Abstract
Zinc finger domains are small DNA-binding modules that can be engineered to bind desired target sequences. Functional transcription factors can be made from these DNA-binding modules, by fusion with an appropriate effector domain. In this study, eight three-zinc-finger proteins (ZFPs) that bound HIV-1 sequences in vitro were engineered into transcription repressors by linking them to the Krüppel-associated box (KRAB) repressor domain (KOX1). One protein, ZFP HIVB-KOX, which bound to a 9-bp region overlapping two Sp1 sites, was found to repress a Tat-activated 5′ LTR cellular HIV-reporter assay to almost basal levels. A related six-finger protein, HIVBA′-KOX, was made to target all three Sp1 sites in the 5′ LTR promoter and efficiently inhibited both basal and Tat-activated transcription in unstimulated and mitogen-stimulated T cells. In contrast, a combination of two unlinked three-finger ZFPs, HIVA′-KOX and HIVB-KOX, which bind over the same region of DNA, resulted in less effective repression. Finally, HIVBA′-KOX was tested for its capacity to block viral replication in a cellular infection assay using the HIV-1 HXB2 strain. This ZFP was found to inhibit HIV-1 replication by 75% compared with control constructs, thus demonstrating the potential of this approach for antiviral therapy.
In recent years, the classical Cys2-His2 zinc-finger motif has been widely used as a scaffold for the construction of customized transcription factors (refs. 1–7; reviewed in refs. 8–10). Engineered zinc-finger proteins (ZFPs) are particularly suited to this purpose not only because they are capable of binding to a wide range of DNA sequences (11–13) but also because finger subunits may be linked together to bind long asymmetric DNA sequences (14, 15). For example, three- to six-finger domains have been constructed by a number of groups and have successfully activated or repressed gene targets in a wide variety of systems (reviewed in refs. 8–10). Although ZFPs have been targeted before to sequences in the HIV promoter (12, 16), the potential of these ZFPs to inhibit a live viral infection has not yet been investigated. In this article and in the accompanying paper (17), we demonstrate inhibition of two clinically relevant viruses.
Currently, more than 35 million people worldwide are infected with HIV, and most will develop AIDS. The most effective treatment to date is a mixture of reverse transcriptase and protease inhibitors, which can significantly reduce the amount of HIV in the blood. However, these drugs can cause severe side effects and are expensive. In addition, viral escape mutants may develop. As an alternative to chemotherapy, DNA vaccines carrying HIV genes have been used in attempts to immunize against HIV (18, 19). Despite such progress in recent years, the need to seek out and develop new therapies remains vitally important.
Designer ZFPs have the potential to exploit several HIV-specific processes, both at the level of transcription and in other nucleic acid–protein interactions. HIV-1 encodes two regulatory proteins, Tat and Rev, which both function through interactions with specific RNA elements in the viral genome. These elements are transactivation response element (TAR) and the Rev response element. Both of these protein–RNA interactions are required for HIV replication and are potential target points for antiviral strategies. Although no studies using ZFPs have targeted the Tat–TAR interaction, there are two examples of engineering zinc fingers that bind the Rev response element (20, 21). However, neither report examined whether these ZFPs were able to inhibit viral replication.
Despite the fact that engineered ZFP transcription factors have been shown to regulate several important endogenous genomic targets from humans (22–24) and mice (25), there has not been a previous example of regulation of an integrated proviral sequence. The engineered ZFPs of this study were designed to bind key DNA sequences within the HIV-1 5′ LTR promoter, including Sp1 sites and the TATA element. To inhibit transcription and, therefore, replication of the virus, our ZFPs were fused to the Krüppel-associated box (KRAB) repression domain from the human protein KOX1, which has been shown to be effective in a different system (26).
By studying the behavior of these ZFP constructs in a variety of cellular assays, we have evaluated the potential of this distinctive class of antiviral agent. In particular, because these ZFPs do not occlude the nuclear factor (NF)-κB-binding sites in the LTR, we have examined the ability of these constructs to inhibit NF-κB-driven transcription in activated T cells. The activation of HIV transcription after the exposure of infected T cells to antigen or mitogen is a key process in disease progression because it results in the reactivation of a latent provirus, which may have been transcriptionally silent for many years. This activation of HIV transcription is mediated by the translocation of active NF-κB to the nucleus and binding to two sites that lie immediately upstream of the Sp1 sites in the LTR. Significantly, we show that engineered ZFPs linked to the KOX1 domain are able to inhibit both basal and Tat-activated transcription from the LTR in unstimulated and stimulated T cells and are also able to inhibit viral replication in cellular infection assays.
