μg of protein from the bacterial lysate supernatant, whereas the filled bars show the 4-nitrophenyl phosphate hydrolysis with 40 μg of the same material. Constitutive phosphatase activity levels and spontaneous transstilbene oxide hydrolysis levels in sEH-free E. coli lysates are indicated by broken and dotted lines, respectively. Notably, the EH activity is perfectly paralleled by the phosphatase activity. The production of active enzyme is maximal at an arabinose concentration of 5 μM. Beyond that, enhanced inclusion body formation leads to a disproportionate increase in the amount of improperly folded recombinant sEH, and the yield in active sEH drops substantially.
Recombinant Expression of Full-Length Human sEH in E. coli Results in Moderate 4-Nitrophenyl Phosphatase Activity.
To decide whether interdomain stabilization, species difference, or phosphatase activity of the C-terminal domain causes the higher activity of the rat holoenzyme as compared with the human N-terminal domain, we expressed the complete human sEH in E. coli. The phosphatase activity of the soluble holoenzyme obtained did not differ significantly from that of the N-terminal domain expressed separately (Table 1). This indicates that a species difference between rat and human, rather than domain stabilization or contribution of the C-terminal domain to the phosphatase activity, is the reason for the observed difference in the turnover rate of 4-nitrophenyl phosphate.
No Indication for Cross Talk Between Domains.
The physical connection between the EH domain and the phosphatase domain in sEH suggested the intriguing idea that the N-terminal domain might possess a regulatory function and so control the rate of epoxide hydrolysis. We therefore compared the kinetics of epoxide hydrolysis in the presence and absence of 4-nitrophenyl phosphate with the native rat sEH. However, no change in the rate constant or Km for epoxide hydrolysis was observed in the presence of the phosphatase substrate (final concentration 3 mM; Km, 0.24 mM). Likewise, the presence of the EH substrate trans-stilbene oxide (final concentration 25 μM; Km, 3 μM), or the inhibitors dicyclohexyl urea (final concentration 100 μM; Ki, 0.03 μM) or 4-fluorochalcone oxide (final concentration 1 mM; Ki, 0.5 μM) did not affect the kinetics of 4-nitrophenyl phosphate hydrolysis. Thus, the two domains seem to operate independently under our experimental conditions. Furthermore, the lack of effect of the inhibitors on the phosphatase activity presents final proof that the phosphatase activity resides exclusively in the N-terminal domain of sEH.
Other Substrates.
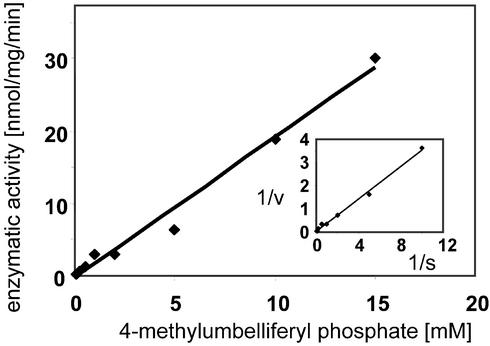
To explore a possible role of the N-terminal domain of sEH in xenobiotic metabolism, we tested the ability of the enzyme to hydrolyze 4-nitrophenyl sulfate. However, despite the similarity of the compound to 4-nitrophenyl phosphate, we could not detect any turnover by human or rat sEH, indicating that the catalytic domain is specifically acting on phosphate esters. As a second generic phosphatase substrate, we used 4-methylumbelliferyl phosphate. Turnover with the rat sEH could easily be detected and increased with the substrate concentration in a linear fashion up to 15 mM, the highest concentration that could be tested. Due to this lack of saturation, neither Km nor kcat could be determined, but the ratio kcat/Km, the indicator of enzyme performance, was calculated to be 3 M−1⋅s−1 from the slope of the Lineweaver–Burk plot (Fig. 5). This value is three orders of magnitude lower than the kcat/Km for 4-nitrophenyl phosphate with the same enzyme. Interestingly, with the human enzyme, the turnover was clearly detectable but too low for a valid quantification (kcat/Km < 0.01 M−1⋅s−1). Thus, the pronounced species difference was observed with two structurally different substrates.
Figure 5.
