Abstract
Reduced brain levels of long chain polyunsaturated fatty acids [arachidonic acid and docosahexanoic acid (DHA)] are observed in elderly subjects and patients with Alzheimer's disease. To determine the effects of n-3 fatty acids on aged rat brain, 2-year-old rats were fed fish oil (27% DHA content) for 1 month, and gene expression analysis and fatty acid and molecular species composition of the major phospholipid species were assessed. No significant alteration could be observed in the fatty acid composition of ethanolamine phosphoglycerides and phosphatidylserines with the exception of DHA, which was slightly higher in brains of rats receiving fish oil. However, a drastic reduction in arachidonic acid in phosphatidylinositoles was observed. The expression of 23 genes was altered in response to fish oil feeding in the hippocampus. The transcription of transthyretin (TTR) was induced by 10-fold as evidenced by microarray analysis and confirmed by real-time quantitative RT-PCR. Expression of IL-1 and NO synthase, which has been implicated in the prevention of neurological diseases, was unaltered. TTR is an amyloid β protein scavenger, so an increase in its expression could prevent amyloid aggregate formation. We believe the beneficial effects of fish oil might be common to other agents, i.e., induce TTR expression, like nicotine and Ginkgo biloba extract.
Low serum docosahexaenoic acid (DHA; 22:6n-3) level is a significant risk factor for the development of cognitive impairment and dementia (1). Low levels of DHA in the brain are associated with onset of sporadic Alzheimer's disease (AD) and cognitive decline during aging (2). DHA was shown to provide protection from impairment of learning ability in AD model rats (3), and it was observed that dietary DHA improved long-term potentiation in dentate gyrus in rats (4). Additionally, several epidemiological studies revealed an association between fish consumption and a low prevalence of AD (5, 6). In rats fed fish oil for a long period synaptic membrane fluidity and maze-learning ability were enhanced with respect to animals fed diets lacking omega 3 fatty acids (7–9). Various mechanisms have been suggested to account for these physiological changes in the brain and retina, as reviewed recently by Kurlack and Stephenson (10), Lauritzen et al. (11), and Salem et al. (12). Briefly, DHA plays a crucial role in membrane order (membrane fluidity), which can influence the function of membrane receptors such as rhodopsin (13, 14); regulation of membrane-bound enzymes (Na/K-dependent ATPase) (15); dopaminergic and serotoninergic neurotransmission (16); signal transduction via effects on inositol phosphates, diacylglycerol, and protein kinase C (17); regulation of the synthesis of eicosanoids derived from arachidonic acid (AA) (10); regulation of gene expression (18–20); regulation of phosphatidylserine levels (21); protection of neural cells from apoptotic death (22, 25); stimulation of neurite outgrowth in PC12 cells (23, 24); selective accumulation of DHA by synaptic growth cones during neuronal development (23, 24); regulation of nerve growth factor (29); and regulation of neuron size (26, 27).
The aim of the present study was to investigate the beneficial effects of dietary fish oil on rat brain at the molecular level and to identify genes exhibiting altered expression associated with learning functions in aged rats.
We have recently shown that dietary n-3 polyunsaturated fatty acids (PUFAs) can modulate the expression of numerous genes in the brain when the animals were kept on a diet rich in fish oil from conception until adulthood (19). In this study, the hippocampi of 2-year-old, essential fatty acid-sufficient rats and animals having rat chow supplemented with fish oil containing omega 3 PUFAs for 1 month were isolated, and fatty acid composition of the major phospholipids, choline, serine, inositol, and ethanolamine phosphoglycerides was determined. Previously, we found several candidate genes with altered expression in brain of young rats in response to long-term fish oil feeding including those controlling synaptic plasticity, cytoskeleton and membrane association, ion channel formation, signal transduction, and energy metabolism. Here, we fed aged rats fish oil for a short period and followed the gene expression alterations in the hippocampus by cDNA microarrays and real-time quantitative RT-PCR (RT-QPCR).
Experimental Procedures
Animals, Diets, Brain, and RNA Preparation.
Four groups of six male Wistar rats were studied. Two groups were fed rat chow supplemented with fish oil [5% eicosapentaenoic acid (EPA) and 27% DHA; total fat content: 8%] and two were kept on normal rat chow (total fat content: 4%). Two-year-old rats were placed on these diets for 4 weeks. Fatty acid composition of the diets is given in Table 1. Individual hippocampus (containing the choroid plexus) samples were dissected, and ≈25 mg of tissue from each animal was pooled into four groups (two oil-fed and two control groups). Brain samples were immediately subjected to RNA preparation. Total RNA was purified from each pool with a NucleoSpin RNA purification kit (Macherey & Nagel) according to the manufacturer's instructions. RNA preparations from each group were pooled, and their quality and quantity were assessed by gel electrophoresis and OD260/OD280 ratios. Total RNA was used for microarray analysis and real-time RT-QPCR.
Table 1.
Fatty acid composition of diets
| Fatty acids, wt% | Control | Fish |
|---|---|---|
| 16:0 | 11.6 | 18.4 |
| 16:1n-7 | 0.9 | 3.2 |
| 18:0 | 2.9 | 7.1 |
| 18:1n-9 | 18.0 | 23.0 |
| 18:1n-7 | — | 0.1 |
| 18:2n-6 | 53.6 | 22.9 |
| 18:3n-3 | 6.4 | 2.0 |
| 20:1n-9 | — | 0.6 |
| 20:4n-6 | 0.1 | 0.8 |
| 20:5n-3 | 0.6 | 2.8 |
| 22:5n-3 | 0.2 | 0.5 |
| 22:6n-3 | 1.1 | 11.2 |
| Lipid cont., wt% | 4 | 8 |
Only the most common fatty acids are listed (calculated from triplicates).
