Abstract
Sir2 is an NAD-dependent histone deacetylase required for transcriptional silencing. To study the mechanism of Sir2 function, we examined the biochemical properties of purified recombinant Drosophila Sir2 (dSir2). First, we performed histone deacetylation assays and found that dSir2 deacetylates a broad range of acetylated lysine residues. We then carried out in vitro transcription experiments and observed that dSir2 does not repress transcription with either naked DNA templates or chromatin assembled from native (and mostly unacetylated) histones. It was possible, however, that repression by dSir2 requires an acetylated histone substrate. We therefore tested the transcriptional effects of dSir2 with native histones that were hyperacetylated by treatment with acetic anhydride. Assembly of the hyperacetylated histones onto DNA yields a soluble histone–DNA complex that differs from canonical nucleosomal chromatin. With this hyperacetylated histone–DNA complex, we observed potent (50- to 100-fold) NAD-dependent transcriptional repression by purified dSir2. In contrast, repression by dSir2 was not observed in parallel experiments in which histones were hyperpropionylated with propionic anhydride. We also found that dSir2 mediates the formation of a nuclease-resistant fast-sedimenting histone–DNA complex in an NAD-dependent manner. Unlike dSir2, the dHDAC1 deacetylase does not strongly repress transcription or generate a nuclease-resistant histone–DNA complex. Furthermore, with yeast Sir2, the transcriptional repression we observe correlates with deacetylation activity in vitro and silencing activity in vivo. These findings suggest that deacetylation by Sir2 causes a conformational change or rearrangement of histones into a transcriptionally repressive chromatin structure.
Chromatin is an integral component of the transcription process (1–9). A variety of factors have been found to affect transcriptional activity by covalent modification of histones or ATP-driven remodeling of chromatin structure. For instance, proteins that acetylate histones are generally involved in transcriptional activation, whereas histone deacetylases (HDACs) generally appear to function in transcriptional repression. In addition, ATP-utilizing nucleosome remodeling factors are able to facilitate DNA-dependent processes in chromatin. At present, many proteins that catalyze histone modification or nucleosome remodeling have been identified. It will thus be important to investigate the biochemical mechanisms by which these factors regulate transcription.
Sir2 is an NAD-dependent HDAC that functions in transcriptional silencing (for recent reviews, see refs. 10–12). In Saccharomyces cerevisiae, Sir2 is required for repression at the silent mating loci (13), telomeres (14, 15), and rDNA (16–18). In addition, yeast Sir2 (ySir2) functions with the Sir3, Sir4, and Rap1 proteins in silencing at the silent mating loci and at telomeres (19–21) but acts with different proteins (including Net1 and Cdc14) in rDNA silencing (22, 23). In addition, ySir2 was found to extend life span, possibly by suppression of rDNA recombination (11, 24, 25). In Drosophila, Sir2 is involved in centromeric heterochromatic silencing but apparently not telomeric silencing (26). In addition, Drosophila Sir2 (dSir2) interacts genetically and physically with the Hairy corepressor protein (26).
Sir2 is a member of a family of proteins (also known as sirtuins) that are conserved from bacteria to humans (27). It appears that there are five Sir2-like proteins (including Sir2 itself) in yeast as well as at least five in Drosophila and seven in humans. Sir2 and related proteins have been observed to exhibit ADP-ribosyltransferase activity (28, 29) as well as NAD-dependent HDAC activity (30–32). The HDAC activity of Sir2 correlates well with the observation that silent chromatin is generally hypoacetylated and that the extent of acetylation at the silent mating loci is inversely related to the level of Sir2 in vivo (33). In addition, mutations that reduce HDAC activity of Sir2 were also found to be defective in silencing (30). Moreover, yeast strains carrying a null mutant allele of NPT1, which is involved in NAD biosynthesis, were observed to possess a reduced intracellular concentration of NAD as well as to exhibit loss of silencing that is comparable to that seen in the absence of Sir2 (32). These and other findings collectively provide strong evidence for a model in which NAD-dependent histone deacetylation by Sir2 is required for transcriptional silencing.
In this study, we examined the biochemical activities of the Sir2 protein. To gain a better understanding of transcriptional repression by Sir2, we analyzed the effects of Sir2-mediated histone deacetylation upon chromatin structure and transcriptional activity. These studies suggest that Sir2 mediates a conformational change or rearrangement in chromatin that results in the formation of a transcriptionally repressive structure.
Materials and Methods
Recombinant Proteins and Other Materials.