Enzyme kinetics of 4-methylumbelliferyl phosphate hydrolysis by rat sEH. Displayed is the dependence of the turnover rate on the substrate concentration. Note the lack of any indication of saturation. Therefore, the rate constant under saturating conditions cannot be determined. However, kcat/Km can be obtained by mathematical transformation of the slope of the Lineweaver–Burk plot (Inset), which itself represents Km/Vmax.
Discussion
In the present study, we show, to our knowledge for the first time, that mammalian sEH possesses phosphatase activity, and that this activity is a property of its N-terminal domain. Comparing the structure of this portion of sEH with other members of the HAD superfamily revealed a perfect spatial conservation of most catalytic residues and a particularly striking similarity to enzymes with known phosphatase activity. Catalysis in the various family members depends on the correct placement of active site groups with respect to the substrate. The catalytic conservation is not necessarily apparent from comparisons of the amino acid sequences alone, because some essential catalytic components can be contributed by different regions of the sequence (and structure). In the case of the mammalian sEH, the catalytic nucleophile is Asp-9, whereas Asp-11, Thr-123, Lys-160, Asp-184, and Asp-185 should be implicated in the binding of a divalent cation and/or the phosphate moiety of the substrate. Accordingly, pure native sEH displays phosphatase activity with the generic substrate 4-nitrophenyl phosphate. The activity resides in the N-terminal domain, as proven by the analysis of the truncated recombinant enzyme compared with the holoenzyme and the lack of effect of sEH inhibitors on phosphatase activity. Catalysis depends on the presence of Asp-9, as evidenced by the inactivity of the Asp-9→Ala mutant.
With the two generic phosphatase substrates used in the present study, the activity of the rat enzyme was orders of magnitudes higher than that of the human enzyme. In view of the fact that the constitutive expression of sEH in the livers of rats is rather low as compared with that in other species (31), including humans, one might speculate that the higher phosphatase activity of the rat enzyme compensates for this low expression level.
In view of the classical role of sEH as a detoxifying enzyme, processing potentially harmful epoxides, the association with a phosphatase activity was unexpected. In most cases, the roles of bifunctional proteins are in some way related, e.g., they have a physical interaction or are members of the same metabolic pathway. This property has allowed them, for example, to be used as “Rosetta stones” in genomics efforts (32). The recently uncovered role of the enzyme in regulatory processes, such as blood pressure regulation (4, 5), suggested that one reasonable role for the phosphatase domain of sEH would be to modulate the activity of the EH domain. In the calcium pump of the sarcoplasmic reticulum, a member of the P-type ATPases, a phosphatase domain that belongs to the HAD superfamily is apparently the driving force for the structural rearrangements that finally result in ion transport (33). By analogy, we hypothesized that the phosphatase domain might regulate the activity of the EH domain via conformational rearrangement. However, neither 4-nitrophenyl phosphate nor ATP (data not shown) affected the EH activity of sEH. Likewise, EH substrates and inhibitors did not influence the phosphatase activity of sEH. Therefore, direct interaction of the two active sites does not seem to take place. This is perhaps not unexpected, given the large distance between the EH and the phosphatase active centers (46 and 41 Å for the intra- and intersubunit distance, respectively; see Fig. 1).
In a second attempt to understand the relationship between the two domains, we sought a substrate of possible toxicological relevance. A group of obvious candidates is the sulfate esters. Sulfate itself is isosteric with phosphate, and some sulfate esters have been shown to be exceptionally active mutagens (34). However, we were unable to detect any sulfatase activity with 4-nitrophenyl sulfate, the sister compound to our proven phosphatase substrate. Thus, a role for the phosphatase domain in xenobiotic metabolism has not been established.
At this stage, it is possible to do little more than speculate about the potential physiological role played by the N-terminal domain of the sEH. However, it was recently observed that inhibitors of the sEH targeting only the activity of the C-terminal domain revealed effects that were markedly less impressive than effects observed in sEH−/− mice (5, 35). Thus, it is tempting to suggest that the phosphatase domain of the sEH plays a synergistic role with the EH in the regulation of the underlying physiological processes. A rather speculative yet intriguing hypothesis toward that direction is that the sEH phosphatase activity might be implicated in the down-regulation of NO synthase (NOS) activity: NOS and sEH have opposite effects on vascular resistance. While NOS generates the classical endothelial-derived relaxing factor NO, sEH breaks down vasoactive arachidonic acid epoxides, which are recognized endothelial-derived hyperpolarization factors. It has recently been reported that endothelial NOS, the isozyme particularly involved in blood pressure regulation, is activated by shear stress through phosphorylation at Ser-1177 (36). Because sEH has been detected in endothelial cells and is thus localized in the same cell type as the endothelial NOS (37), it is a very interesting candidate for a Ser-1177 phosphatase, counteracting the shear stress-induced endothelial NOS activation.