Analysis of Phospholipids.
Lipids were extracted from individual hippocampi as described (19). Fatty acid composition of phospholipid classes obtained by TLC was determined by gas chromatography with a FFAP column (30 m, 0.25 mm i.d., Supelco). Molecular species composition of ethanolamine phosphoglycerides, separated by TLC (19), was determined by reversed-phase HPLC with a Supelcosyl LC-18 column (2.1 i.d. 25 cm, Supelco) and acetonytrile/isopropanol (85:15, vol/vol) as solvent, after segregation of the anthroxylated diradyl glycerides into diacyl, alkylacyl, and alkenylacyl subclasses as recommended by Takamura and Kito (28).
Construction of Microarrays.
Construction and use of microarrays were performed as described (19). Briefly, 3,200 amplified cDNA inserts from rat brain, liver, and periferial ganglion libraries were arrayed in duplicate on amino-silanized slides (Sigma) with a MicroGrid Total Array System spotter (BioRobotics, Cambridge, U.K.). The diameter of each spot was ≈250 μm. After printing, DNA was UV-crosslinked to the slides with 700 mJ energy (Stratalinker, Stratagene). Before hybridization, the slides were blocked in 1× SSC, 0.2% SDS, 1% BSA for 30 min at 42°C, rinsed in water, and dried.
Microarray Probe Preparation and Hybridization.
Three micrograms of total RNA from each sample was amplified by a linear antisense RNA amplification method and labeled with Cy3 or Cy5 fluorescent dyes during reverse transcription as described (30). Briefly, 2.5 μg of amplified RNA was labeled with Cy3-dCTP or Cy5-dCTP (NEN) during reverse transcription in 20 μl total volume. After a 2 h incubation at 37°C, heteroduplexes were purified as described (31) and denaturated, and the mRNA was hydrolyzed with NaOH for 15 min at 37°C and neutralized. The labeled cDNA was purified with a PCR purification kit (Macherey & Nagel) according to the manufacturer's instructions. Probes generated from the control and oil-fed samples were mixed, dissolved in 15 μl of hybridization buffer (50% formamide/5× SSC/0.1% SDS/100 μg/ml salmon sperm DNA), and applied onto the array after denaturation by heating for 1 min at 90°C. After hybridization at 42°C for 20 h in a humid hybridization chamber, slides were washed as described (19).
Scanning and Data Analysis.
Each array was scanned under a green laser (532 nm; for Cy3 labeling) or a red laser (660 nm; for Cy5 labeling) by using a ScanArray Lite (GSI Lumonics, Billerica, MA) scanning confocal fluorescent scanner with 10-μm resolution. Image analysis was performed with SCANALYZE2 software (www.microarrays.org/software.html). Analysis of replica spots and experiments, local background corrections, and statistical analysis were performed as described (19). Significant spots have >0.55 CHGTB2 values in both Cy3 and Cy5 channels. These values mean the fraction of pixels in the spot is >1.5 times the background, which can be used to filter out weak spots and spots where the intensity comes from a few bright pixels having high mean intensities but low values. Each experiment was performed twice with both fluorescent dyes for labeling control and sample to reduce the number of false-positive or false-negative ratios deriving from possible uneven incorporation of the dyes during labeling or from other experimental variables introduced by hybridization, washing conditions, or array features.
Real-Time RT-QPCR.
Real-time RT-QPCR was performed on a RotorGene 2000 instrument (Corbett Research, Sydney) with gene-specific primers (designed with the software primerexpress, Applied Biosystems) and SYBRGreen I protocol to confirm the gene expression changes observed with microarrays. Twenty micrograms of total RNA from each pool was reverse-transcribed in the presence of poly(dT) sequences in a total volume of 20 μl. After dilution of the mix with 30 μl of water, 1 μl of this mix was used as template in the RT-QPCR. All of the reactions were performed four times with the following cycling protocol: 360-s heat start at 95°C, 45 cycles of denaturation at 95°C for 25 s, annealing at 59°C for 30 s, and extension at 72°C for 20 s. Fluorescence detection was performed at 72°C. Relative expression ratios were normalized to β-actin. The PCR primers used were as follows: β-actin, forward primer, 5′-TTCAACACCCCAGCCATGT-3′, reverse primer, 5′-GCATACAGGGACAACACAGCC-3′; transthyretin (TTR), forward primer, 5′-GGCTCACCACAGATGAGAAGTTC-3′, reverse primer, 5′-ACAAATGGGAGCTACTGCTTTGGC-3′; IL-1α, forward primer, 5′-GCAGCTTTCCCAGAGCTGTT-3′, reverse primer, 5′-GCAGTCCCCG TGCCAG-3′; IL-1β, forward primer, 5′-CTTCCCCAGGACATGCTAGG-3′, reverse primer, 5′-CAAAGGCTTCCCCTGGAGAC-3′; and NO synthase (NOS), forward primer, 5′-AATGCGGAAGGTCATGGC-3′, reverse primer, 5′-CGACTTTCCTGTC TCAGTAGCAAA-3′.
Results
Fatty Acid Composition of the Hippocampus.