A construct that encodes full-length dSir2 with a C-terminal His-6 tag was prepared from a cDNA (LD13904; Research Genetics, Huntsville, AL) of Drosophila SIRT1 (GenBank accession no. AF068758). The corresponding His-tagged dSir2 protein was synthesized in Sf9 cells by using a baculovirus expression system (Bac to Bac; GIBCO/BRL) and purified by Ni(II) affinity chromatography (Ni-NTA resin; Qiagen, Chatsworth, CA). A construct that encodes full length ySir2 with an N-terminal GST tag (pDM111; ref. 23) and derivatives of this construct (GST-Sir2-H364Y and GST-Sir2-G270E) were expressed and purified as described (34). NF-κB p65, Sp1, and Drosophila HDAC1 (dHDAC1) proteins were prepared as described (35). Polyclonal rabbit antibodies against dSir2 were generated against full-length recombinant dSir2 protein purified under denaturing conditions. The HDAC inhibitors FR901228 and trichostatin A were used as described (35).
Core Histones.
Native core histones were purified from Drosophila (36). Hyperacetylated core histones were prepared as follows. Core histones (1 mg/ml) were dialyzed into acetylation buffer (50 mM NaHCO3, pH 8/2 M NaCl) and then acetylated with acetic anhydride (10 mM final concentration; added as the neat liquid) on ice for 30 min. The reaction was quenched by the addition of 50 mM Tris⋅HCl (pH 8.0), and the histones were subjected to extensive dialysis (against 10 mM Hepes/K+, pH 7.6/1 mM EDTA/10 mM KCl/10% glycerol) to remove unreacted acetic anhydride. Propionylated histones were prepared in an analogous manner with propionic anhydride.
Assays.
HDAC reactions were performed by incubation of purified dSir2 (100 nM) or dHDAC1 (100 nM) with hyperacetylated core histones (10 μg/ml, 0.75 μM in histone polypeptides) for 30 min at 30°C in buffer (50 μl of total reaction volume) consisting of 50 mM Hepes, K+, pH 7.6, 50 mM NaCl, 0.1 mM EDTA, 1 mM DTT, and 5% glycerol. NAD was used at 100 μM final concentration. The reactions were terminated by the addition of SDS gel loading buffer and then subjected to 15% polyacrylamide–SDS electrophoresis. Total core histones were detected by staining with Coomassie brilliant blue R-250. The deacetylation of specific lysine residues in the core histones was detected by Western blot analysis with antibodies that recognize the acetylated (but not unacetylated) form of the residues. Antibodies against acetylated histone H3 (Ac-K9; Ac-K14; Ac-K9,14) and acetylated histone H4 (Ac-K5; Ac-K8; Ac-K12; Ac-K16; Ac-K5,8,12,16) were from Upstate Biotechnology, Lake Placid, NY. Antibodies against acetylated lysines, acetylated histone H2B (Ac-K5; Ac-K20), and acetylated histone H3 (Ac-K18; Ac-K23) were from Cell Signaling Technology (Beverly, MA). ADP-ribosyltransferase assays were performed by incubation of dSir2, 32P-NAD, and core histones (28, 29). dSir2-mediated transfer of 32P to the core histones was not detected by SDS-polyacrylamide gel electrophoresis and autoradiography (data not shown).
Chromatin Assembly and Analysis.
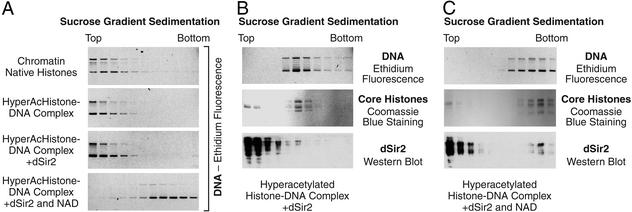
The plasmid pHIV (35), which contains the promoter region of the HIV-1 long terminal repeat, was used as the DNA template. Transcription of chromatin assembled with purified ACF and NAP-1 was carried out as described previously (37). Quantitation of the data was carried out with a PhosphorImager (Molecular Dynamics). All reaction conditions were performed in duplicate. Sedimentation analyses were carried out with linear 20–50% (wt/vol) sucrose gradients in a Beckman SW55 rotor (Beckman Coulter) (Fig. 4A: 25,000 rpm, 3 h; Fig. 4B: 45,000 rpm, 6 h; Fig. 4C: 25,000 rpm, 3.5 h). Each experiment was performed a minimum of two independent times to ensure the reproducibility of the results.
Figure 4.