Acknowledgments
This work was supported by the German Research Foundation.
Abbreviations
- EH
epoxide hydrolase
- sEH
soluble EH
- HAD
haloacid dehalogenase
References
- 1.Arand M, Oesch F. In: Enzyme Systems That Metabolise Drugs and Other Xenobiotics. Ioannides C, editor. New York: Wiley; 2002. pp. 459–483. [Google Scholar]
- 2.Oesch F. Xenobiotica. 1973;3:305–340. doi: 10.3109/00498257309151525. [DOI] [PubMed] [Google Scholar]
- 3.Moghaddam M F, Grant D F, Cheek J M, Greene J F, Williamson K C, Hammock B D. Nat Med. 1997;3:562–566. doi: 10.1038/nm0597-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu Z, Xu F, Huse L M, Morisseau C, Draper A J, Newman J W, Parker C, Graham L, Engler M M, Hammock B D, et al. Circ Res. 2000;87:992–998. doi: 10.1161/01.res.87.11.992. [DOI] [PubMed] [Google Scholar]
- 5.Sinal C J, Miyata M, Tohkin M, Nagata K, Bend J R, Gonzalez F J. J Biol Chem. 2000;275:40504–40510. doi: 10.1074/jbc.M008106200. [DOI] [PubMed] [Google Scholar]
- 6.Fisslthaler B, Popp R, Kiss L, Potente M, Harder D R, Fleming I, Busse R. Nature. 1999;401:493–497. doi: 10.1038/46816. [DOI] [PubMed] [Google Scholar]
- 7.Knehr M, Thomas H, Arand M, Gebel T, Zeller H D, Oesch F. J Biol Chem. 1993;268:17623–17627. [PubMed] [Google Scholar]
- 8.Grant D F, Storms D H, Hammock B D. J Biol Chem. 1993;268:17628–17633. [PubMed] [Google Scholar]
- 9.Beetham J K, Tian T, Hammock B D. Arch Biochem Biophys. 1993;305:197–201. doi: 10.1006/abbi.1993.1411. [DOI] [PubMed] [Google Scholar]
- 10.Arand M, Grant D F, Beetham J K, Friedberg T, Oesch F, Hammock B D. FEBS Lett. 1994;338:251–256. doi: 10.1016/0014-5793(94)80278-5. [DOI] [PubMed] [Google Scholar]
- 11.Lacourciere G M, Armstrong R N. Chem Res Toxicol. 1994;7:121–124. doi: 10.1021/tx00038a001. [DOI] [PubMed] [Google Scholar]
- 12.Hammock B D, Pinot F, Beetham J K, Grant D F, Arand M E, Oesch F. Biochem Biophys Res Commun. 1994;198:850–856. doi: 10.1006/bbrc.1994.1121. [DOI] [PubMed] [Google Scholar]
- 13.Borhan B, Jones A D, Pinot F, Grant D F, Kurth M J, Hammock B D. J Biol Chem. 1995;270:26923–26930. doi: 10.1074/jbc.270.45.26923. [DOI] [PubMed] [Google Scholar]
- 14.Arand M, Wagner H, Oesch F. J Biol Chem. 1996;271:4223–4229. doi: 10.1074/jbc.271.8.4223. [DOI] [PubMed] [Google Scholar]
- 15.Beetham J K, Grant D, Arand M, Garbarino J, Kiyosue T, Pinot F, Oesch F, Belknap W R, Shinozaki K, Hammock B D. DNA Cell Biol. 1995;14:61–71. doi: 10.1089/dna.1995.14.61. [DOI] [PubMed] [Google Scholar]
- 16.Koonin E V, Tatusov R L. J Mol Biol. 1994;244:125–132. doi: 10.1006/jmbi.1994.1711. [DOI] [PubMed] [Google Scholar]
- 17.Argiriadi M A, Morisseau C, Hammock B D, Christianson D W. Proc Natl Acad Sci USA. 1999;96:10637–10642. doi: 10.1073/pnas.96.19.10637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karplus K, Barrett C, Hughey R. Bioinformatics. 1998;14:846–856. doi: 10.1093/bioinformatics/14.10.846. [DOI] [PubMed] [Google Scholar]