Fatty acid composition of major phospholipids in the hippocampus of the experimental animals is shown in Table 2. No significant alteration could be observed in the fatty acid composition of choline phosphoglyceride and phosphatidylserine (Table 2). As seen in Table 2, the fatty acid composition of ethanolamine phosphoglycerides was almost identical in the two hippocampus samples (control and animals fed fish oil) with the exception of DHA, which is slightly higher in brains of animals receiving fish oil. However, fish oil feeding brought about a significant reduction in AA and a slight but not significant increase in palmitic and stearic acids in the phosphatidylinositol fraction.
Table 2.
Fatty acid composition of phosphatidylcholine, phosphatidylserine, phosphatidylethanolamine, and phosphatidylinositol in relation to diet in rat hyppocampus
| Fatty acid | Phosphatidylcholine
|
Phosphatidylserine
|
Phosphatidylethanolamine
|
Phosphatidylinositol
|
||||
|---|---|---|---|---|---|---|---|---|
| CTR | FO | CTR | FO | CTR | FO | CTR | FO | |
| DMA 16:0 | — | — | — | — | 3.90 ± 0.69 | 3.56 ± 0.47 | — | — |
| 16:0 | 42.29 ± 4.14 | 44.43 ± 2.99 | 6.98 ± 1.61 | 4.73 ± 1.15 | 7.15 ± 0.91 | 7.03 ± 1.22 | 18.29 ± 1.80 | 22.82 ± 1.72 |
| 16:1n-7 | 1.01 ± 0.28 | 1.21 ± 0.18 | 0.85 | 1.02 ± 0.13 | 2.08 ± 0.65 | 1.65 ± 1.08 | 1.52 ± 0.45 | 2.10 ± 0.59 |
| DMA 18:0 | — | — | — | — | 5.14 ± 1.14 | 5.24 ± 0.99 | — | — |
| DMA 18:1n-9 | — | — | — | — | 1.69 ± 0.57 | 1.85 ± 0.43 | — | — |
| DMA 18:1n-7 | — | — | — | — | 2.53 ± 0.28 | 2.41 ± 0.38 | — | — |
| 18:0 | 14.38 ± 1.21 | 14.34 ± 0.97 | 44.50 ± 2.99 | 42.18 ± 3.20 | 16.60 ± 1.39 | 16.06 ± 1.08 | 37.79 ± 2.70 | 44.30 ± 2.49 |
| 18:1n-9 | 22.46 ± 1.19 | 23.04 ± 1.61 | 17.55 ± 1.10 | 16.66 ± 1.92 | 11.55 ± 2.16 | 12.30 ± 1.64 | 8.00 ± 2.01 | 6.76 ± 1.10 |
| 18:1n-7 | 5.43 ± 0.59 | 5.33 ± 0.70 | 1.38 ± 0.49 | 0.68 ± 0.59 | 1.83 ± 0.95 | 2.00 ± 0.56 | — | — |
| 18:2n-6 | 1.27 ± 0.30 | 1.35 ± 0.36 | 0.30 ± 0.15 | 0.86 ± 0.60 | 0.47 ± 0.21 | 0.29 | 0.90 | 2.03 ± 1.03 |
| 20:1n-9 | — | — | 0.66 ± 0.54 | 1.62 ± 0.64 | 2.42 ± 0.98 | 2.82 ± 1.05 | — | — |
| 20:4n-6 | 7.57 ± 1.32 | 6.18 ± 0.98 | 3.38 ± 0.56 | 3.48 ± 0.59 | 12.42 ± 1.64 | 10.62 ± 1.48 | 26.82 ± 3.41 | 17.12 ± 2.58 |
| 22:4n-6 | — | — | 2.69 ± 0.49 | 2.79 ± 0.14 | 5.98 ± 0.89 | 5.11 ± 0.75 | — | — |
| 22:6n-3 | 2.58 ± 0.52 | 2.98 ± 0.58 | 16.54 ± 0.87 | 17.13 ± 1.16 | 15.11 ± 1.10 | 17.22 ± 0.40 | — | — |
| Rest | 3.01 ± 1.99 | 1.14 ± 2.19 | 5.17 ± 2.72 | 9.53 ± 1.78 | 11.13 ± 1.29 | 11.84 ± 1.16 | 6.48 ± 2.61 | 4.87 ± 2.15 |
| PUFA/MUFA | 0.39 | 0.35 | 1.12 | 1.21 | 1.53 | 1.44 | 2.90 | 2.16 |
| PUFA/UFA | 0.28 | 0.26 | 0.52 | 0.54 | 0.60 | 0.59 | 0.74 | 0.60 |
| MUFA/UFA | 0.71 | 0.73 | 0.47 | 0.45 | 0.39 | 0.40 | 0.25 | 0.28 |
FO, fish oil; CTR, control; DMA, dimethyl acetal; MUFA, monounsaturated fatty acid, UFA, unsaturated fatty acid. All measurements were performed three times.