Deacetylation of histones by dSir2 and NAD yields a fast-sedimenting histone–DNA species. (A) Sucrose gradient sedimentation analysis. The indicated components were used in the same relative proportions as in the transcription reactions (Fig. 2). The migration of DNA was monitored by agarose gel electrophoresis and staining with ethidium bromide. (B) Sucrose gradient sedimentation analysis was carried out with template DNA assembled with hyperacetylated histones and dSir2, as in Fig. 2D. DNA was visualized by ethidium fluorescence, core histones were monitored by staining with Coomassie blue, and dSir2 was detected by Western blot analysis. (C) Sucrose gradient sedimentation analysis was performed as in B, except that NAD was also included in the assembly reaction.
Results and Discussion
dSir2 Catalyzes the Deacetylation of a Wide Range of Lysine Residues in Core Histone Tails.
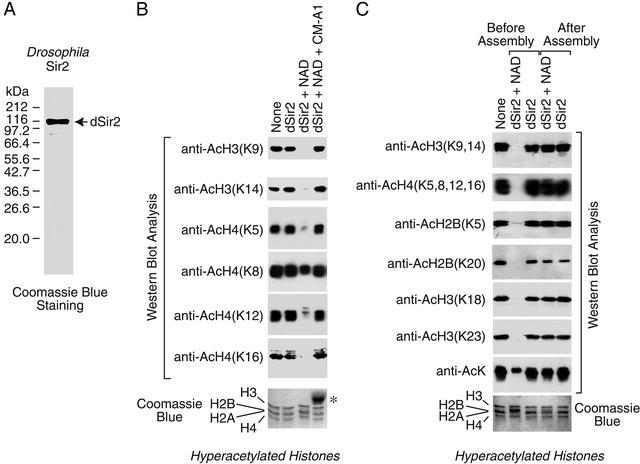
In this work, we focused primarily on the biochemical properties of Drosophila Sir2 (dSir2). To this end, we synthesized full-length dSir2 in Sf9 cells with a C-terminal His-6 tag and purified the recombinant protein to near homogeneity by Ni(II) affinity chromatography (Fig. 1A). We then tested the ability of the purified protein to catalyze the deacetylation of histones. In these experiments, we prepared hyperacetylated core histones by treatment of purified native Drosophila core histones with acetic anhydride (35, 38). This reaction was carried out in 2 M NaCl to promote the maintenance of the octameric form of the core histones. Unreacted acetic anhydride was removed by extensive dialysis, and the resulting hyperacetylated histones were used as the substrate for the deacetylation reaction. The extent of deacetylation at individual lysine residues was monitored by Western blot analysis with antibodies that recognize the acetylated form of specific lysine residues in the core histone tails. Consistent with previous findings (26), purified dSir2 has NAD-dependent HDAC activity (Fig. 1B). In addition, dSir2 did not deacetylate histones if NADH, NADP, or NADPH were used instead of NAD; was not able to deacetylate hyperacetylated BSA; did not exhibit any detectable ADP ribosyltransferase activity; and was not affected by the HDAC inhibitors trichostatin A (at 2.5 μM) or FR901228 (at 1 μM) (data not shown).
Figure 1.
dSir2 catalyzes the deacetylation of a broad range of lysine residues in core histone tails. (A) Synthesis and purification of dSir2. His-6-tagged, full-length dSir2 was purified and then subjected to 8% polyacrylamide-SDS gel electrophoresis and staining with Coomassie brilliant blue R-250. (B) Deacetylation by dSir2 requires NAD and is inhibited by coumermycin A1. Purified Drosophila core histones were hyperacetylated with acetic anhydride and then used as substrates for deacetylation by dSir2. Where indicated, dSir2 (100 nM), NAD (100 μM), and coumermycin A1 (100 μM) were included in the reactions. The extent of deacetylation at the indicated lysine residues was monitored by Western blot analysis with antibodies that specifically recognize the acetylated form of the residues. The amount of total histone polypeptides in the reaction mixtures was monitored by 15% polyacrylamide-SDS gel electrophoresis and staining with Coomassie brilliant blue R-250. The asterisk denotes a contaminant in the preparation of coumermycin A1. (C) dSir2 deacetylates histones when added prior to chromatin assembly, but not subsequent to chromatin assembly. Chromatin assembly reactions were performed with hyperacetylated histones, ACF, NAP-1, plasmid DNA, and ATP. dSir2 (100 nM) and NAD (100 μM) were added, as indicated, either before or after the chromatin assembly reactions. The deacetylation of specific lysine residues was detected by Western blot analysis as in B.
ySir2 deacetylates K9 and K14 of histone H3 and K5, K12, and K16 of histone H4 (30, 31). We found that dSir2 catalyzes the deacetylation of K9 and K14 of H3 and K5, K12, and K16 of H4 as well as K18 and K23 of H3 and K5 and K20 of H2B (Fig. 1 B and C). The only lysine residue that was observed to be weakly deacetylated by dSir2 is K8 of histone H4 (Fig. 1B). In addition, deacetylation by dSir2 is completely inhibited by 200 μM coumermycin A1 (Fig. 1B). Titration experiments revealed that 50% inhibition of dSir2 deacetylase activity is achieved at about 10 μM coumermycin A1 (data not shown).