- 20.Holm L, Sander C. Nucleic Acids Res. 1999;27:244–247. doi: 10.1093/nar/27.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 22.Harris M, Jones T A. Acta Crystallogr D. 2001;57:1201–1203. doi: 10.1107/s0907444901007697. [DOI] [PubMed] [Google Scholar]
- 23.Wixtrom R N, Silva M H, Hammock B D. Anal Biochem. 1988;169:71–80. doi: 10.1016/0003-2697(88)90256-4. [DOI] [PubMed] [Google Scholar]
- 24.Müller F, Arand M, Frank H, Seidel A, Hinz W, Winkler L, Hänel K, Blee E, Beetham J K, Hammock B D, Oesch F. Eur J Biochem. 1997;245:490–496. doi: 10.1111/j.1432-1033.1997.00490.x. [DOI] [PubMed] [Google Scholar]
- 25.Chomczynski P, Sacchi N. Anal Biochem. 1986;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 26.Arand M, Hemmer H, Dürk H, Baratti J, Archelas A, Furstoss R, Oesch F. Biochem J. 1999;344:273–280. doi: 10.1042/0264-6021:3440273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hisano T, Hata Y, Fujii T, Liu J Q, Kurihara T, Esaki N, Soda K. J Biol Chem. 1996;271:20322–20330. doi: 10.1074/jbc.271.34.20322. [DOI] [PubMed] [Google Scholar]
- 28.Morais M C, Zhang W, Baker A S, Zhang G, Dunaway-Mariano D, Allen K N. Biochemistry. 2000;39:10385–10396. doi: 10.1021/bi001171j. [DOI] [PubMed] [Google Scholar]
- 29.Ridder I S, Rozeboom H J, Kalk K H, Janssen D B, Dijkstra B W. J Biol Chem. 1997;272:33015–33022. doi: 10.1074/jbc.272.52.33015. [DOI] [PubMed] [Google Scholar]
- 30.Wang W, Kim R, Jancarik J, Yokota H, Kim S-H. Structure (London) 2000;9:65–71. doi: 10.1016/s0969-2126(00)00558-x. [DOI] [PubMed] [Google Scholar]
- 31.Ota K, Hammock B D. Science. 1980;207:1479–1481. doi: 10.1126/science.7361100. [DOI] [PubMed] [Google Scholar]
- 32.Marcotte E M, Pellegrini M, Ng H L, Rice D W, Yeates T O, Eisenberg D. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 33.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 34.Glatt H. FASEB J. 1997;11:314–321. doi: 10.1096/fasebj.11.5.9141497. [DOI] [PubMed] [Google Scholar]
- 35.Fornage M, Hinojos C A, Nurowska B W, Boerwinkle E, Hammock B D, Morisseau C H, Doris P A. Hypertension. 2002;40:485–490. doi: 10.1161/01.hyp.0000032278.75806.68. [DOI] [PubMed] [Google Scholar]
- 36.Dimmeler S, Fleming I, Fisslthaler B, Hermann C, Busse R, Zeiher A M. Nature. 1999;399:601–605. doi: 10.1038/21224. [DOI] [PubMed] [Google Scholar]
- 37.Fang X, Kaduce T L, Weintraub N L, Harmon S, Teesch L M, Morisseau C, Thompson D A, Hammock B D, Spector A A. J Biol Chem. 2001;276:14867–14874. doi: 10.1074/jbc.M011761200. [DOI] [PubMed] [Google Scholar]
- 38.Parsons J F, Lim K, Tempczyk A, Krajewski W, Eisenstein E, Herzberg O. Proteins. 2002;46:393–404. doi: 10.1002/prot.10057. [DOI] [PMC free article] [PubMed] [Google Scholar]