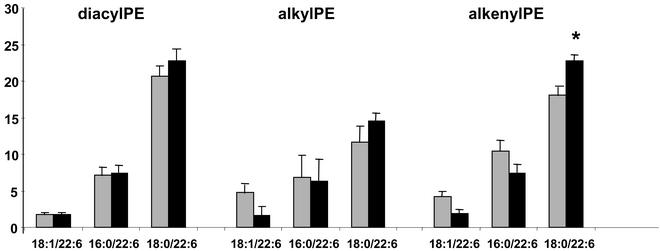
We found little change in brain DHA, which is in good accordance with previous findings (19, 32) and the observation that even for omega 3-deficient animals repletion with omega 3 took a very long time (>10 weeks or longer) (33). Still, this finding was reflected in the molecular species composition of the three subclasses of phosphatidylethanolamines. Levels of species carrying 18:0 in the sn-1 position and DHA in the sn-2 position of the glycerol backbone were higher than in the corresponding control species. Interestingly the 18:1/22:6 and 16:0/22:6 alkyl and alkenylacyl species were represented in lesser amounts in brains of fish oil-fed rats (Fig. 1). Fish oil feeding brought about a reduction in AA containing 18:1, 16:0, and 18:0 in position sn-1 of the glycerol backbone in the case of diacyl species and 18:0 in the case of alkylacyl and alkenylacyl species. In contrast, there was some increase in levels of alkenylacyl 18:0/20:4 and 16:0/20:4 species (data not shown). This finding indicates that fish oil (DHA) differentially affects the metabolism of phosphatidylethanolamine molecular species. Choline phosphoglycerides did not respond to the dietary intervention.
Figure 1.
DHA containing ethanolamine phosphoglyceride molecular species in rat hippocampus. Each bar is the average of five independent determinations. Gray bars represent data of control group; black bars represent values obtained from hippocampi of animals fed fish oil (*, P < 0.05).
Gene Expression Analysis of Rat Hippocampus After Fish Oil Feeding.
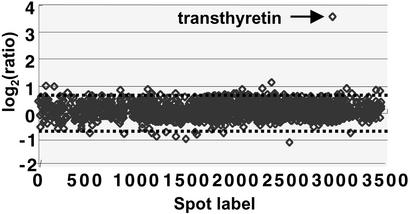
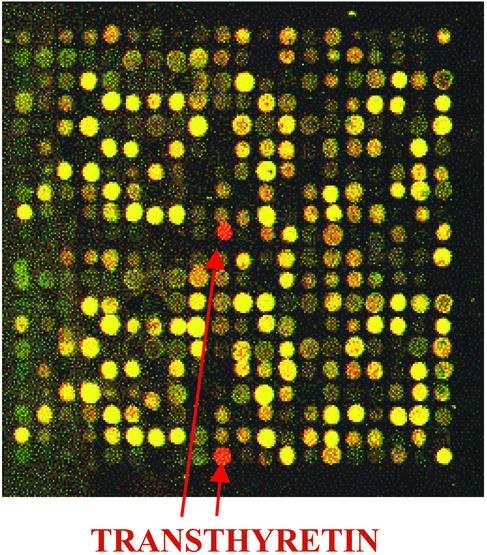
For microarray analysis we used rat-specific cDNA microarrays containing 3,200 gene-specific samples in duplicate. We found that even short-term administration of fish oil triggered changes in the gene expression profile. From 3,200 genes 1,443 had significant intensity values (see Experimental Procedures for statistical calculations and Fig. 3 for scatter plot image of the average ratios of individual gene samples of fish oil feeding vs. control) and only 0.75% had altered expression. Eleven genes were up-regulated and 12 genes were down-regulated in the hippocampi of fish oil-fed animals, although only slight differences could be detected. Among the repressed genes were apolipoprotein C, one heat shock protein gene (hsp86), two membrane associated protein genes, and a possible transcriptional regulator (Table 3). Among the induced genes were three membrane-associated protein genes, a nuclear factor, and the mitochondrial creatine kinase. There was only one gene that showed a marked change in its transcription; the expression of TTR exhibited a dramatic 11.75-fold induction (9.5- and 14-fold induction for replicate experiments) (Figs. 2 and 3 and Table 3).
Figure 3.
Scatter plot of log2 average ratio of gene expression of rat brains obtained from fish oil-fed and control animal groups. Average ratios were calculated from four data spots generated by two independent experiments using two different pools of animals with the same treatment. Between the dashed lines is indicated the interval −1.75- to 1.75-fold regulation (corresponding to log2 = 0.8) in which changes in expression were considered insignificant.
Table 3.
Effects of fish oil on rat hippocampus gene expression
| Gene product | GenBank accession no. | Ratio* | SD |
|---|---|---|---|
| Induced expression | |||
| Intestinal membrane A4 protein | BE349686 | 1.73 | 0.23 |
| High mobility group 2 protein | H35214 | 1.77 | 0.08 |
| Nuclear transport factor 2 homolog | X91651 | 1.78 | 0.1 |
| EST | AA052369 | 1.79 | 0.11 |
| CD39 antigen-like 4 | AW544751 | 1.81 | 0.09 |
| Similar to BAT2 | BE329313 | 1.86 | 0.11 |
| EST | AA245586 | 1.86 | 0.12 |
| Mitochondrial creatine kinase | X59736 | 1.95 | 0.19 |
| Hypothetical | AA221692 | 1.98 | 0.21 |
| ER lumen protein retaining receptor 2 | W46090 | 2.15 | 0.23 |
| TTR | NM_012681 | 11.75 | 2.25 |
| Repressed expression | |||
| Makorin RING zinc-finger protein 2 | BG016106 | 0.47 | 0.26 |
| Lactate dehydrogenase homolog | AW140623 | 0.51 | 0.28 |
| Anaphylatoxin C3a receptor | U86379 | 0.54 | 0.05 |
| EST | AA270674 | 0.55 | 0.09 |
| EST | AA230444 | 0.57 | 0.08 |
| Apolipoprotein Cl | NM_012824 | 0.58 | 0.14 |
| Toll-associated serine protease | AF057025 | 0.59 | 0.01 |
| EST | AW544503 | 0.59 | 0.09 |
| RecQ protein-like | W14898 | 0.60 | 0.15 |
| K-Cl cotransporter 3b | U55816 | 0.60 | 0.06 |
| EST | AW536066 | 0.61 | 0.11 |
| Heat shock protein, 86 kDa | AJ297736 | 0.61 | 0.03 |
Calculated from four independent values obtained from two independent experiments.