Transcriptional Repression by dSir2 in Vitro Requires NAD and Hyperacetylated Histones.
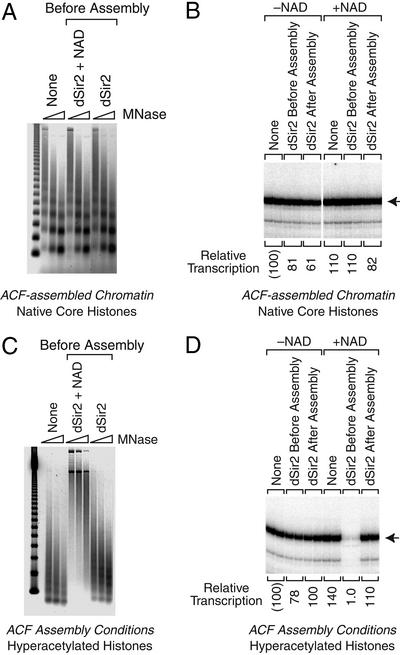
Next, we investigated the effect of dSir2 on transcription in vitro. In these experiments, transcription reactions were performed with the HIV-1 long terminal repeat promoter in conjunction with purified Sp1 and NF-κB p65 activator proteins. With naked DNA templates, we did not observe any transcription repression by dSir2 and NAD (data not shown). We then tested the effect of dSir2 on transcription of chromatin templates. To this end, we assembled chromatin with purified recombinant ACF, purified recombinant NAP-1, and purified native core histones from Drosophila embryos (39). The assembly reactions were carried out in the absence or presence of dSir2 and NAD. Micrococcal nuclease digestion analysis of the chromatin indicated that dSir2 does not affect the assembly of the purified native histones into chromatin (Fig. 2A). In addition, there was no detectable effect of dSir2 on transcription from the chromatin assembled with native core histones (Fig. 2B). Thus, dSir2 that is active for histone deacetylation is not able to repress transcription in vitro with either naked DNA or chromatin that is assembled from native histones.
Figure 2.
Transcriptional repression by dSir2 requires NAD and hyperacetylated histones. (A) dSir2 and NAD do not disrupt the periodicity of nucleosomes assembled with ACF and NAP-1. Chromatin was assembled with ACF, NAP-1, purified native Drosophila core histones, ATP, and pHIV template DNA. Where indicated, purified dSir2 (100 nM) and NAD (100 μM) were added before chromatin assembly. The resulting samples were subjected to micrococcal nuclease (MNase) digestion analysis. (B) dSir2 does not inhibit transcription of chromatin assembled with native (and mostly unacetylated) core histones. Aliquots of the same chromatin samples used in A were subjected to in vitro transcription analysis. Sp1 (10 nM) and NF-κB p65 (100 nM) were added to the chromatin templates, and transcription was carried out with a HeLa nuclear extract. Where indicated, purified dSir2 (100 nM) and NAD (100 μM) were added either before or after chromatin assembly. The resulting transcripts were detected by primer extension analysis. Transcriptional activity is reported as relative to that observed with in the absence of dSir2 and NAD, which is designated as “(100).” (C) dSir2 and NAD mediate the formation of a nuclease-resistant structure with hyperacetylated histones. Chromatin assembly and micrococcal nuclease digestion analysis was performed as in A, except that hyperacetylated core histones were used instead of native core histones. (D) dSir2 is a potent NAD-dependent repressor of transcription with hyperacetylated core histones. Aliquots of the same chromatin samples used in C were subjected to in vitro transcription analysis as in B.
It is important to note that the native histones from Drosophila embryos are mostly unacetylated. Hence, it was possible that the lack of transcriptional repression by dSir2 was due to the absence of histone acetylation. We therefore carried out experiments analogous to those in Fig. 2 A and B, except that we used either acetylated HeLa histones (purified from butyrate-treated HeLa cells) or histones that were acetylated in vitro with purified p300. We did not, however, observe >2-fold repression by dSir2 under either of these conditions (data not shown).