Figure 2.
Hybridization of a mixture of Cy3-labeled cDNA (derived from control sample) and Cy5-labeled cDNA (from fish oil-fed sample) on a 3,200 rat-specific cDNA microarray. Of 16 subarrays, only one, which contains 200 gene samples in duplicate, is shown.
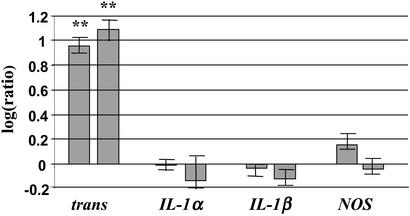
To confirm the differential expression of genes obtained by microarray analysis in hippocampi of rats fed a fish oil-rich diet, we analyzed several genes by real-time RT-QPCR. The induction of TTR expression obtained from cDNA microarray data were confirmed by real-time RT-QPCR. With this method the average induction was 9.1-fold (±1.2 SD) for one treated group and 12.2-fold (±2.1 SD) for the other as compared with control groups in four replicate experiments (Fig. 4).
Figure 4.
Relative real-time QRT-PCR analysis of cDNA samples (≈35-ng template in each PCR) prepared from hippocampus RNA of fish oil-fed and control animal groups. Bar diagram shows the quantitative ratios of TTR, IL-1α, IL1-β, and NOS. Ratios were calculated from relative expression values (normalized to actin) of samples obtained from rats fed fish oil and animals kept on normal rat chow. Ratio 1 (0 in logaritmic scale) means no change in transcription caused by fish oil feeding. All of the reactions were performed four times. **, P < 0.01.
It was hypothesized that fish oil could suppress the expression of IL-1 production and/or induce the synthesis of the endothelial NO in the brain. In our system neither IL-1α, IL-1β, nor NOS genes showed changes in mRNA levels upon fish oil consumption (Fig. 4).
In all cases, relative transcript ratios were determined by normalizing Ct values to β-actin expression, and all measurements were performed four times.
Discussion
We have recently showed that dietary PUFA can alter the expression of numerous genes in rat brain when animals were fed for a long period (19). Here, we fed 2-year-old rats fish oil (rich in EPA and DHA) for 1 month and found that the expression of some genes altered in the hippocampus with cDNA microarray technology. In this study only a few genes (0.75% of the total genes with significant values) exhibited change in their transcription, whereas in the previous study conducted over a 2-month period there were eight times more genes modulated by the omega 3 fatty acid feeding protocol. This comparison suggests that there is an age-related and time-dependent role of dietary n-3 fatty acids in relation to modulation of gene expression.
During the short-term fish oil consumption the expression of TTR gene was induced in a marked degree in aged rat brain. Its expression increased by ≈10-fold.
TTR is a tetrameric thyroid hormone transport protein that is secreted by hepatocytes into the serum and by the epithelium of the choroid plexus into the cerebral spinal fluid. In the body, a system of plasma proteins, including TTR, binds thyroid hormones and prevents their direct partition into cell membranes (the evolution, function, and distribution of TTR is reviewed in refs. 34 and 35). TTR has also been implicated in the transport and deposition of amyloid β peptide (Aβ) (36–38). Deposition of Aβ is involved in the pathogenesis of AD (39, 40). Amyloid aggregates are formed from the soluble form of Aβ, which is a normal metabolic product detectable in the ventricular cerebral spinal fluid and serum of normal and AD subjects. Several experimental models of transfected cells and transgenic animals are used to analyze the mechanisms by which Aβ is formed and aggregates to trigger the neuronal degeneration and apoptosis, which finally cause AD (39–42). Aβ forms amyloid-like fibrils in vitro, which can be inhibited by normal human cerebral spinal fluid and the purified TTR (36, 37). Coexpression of β-peptide and TTR in Caenorhabditis elegans was shown to lead to a significant reduction of insoluble β-amyloid plaques (38). Recently, increased TTR expression has been observed in APPSW transgenic mice (overexpressing a mutant form of human amyloid precursor protein) with lack of neurodegeneration. The authors proposed that neuroprotection in these mice is caused by early gene expression changes of some genes, including the TTR gene (43). Moreover, an inverse relationship has been found between TTR level in cerebral spinal fluid and the severity of dementia in AD patients (44).
Induction of the expression and secretion of TTR has been demonstrated after administration of nicotine (45) and leaf extracts of G. biloba (46). Epidemiological studies have shown that nicotine reduces the risk for AD (47). Nicotine increased TTR mRNA levels in a time- and dose-dependent manner in rat choroid plexus. It was proposed that the positive effect of nicotine is caused by elevated TTR concentrations retarding the aggregation of amyloids in the brain (45). Ginkgo leaf extract is commonly used to combat a variety of neurological disorders, including AD (48). In high-density oligonucleotide microarrays a 16-fold increase in TTR mRNA level in mice hippocampus was found after 4 weeks of administration of the extract (46).
Previously, we showed that the ATP-generating machinery of brain responded to n-3 fatty acids most intensively. Here we also found the induction of mitochondrial creatine kinase transcription. Decreased expression of the apolipoprotein (apo) C1 gene was also observed in response to a fish oil-rich diet, although the repression was moderate. The H2 allele of apoC-I is associated with AD, and higher ApoC-I protein levels were shown in hippocampus of patients; however, lower concentrations of apoC-I mRNA could be detected (49). A slight repression (1.7-fold) of hsp86 transcription could be detected in the hippocampus of the fish oil-fed animals. In AD brains, levels of heat shock proteins were increased in both the cytosolic and membranous fractions, and chaperonine HSP90 was colocalized with amyloid plaques (50). However, there might be some correlation between the altered expression of these genes and neurological functions; the magnitude of these changes are not significant.
It has been hypothesized that fish oil could exert its beneficial effect by suppressing IL-1 production and/or stimulating endothelial NO production in the brain (51–53). However, in our system neither IL-1α, IL-1β, nor NOS genes showed changes in transcription upon fish oil consumption. Here, we offer an alternative explanation: n-3 PUFAs stimulate TTR synthesis, which could counteract the appearance of insoluble amyloid aggregates.
An open question is whether the magnitude increase in TTR mRNA synthesis can be explained by the DHA-induced alteration in molecular composition of hippocampal membranes. It is noteworthy that DHA accumulated in ethanolamine but not in choline phosphoglycerides (Fig. 1). The same result was described in brain hemispheres in a previous study where a number of genes but not TTR were induced (19). Ethanolamine phosphoglycerides are very abundant in the inner leaflet of the membrane bilayer and accumulation of several 18:0/22:6 species with a concomitant reduction of several AA-containing species might have affected molecular organization and related functions of this membrane layer. Changes in physico-chemical properties of membrane could trigger alteration of numerous genes. However, the specificity of this induction (one significant induction of 3,200 genes) suggests that the observed effect might be independent from the observed alterations in membrane composition. Different moieties in phosphatidylethanolamines changed association of G proteins and other peripheral proteins with membranes and thus modulated cell signaling (54). In this work we also found alterations in fatty acid composition of phosphatidylethanolamines, which also could affect cell signaling events.
It is interesting that extract of G. biloba is also an effective TTR inducer (46). We believe that it is unlikely that this extract affects membrane lipid composition in a fashion similar to fish oil.
Another explanation for the specific TTR-inducing effect of n-3 fatty acids could be based on the stimulation of a specific transcription factor, which could regulate TTR transcription. The hepatocyte nuclear factor 6 (HNF-6) binds the original HNF-3 site of the TTR promoter (−94 to −106) and activates its transcription (55). This promoter shares sequence homology with the promoter region of fatty acid binding protein (55). Thus the presence of fatty acids could induce the expression of genes coding for fatty acid binding protein and TTR at the same time; however, the actual signal that triggers the response remains unclear.
Data suggest, that natural TTR inducers, such as Ginkgo extract or fish oil could be effective for the prevention of AD. We propose that increasing TTR expression, and thereby facilitating the clearance of Aβ from brain tissue, is a plausible common mechanism for their protective effect. Further studies will be required to fully understand the molecular background of the protective efficacy of fish oil in AD and other neurological diseases such as Parkinson's disease. Although, DHA has been shown to play an important role in learning and memory (6–8) and DHA supplementation alone was efficient in improving learning ability of AD model rats (3), other essential fatty acids are also believed to exert positive effects on brain functions (56). Because our fish oil contained EPA, it is not evident whether EPA also contributed to this effect. Therefore, in the future it would be worth investigating the effects of EPA and DHA alone on modulating TTR gene expression in the hippocamppus.
Acknowledgments
We thank A. Zvara and L. Hackler, Jr., for their help with microarray methodology and Z. Nemes for correcting the manuscript. We are grateful to A. Sinclair for revising and critically evaluating the manuscript. This work was supported by a grant from the Hungarian National Research and Development Initiative (1/040/2001), Hungarian Scientific Research Grants OTKA T-026451 and OTKA F-042850, and a grant from the European Community (LL, QLRT-2001-00172). Fish oil was a generous gift of BLT Berg Lipidtech (Aale, Norway).