To investigate further the potential relation between histone acetylation and dSir2-mediated repression, we tested the transcriptional effects of dSir2 with native histones that were hyperacetylated by treatment with acetic anhydride. [These hyperacetylated histones are identical to those used in the deacetylase assays (Fig. 1 B and C).] Incubation of the hyperacetylated histones with ACF and NAP-1 under chromatin assembly conditions yields a histone–DNA complex that differs from canonical nucleosomal chromatin (Fig. 2C, left lanes). The addition of dSir2 and NAD to the hyperacetylated histones prior to the chromatin assembly reactions led to the formation of a nuclease-resistant histone–DNA complex (Fig. 2C, center lanes) that is transcriptionally repressed (Fig. 2D). [In the absence of dSir2, the level of transcription from the hyperacetylated histone–DNA complex is comparable to that seen with chromatin (as in Fig. 2B).] This nuclease-resistant transcriptionally repressive complex was not formed by dSir2 alone in the absence of NAD (Fig. 2 C and D). In addition, when dSir2 and NAD were added subsequent to the chromatin assembly reactions under conditions where histone deacetylation does not occur (Fig. 1C), transcriptional repression was not observed (Fig. 2D). Thus, as seen by comparison of Fig. 2 B and D, transcriptional repression by dSir2 in vitro requires NAD and acetylated histones. There is also a correlation between histone deacetylation, the formation of a nuclease-resistant complex, and dSir2-dependent transcriptional repression (Figs. 1C and 2 C and D).
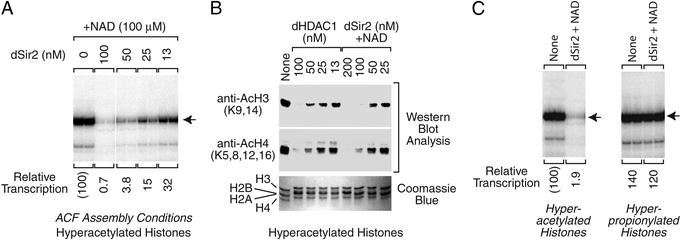
To determine the amount of dSir2 that is required to observe efficient transcriptional repression, we carried out a series of transcription reactions with varying concentrations of dSir2. These experiments indicated that the magnitude of repression increases with dSir2 concentration (Fig. 3A). At 100 nM dSir2, we typically observed ≈100-fold repression. This concentration of dSir2 correlates to approximately two molecules of dSir2 per core histone octamer. We also examined the amount of histone deacetylation that occurs with varying concentrations of dSir2 (Fig. 3B) and found that there is a rough correlation between the extent of dSir2-mediated deacetylation and transcriptional repression. (As seen later, however, histone deacetylation by dHDAC1 does not correlate with transcriptional repression.)
Figure 3.
Characterization of transcriptional repression by dSir2 in vitro. (A) Effect of dSir2 concentration on transcriptional repression. Reactions were performed as in Fig. 2D with the indicated final concentrations of purified dSir2. (B) The extent of histone deacetylation by dSir2 roughly correlates with the amount of transcriptional repression. HDAC assays were performed, as in Fig. 1, with the indicated final concentrations of dSir2 or dHDAC1. (C) dSir2 does not repress transcription with hyperpropionylated core histones. Transcription reactions were carried out as in Fig. 2D. The properties of hyperpropionylated core histones (prepared by incubation of core histones with propionic anhydride) were compared with those of hyperacetylated core histones (prepared with acetic anhydride).
We also considered the possibility that dSir2 irreversibly modifies the DNA template into a transcriptionally inactive form. To address this hypothesis, we carried out assembly reactions with dSir2 and NAD, as in Fig. 2D, and then deproteinized the template DNA prior to in vitro transcription analysis. The resulting DNA templates were transcribed with an efficiency that is comparable to that of naked DNA (data not shown). It is thus unlikely that transcriptional repression by dSir2 is due to an irreversible modification of the DNA template.
Transcriptional Repression by dSir2 Does Not Occur with Propionylated Histones.
Because of the use of acetic anhydride as the acetylating agent, we sought to test whether the observed transcriptional effects were an unforeseen consequence of the treatment of histones with a carboxylic acid anhydride. To investigate this possibility, we carried out transcription reactions with histones that were hyperpropionylated with propionic anhydride. If the dSir2 repressive effects were due to treatment of histones with a carboxylic acid anhydride, then we would observe transcriptional repression with both the hyperacetylated histones and the hyperpropionylated histones. We observed, however, that dSir2 mediates transcriptional repression with the hyperacetylated histones but not with the hyperpropionylated histones (Fig. 3C). Hence, modification of histones with a carboxylic acid anhydride does not result in a change in the general properties of the histones that leads to dSir2-mediated repression. Instead, transcriptional repression by dSir2 in vitro is specific for acetylated histones.
Deacetylation of Histones by dSir2 Generates a Fast-Sedimenting Nucleoprotein Complex.
To investigate the nature of the transcriptionally repressive histone–DNA complex that is formed by deacetylation by dSir2, we carried out sucrose gradient sedimentation analyses. In these experiments, we observed that chromatin assembled with native histones sediments at approximately the same rate as hyperacetylated histone–DNA complexes that are formed under parallel conditions (Fig. 4A, upper two gels). [Notably, chromatin samples prepared with native versus hyperacetylated histones exhibit different micrococcal nuclease digestion patterns (Fig. 2 A and C) but sediment at a similar rate.] In addition, the rate of sedimentation of the hyperacetylated histone–DNA complexes does not change on the addition of dSir2 alone (Fig. 4A, third gel). However, when the hyperacetylated histones were incubated with both dSir2 and NAD, the resulting histone–DNA complexes exhibited a faster rate of sedimentation (Fig. 4A, bottom gel). Thus, dSir2 and NAD mediate the formation of a transcriptionally repressed (Fig. 2D), nuclease-resistant (Fig. 2C), and fast-sedimenting (Fig. 4A) complex with hyperacetylated histones and DNA.
We further examined whether dSir2 remains associated with the fast-sedimenting histone–DNA complex. To this end, we performed sucrose gradient sedimentation and Western blot analyses. In the absence of NAD, dSir2 does not cosediment with the hyperacetylated histone–DNA complex (Fig. 4B). These results indicate that dSir2 does not associate stably with the hyperacetylated histone–DNA complex. In the presence of NAD, a small fraction (≈1%) of the dSir2 cosediments with the histone–DNA complex (Fig. 4C). Thus, approximately one molecule of dSir2 per 50 core histone octamers remains associated with the histone–DNA complex. These results suggest that the change in the sedimentation properties of the dSir2+NAD-treated complexes is due to a conformational change or rearrangement of the histone–DNA complex rather than the stoichiometric association of dSir2.
Deacetylation of Histones by dHDAC1 Does Not Inhibit Transcription.
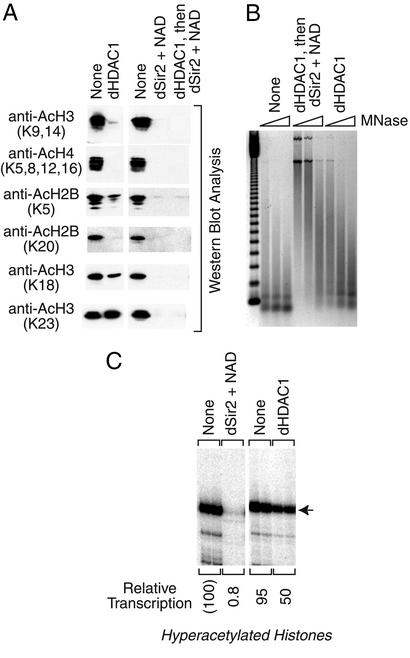
To investigate whether transcriptional repression by dSir2 is due to the deacetylation of histones, we compared the properties of dSir2 with those of the dHDAC1 deacetylase. First, we found that the specific activity of deacetylation by dHDAC1 is similar to that by dSir2 (Fig. 3B). In our standard reaction conditions, we found that dHDAC1 and dSir2 are each able to catalyze the near complete deacetylation of histones at 100 nM concentration (Fig. 3B). We then tested the ability of dHDAC1 and dSir2 to deacetylate specific lysine residues and found that there are some differences in their substrate specificities (Fig. 5A). For instance, dSir2 is more effective at deacetylation of K18 and K23 of H3 than dHDAC1. We also carried out micrococcal nuclease digestion analyses of hyperacetylated histone–DNA complexes that were assembled in the presence of dHDAC1. In these experiments, dHDAC1 did not mediate the formation of a nuclease-resistant histone–DNA complex (Fig. 5B), in contrast to that seen with dSir2 (Figs. 2C and 5B). Thus, dHDAC1 is active for the deacetylation of histones, but does not generate a nuclease-resistant structure with the hyperacetylated histones.
Figure 5.