Abbreviations
- DHA
docosahexaenoic acid
- AD
Alzheimer's disease
- PUFA
polyunsaturated fatty acid
- RT-QPCR
quantitative RT-PCR
- TTR
transthyretin
- Aβ
amyloid β peptide
- EPA
eicosapentaenoic acid
- AA
arachidonic acid
- NOS
NO synthase
References
- 1.Conquer J A, Tierney M C, Zecevic J, Bettger W J, Fisher R H. Lipids. 2000;35:1305–1312. doi: 10.1007/s11745-000-0646-3. [DOI] [PubMed] [Google Scholar]
- 2.Farooqui A A, Horrocks L A. J Mol Neurosci. 2001;16:263–272. doi: 10.1385/jmn:16:2-3:263. [DOI] [PubMed] [Google Scholar]
- 3.Hashimoto M, Hossain S, Shimada T, Sugioka K, Yamasaki H, Fuji Y, Ishibashi Y, Oka J-I, Shido O. J Neurochem. 2002;81:1084–1091. doi: 10.1046/j.1471-4159.2002.00905.x. [DOI] [PubMed] [Google Scholar]
- 4.McGahon B M, Martin D S D, Horrobin D F, Lynch M A. Neuroscience. 1999;94:305–314. doi: 10.1016/s0306-4522(99)00219-5. [DOI] [PubMed] [Google Scholar]
- 5.Horrocks L A, Yeo Y K. Pharmacol Res. 1999;40:211–225. doi: 10.1006/phrs.1999.0495. [DOI] [PubMed] [Google Scholar]
- 6.Grant W B, Campbell A, Itzhaki R F, Savory J. J Alzheimers Dis. 2002;3:179–189. doi: 10.3233/jad-2002-4308. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki H, Park S J, Tamura M, Ando S. Mech Ageing Dev. 1998;101:119–128. doi: 10.1016/s0047-6374(97)00169-3. [DOI] [PubMed] [Google Scholar]
- 8.Lim S, Suzuki H. J Nutr. 2001;131:319–324. doi: 10.1093/jn/131.2.319. [DOI] [PubMed] [Google Scholar]
- 9.Sugimoto Y, Taga C, Nishiga M, Fujiwara M, Konishi F, Tanaka K, Kamei C. Biol Pharm Bull. 2002;25:1090–1092. doi: 10.1248/bpb.25.1090. [DOI] [PubMed] [Google Scholar]
- 10.Kurlack L O, Stephenson T J. Arch Dis Child Fetal Neonatal Ed. 1999;80:148–154. doi: 10.1136/fn.80.2.f148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lauritzen L, Hansen H S, Jorgensen M H, Michaelson K F. Prog Lipid Res. 2001;40:1–94. doi: 10.1016/s0163-7827(00)00017-5. [DOI] [PubMed] [Google Scholar]
- 12.Salem N, Litman B, Kim H-Y, Gawrisch K. Lipids. 2001;36:945–959. doi: 10.1007/s11745-001-0805-6. [DOI] [PubMed] [Google Scholar]
- 13.Litman B J, Niu S L, Polozova A, Mitchell D C. J Mol Neurosci. 2001;16:237–242. doi: 10.1385/JMN:16:2-3:237. [DOI] [PubMed] [Google Scholar]
- 14.Feller S E, Gawrisch K, MacKerell A D. J Am Chem Soc. 2002;124:318–326. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 15.Bourre J M, Francois M, Youyou A, Dumont O, Piciotti M, Pascal G, Durand G. J Nutr. 1989;119:1880–1892. doi: 10.1093/jn/119.12.1880. [DOI] [PubMed] [Google Scholar]
- 16.Zimmer L, Dellion-Vaancassel S, Durand G, Guilloteau D, Bodard S, Besnard J C, Chalon S. J Lipid Res. 2000;41:32–40. [PubMed] [Google Scholar]
- 17.Vaidyanathan V V, Rao K V R, Sastry P S. Neurosci Lett. 1994;179:171–174. doi: 10.1016/0304-3940(94)90961-x. [DOI] [PubMed] [Google Scholar]
- 18.Rojas C V, Greiner R S, Martinez J I, Salem N, Uauy R. Lipids. 2002;37:367–374. doi: 10.1007/s1145-002-0904-4. [DOI] [PubMed] [Google Scholar]
- 19.Kitajka K, Puskas L G, Zvara A, Hackler L, Barcelo-Coblijn G, Yeo Y K, Farkas T. Proc Natl Acad Sci USA. 2002;99:2619–2624. doi: 10.1073/pnas.042698699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Urquiza A M, Liu S, Sjoberg M, Zetterstrom R H, Griffiths W, Sjovall J, Perlmann T. Science. 2000;290:2140–2144. doi: 10.1126/science.290.5499.2140. [DOI] [PubMed] [Google Scholar]
- 21.Garcia M C, Ward G, Ma Y-C, Salem N, Kim H-Y. J Neurochem. 1998;70:24–30. doi: 10.1046/j.1471-4159.1998.70010024.x. [DOI] [PubMed] [Google Scholar]
- 22.Akbar M, Kim H-Y. J Neurochem. 2002;82:655–665. doi: 10.1046/j.1471-4159.2002.01015.x. [DOI] [PubMed] [Google Scholar]
- 23.Ikemoto A, Kobayashi T, Watanabe S, Okuyama H. Neurochem Res. 1997;22:671–678. doi: 10.1023/a:1027393724676. [DOI] [PubMed] [Google Scholar]
- 24.Martin R E. J Neurosci Res. 1998;54:805–813. doi: 10.1002/(SICI)1097-4547(19981215)54:6<805::AID-JNR8>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 25.Lauritzen I, Blondeau N, Heurteaux C, Windmann C, Romey G, Lazdunski M. EMBO J. 2000;19:1784–1793. doi: 10.1093/emboj/19.8.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmad A, Moriguchi T, Salem N. Ped Neurol. 2002;26:210–218. doi: 10.1016/s0887-8994(01)00383-6. [DOI] [PubMed] [Google Scholar]
- 27.Ahmad A, Murthy M, Greiner R S, Moriguchi T, Salem N. Nutr Neurosci. 2002;5:103–113. doi: 10.1080/10284150290018973. [DOI] [PubMed] [Google Scholar]