Histone deacetylation by Drosophila HDAC1 does not repress transcription or generate a nuclease-resistant histone-DNA complex. In these experiments, Drosophila HDAC1 (dHDAC1) and dSir2 were each used at a final concentration of 100 nM. (A) Deacetylation of core histones by dHDAC1. Histone deacetylation reactions were carried out as in Fig. 1B. (B) dHDAC1 does not generate a nuclease-resistant structure. Reactions were carried out as in Fig. 2C. The histones that were treated with both dHDAC1 and dSir2 (middle lanes) were first incubated with dHDAC1 for 30 min before the addition of dSir2 and NAD at the onset of the chromatin assembly reaction. (C) dHDAC1 does not repress transcription with hyperacetylated histones. Reactions were performed as in Fig. 2D.
We had previously observed that dHDAC1 does not repress transcription when used in conjunction with native core histones (35). In these studies, we investigated whether dHDAC1 is able to repress transcription when used in conjunction with the hyperacetylated core histones. Unlike dSir2, dHDAC1 does not strongly repress transcription from the hyperacetylated histone–DNA complexes (Fig. 5C). Hence, the deacetylation of histones by dHDAC1 does not lead to the formation of a nuclease-resistant complex and the loss of transcriptional activity. It is possible that transcriptional repression by dSir2 is due to its ability to deacetylate a critical lysine residue that cannot be deacetylated by dHDAC1. However, chromatin that is assembled with native (and mostly unacetylated) histones is not repressed by either dSir2 (Fig. 2B) or dHDAC1 (35). Therefore, it appears more likely that repression by dSir2 is due to a function that is present in dSir2, but not in dHDAC1, that mediates the rearrangement of histone–DNA interactions.
Transcriptional Repression in Vitro Reflects Silencing Function in Vivo.
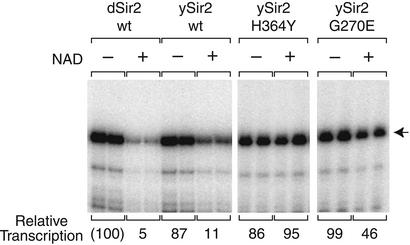
To evaluate further the relation between Sir2 deacetylase activity and transcriptional repression, we tested wild-type and mutant versions of recombinant ySir2 in our transcription assay (Fig. 6). Paralleling results with dSir2, wild-type ySir2 represses transcription in an NAD-dependent manner. In contrast, ySir2-H364Y, a catalytically inactive mutant that is globally defective for silencing in vivo (29, 30), is unable to repress transcription in vitro. Furthermore, ySir2-G270E, a mutant with 60% activity and partial silencing defects in yeast (34), is ≈50% active in repression. Thus, the transcriptional repression we observe correlates with ySir2 catalytic activity in vitro and extent of silencing function in vivo.
Figure 6.
Transcriptional repression by ySir2 in vitro reflects silencing activity in vivo. Reactions were carried out as in Fig. 2D (“Sir2 before assembly”) with dSir2 (100 nM) or the indicated ySir2 proteins (150 nM).
Summary
In this work, we observed potent NAD-dependent transcriptional repression by purified recombinant Sir2 in vitro. dSir2 represses transcription with histones that are hyperacetylated by acetic anhydride, but not with native (and mostly unacetylated) histones or with histones that are hyperpropionylated by propionic anhydride. We also found that dSir2 mediates the formation of a nuclease-resistant fast-sedimenting histone–DNA complex in an NAD-dependent manner. Unlike dSir2, the dHDAC1 deacetylase does not strongly repress transcription or generate a nuclease-resistant histone–DNA complex. Notably, mutant versions of ySir2 that are defective for silencing in vivo are also defective for transcriptional repression in the in vitro assay.
One consideration is the structure of the hyperacetylated histone–DNA complexes. These histone–DNA complexes do not appear to consist of canonical nucleosomes. Yet, at the same time, it is not known whether Sir2-mediated repression involves chromatin that consists of canonical nucleosomes. Upon treatment with dSir2 and NAD, the hyperacetylated histone–DNA complex undergoes a sharp transition to a nuclease-resistant fast-sedimenting histone–DNA complex. This altered complex contains only small amounts of dSir2 (about one molecule of dSir2 per 50 histone octamers), and thus it appears that dSir2 mediates a conformational change or rearrangement in the histone–DNA complex during deacetylation.
In conclusion, these studies describe transcriptional repression by Sir2 in vitro. The requirement for NAD and histone acetylation for repression, as well as the sensitivity to catalytic defects that are associated with silencing defects, suggests that features of this process, as seen in vitro, reflect the nature of Sir2-regulated silencing in vivo. In addition, the ability of dSir2 to mediate a conformational change or rearrangement in protein complexes may be relevant to the function of other Sir2-related proteins.
Acknowledgments
We thank Tammy Juven-Gershon, Karl Haushalter, Arthur Hsu, Tom Boulay, Buyung Santoso, and Vassili Alexiadis for critical reading of the manuscript. This work was supported by grants from the National Institutes of Health to J.T.K. (GM 46995) and L.P. (GM 54778). X.H.P. is a Fellow of the Leukemia and Lymphoma Society.
Abbreviations
- HDAC
histone deacetylase
- dSir2
Drosophila Sir2
- ySir2
Saccharomyces cerevisiae Sir2
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Brown C E, Lechner T, Howe L, Workman J L. Trends Biochem Sci. 2000;25:15–19. doi: 10.1016/s0968-0004(99)01516-9. [DOI] [PubMed] [Google Scholar]
- 2.Strahl B D, Allis C D. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 3.Peterson C L, Workman J L. Curr Opin Genet Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 4.Flaus A, Owen-Hughes T. Curr Opin Genet Dev. 2001;11:148–154. doi: 10.1016/s0959-437x(00)00172-6. [DOI] [PubMed] [Google Scholar]
- 5.Fry C J, Peterson C L. Curr Biol. 2001;11:R185–R197. doi: 10.1016/s0960-9822(01)00090-2. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Reinberg D. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 7.Becker P B, Hörz W. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 8.Emerson B. Cell. 2002;109:267–270. doi: 10.1016/s0092-8674(02)00740-7. [DOI] [PubMed] [Google Scholar]
- 9.Narlikar G J, Fan H-Y, Kingston R E. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 10.Gottschling D E. Curr Biol. 2000;10:R708–R711. doi: 10.1016/s0960-9822(00)00714-4. [DOI] [PubMed] [Google Scholar]
- 11.Guarente L. Genes Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- 12.Dutnall R N, Pillus L. Cell. 2001;105:161–164. doi: 10.1016/s0092-8674(01)00305-1. [DOI] [PubMed] [Google Scholar]
- 13.Rine J, Herskowitz I. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 15.Aparicio O M, Billington B L, Gottschling D E. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 16.Bryk M, Banerjee M, Murphy M, Knudsen K E, Garfinkel D J, Curcio M J. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- 17.Fritze C E, Verschueren K, Strich R, Esposito R E. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith J S, Boeke J D. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 19.Moretti P, Freeman K, Coodly L, Shore D. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 20.Moazed D, Kistler A, Axelrod A, Rine J, Johnson A D. Proc Natl Acad Sci USA. 1997;94:2186–2191. doi: 10.1073/pnas.94.6.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 22.Shou W, Seol J H, Shevchenko A, Baskerville C, Moazed D, Chen Z W S, Jang J, Shevchenko A, Charbonneau H, Deshaies R J. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 23.Straight A F, Shou W, Dowd G J, Turck C W, Deshaies R J, Johnson A D, Moazed D. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- 24.Kaeberlein M, McVey M, Guarente L. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottlieb S, Esposito R E. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 26.Rosenberg M I, Parkhurst S M. Cell. 2002;109:447–458. doi: 10.1016/s0092-8674(02)00732-8. [DOI] [PubMed] [Google Scholar]
- 27.Brachmann C B, Sherman J M, Devine S E, Cameron E E, Pillus L, Boeke J D. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 28.Frye R A. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 29.Tanny J C, Dowd G J, Huang J, Hilz H, Moazed D. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- 30.Imai S, Armstrong C M, Kaeberlein M, Guarente L. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 31.Landry J, Sutton A, Tafrov S T, Heller R C, Stebbins J, Pillus L, Sternglanz R. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith J S, Brachmann C B, Celic I, Kenna M A, Muhammad S, Starai V J, Avalos J L, Escalante-Semerena J C, Grubmeyer C, Wolberger C, Boeke J D. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 34.Garcia S N, Pillus L. Genetics. 2002;162:721–736. doi: 10.1093/genetics/162.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang X, Kadonaga J T. J Biol Chem. 2001;276:12497–12500. doi: 10.1074/jbc.C100034200. [DOI] [PubMed] [Google Scholar]
- 36.Bulger M, Kadonaga J T. Methods Mol Genet. 1994;5:241–262. [Google Scholar]
- 37.Jiang W, Nordeen S K, Kadonaga J T. J Biol Chem. 2000;275:39819–39822. doi: 10.1074/jbc.C000713200. [DOI] [PubMed] [Google Scholar]
- 38.Hebbes T R, Thorne A W, Crane-Robinson C. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ito T, Levenstein M E, Fyodorov D V, Kutach A K, Kobayashi R, Kadonaga J T. Genes Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]