- 28.Takamura H, Kito M. J Biochem (Tokyo) 1991;109:436–439. doi: 10.1093/oxfordjournals.jbchem.a123399. [DOI] [PubMed] [Google Scholar]
- 29.Ikemoto A, Nitta A, Furukawa, Ohishi M, Nakamura A, Fujii Y, Okuyama H. Neurosci Lett. 2000;285:99–102. doi: 10.1016/s0304-3940(00)01035-1. [DOI] [PubMed] [Google Scholar]
- 30.Puskas L G, Zvara A, Hackler L, Jr, van Hummelen P. BioTechniques. 2002;32:1330–1340. doi: 10.2144/02326mt04. [DOI] [PubMed] [Google Scholar]
- 31.Puskas L G, Hackler L, Jr, Kovacs G, Kupihar Z, Zvara A, Micsik T, van Hummelen P. Anal Biochem. 2002;305:279–281. doi: 10.1006/abio.2002.5640. [DOI] [PubMed] [Google Scholar]
- 32.Abedin L, Lien E L, Vingrys A J, Sinclair A J. Lipids. 1999;34:475–842. doi: 10.1007/s11745-999-0387-3. [DOI] [PubMed] [Google Scholar]
- 33.Weisinger H S, Vingrys A J, Sinclair A J. Lipids. 1995;30:471–473. doi: 10.1007/BF02536307. [DOI] [PubMed] [Google Scholar]
- 34.Prapunpoj P, Richardson S J, Schreiber G. Am J Physiol. 2002;283:R885–R896. doi: 10.1152/ajpregu.00042.2002. [DOI] [PubMed] [Google Scholar]
- 35.Schreiber G, Richardson S J. Comp Biochem Physiol. 1997;116:137–160. doi: 10.1016/s0305-0491(96)00212-x. [DOI] [PubMed] [Google Scholar]
- 36.Tsuzuki K, Fukatsu R, Yamaguchi H, Tateno M, Imai K, Fujii N, Yamauchi T. Neurosci Lett. 2000;281:171–174. doi: 10.1016/s0304-3940(00)00834-x. [DOI] [PubMed] [Google Scholar]
- 37.Golabek A, Marques M A, Lalowski M, Wisniewski T. Neurosci Lett. 1995;191:79–82. doi: 10.1016/0304-3940(95)11565-7. [DOI] [PubMed] [Google Scholar]
- 38.Link C D. Proc Natl Acad Sci USA. 2000;92:9368–9372. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambamurti K, Greig N H, Lahiri D K. Neuromol Med. 2002;1:1–31. doi: 10.1385/NMM:1:1:1. [DOI] [PubMed] [Google Scholar]
- 40.Carter J, Lippa C F. Curr Mol Med. 2001;1:733–737. doi: 10.2174/1566524013363177. [DOI] [PubMed] [Google Scholar]
- 41.Isacson O, Seo H, Lin L, Albeck D, Granholm A C. Trends Neurosci. 2002;25:79–84. doi: 10.1016/s0166-2236(02)02037-4. [DOI] [PubMed] [Google Scholar]
- 42.Mattson M P. Nat Rev Mol Cell Biol. 2000;1:120–129. doi: 10.1038/35040009. [DOI] [PubMed] [Google Scholar]
- 43.Stein T D, Johnson J A. J Neurosci. 2002;22:7380–7388. doi: 10.1523/JNEUROSCI.22-17-07380.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Serot J M, Christmann D, Dubost T, Couturier M. J Neurol Neurosurg Psychiatry. 1997;63:506–508. doi: 10.1136/jnnp.63.4.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M D, Kane J K, Matta S G, Blaner W S, Sharp B M. J Neurosci. 2000;20:1318–1323. doi: 10.1523/JNEUROSCI.20-04-01318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe C M, Wolffram S, Ader P, Rimbach G, Packer L, Maguire J J, Schultz P G, Gohil K. Proc Natl Acad Sci USA. 2001;98:6577–6580. doi: 10.1073/pnas.111126298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Merchant C, Tang M X, Albert S, Manly J, Stern Y, Mayeux R. Neurology. 1999;52:1408–1412. doi: 10.1212/wnl.52.7.1408. [DOI] [PubMed] [Google Scholar]
- 48.DeFeudis F V, Drieu K. Curr Drug Targets. 2000;1:25–58. doi: 10.2174/1389450003349380. [DOI] [PubMed] [Google Scholar]
- 49.Petit-Turcotte C, Stohl S M, Beffert U, Cohn J S, Aumont N, Tremblay M, Dea D, Yang L, Poirier J, Shachter N S. Neurobiol Dis. 2001;8:953–963. doi: 10.1006/nbdi.2001.0441. [DOI] [PubMed] [Google Scholar]
- 50.Kakimura J, Kitamura Y, Takata K, Umeki M, Suzuki S, Shibagaki K, Taniguchi T, Nomura Y, Gebicke-Haerter P J, Smith M A, et al. FASEB J. 2002;16:601–603. doi: 10.1096/fj.01-0530fje. [DOI] [PubMed] [Google Scholar]
- 51.Das U N. Prostaglandins Leukotrienes Essent Fatty Acids. 2000;63:351–362. doi: 10.1054/plef.2000.0226. [DOI] [PubMed] [Google Scholar]
- 52.McCarty M F. Med Hypotheses. 1999;53:369–374. doi: 10.1054/mehy.1998.0783. [DOI] [PubMed] [Google Scholar]
- 53.Peers R J. Med J Aust. 1990;153:563–564. [PubMed] [Google Scholar]
- 54.Escriba P V, Ozaita A, Ribas C, Miralles A, Fodor E, Farkas T, Garcia-Sevilla J A. Proc Natl Acad Sci USA. 1997;94:11375–11380. doi: 10.1073/pnas.94.21.11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samadani U, Costa R H. Mol Cell Biol. 1996;16:6273–6784. doi: 10.1128/mcb.16.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Puri B K, Bydder G M, Counsell S J, Corridan B J, Richardson A J, Hajnal J V, Appel C, Mckee H M, Vaddadi K S, Horrobin D F. NeuroReport. 2002;13:123–126. doi: 10.1097/00001756-200201210-00029. [DOI] [PubMed] [Google Scholar]