Materials and Methods
Construction of Three- and Six-Zinc-Finger Proteins.
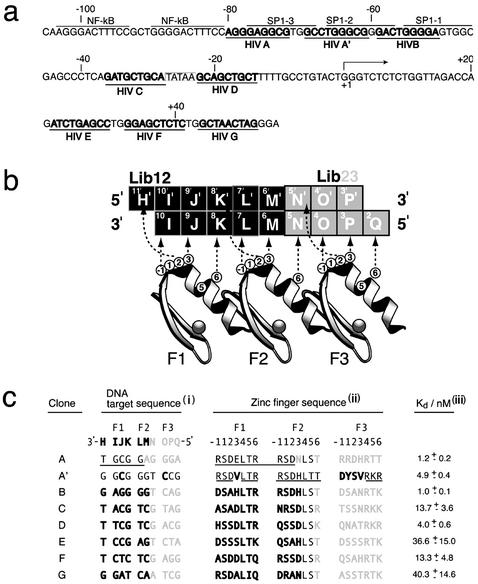
Seven three-finger proteins were constructed by using the bipartite complementary system to bind specific sequences in the HIV-1 LTR (12). These ZFPs were named ZFP HIVA–HIVG (12). ZFP A′ protein was selected from a limited library, which was generated by mutation of the Zif268 WT sequence by using the primers SfiVal3 and NotGCC. SfiVal3 encoded a Val at position 3 in helix 1 so that a T or C could be bound in the final triplet i.e., 5′-GCCTGGG(C/T)G, and NotGCC randomized amino acids at positions −1, 1, and 3 of helix 3, incorporating His or Asp in position −1, Tyr, His, Ser, or Pro in position 2, and Val, Ala, Glu, Leu, or Ser in position 3. This library was cloned into the filamentous phage cloning vector fdtet and screened against binding sites, as described (7). ELISA verified that the protein could bind 5′-GCCTGGGCG and also 5′-GCCTGGGTG, which is a natural variation in HIV-1 (27).
The six- and nine-finger ZFPs were constructed by fusing the constituent three-finger domains by using peptide linkers. Clones HIVA′ and HIVA were joined by using the linker TGGSGGSGERP to make HIVA′A. Clones HIVB and HIVA were coupled to make BA by using the linker LRQKDG(GSG)5GERP. HIVB and HIVA′ were linked by using the peptide sequence TGGSGERP to make HIVBA′ (15, 28). Similarly, HIVFE, HIVGF, and HIVGFE were constructed from the appropriate three-finger domains by using the extended linker peptide LRQKDGERP (29).
Cloning of Zinc-Finger Constructs with Repressor Domains.
The engineered ZFPs were cloned into a modified pcDNA3.1(−) mammalian expression vector (Invitrogen). This vector, pKOX, incorporates a 7-aa nuclear localization sequence from simian virus 40 large T-antigen (30), a KRAB repressor domain from human KOX1 (31), and a C-terminal 10-aa sequence of the c-Myc 9E10 epitope (32), cloned into the EcoRI and BamHI restriction sites. Zinc-finger-coding regions were amplified by PCR from fdtet vector to contain a 5′ XbaI restriction site, a Kozak sequence, and a 3′ EcoRI site. The ZFP constructs were digested with XbaI and EcoRI and ligated into similarly digested pKOX. The plasmids were named pHIVA-KOX, pHIVA′-KOX, pHIVB-KOX, etc.
Construction of Reporter Vectors.
The chloramphenicol acetyltransferase (CAT) reporter plasmid, D5-3-3, contains the CAT gene under the direct control of HIV-1 LTR sequences derived from the NL4 proviral clone (33). The luciferase reporter plasmid was generated by cloning the EcoRV–HindIII fragment of D5-3-3, containing the LTR, into pGL3-basic (Promega) digested with SmaI and HindIII, to create LTR-FF (34).
Transient Assays Using the CAT Reporter Vector.
COS7 cells were cultured at 37°C, under a 5% CO2/95% air atmosphere in DMEM containing penicillin (100 units/ml), streptomycin (100 μg/ml), and 2 mM l-glutamine, supplemented with 10% FCS. Cells were resuspended in PBS at 107 cells per ml, and 0.7 ml was transfected with transfection mix by electroporation. The transfection mix comprises 10 μg of CAT reporter plasmid, 0.1 μg of C63-4-1, a plasmid that expresses Tat from a Moloney virus LTR promoter (35), and 10 μg of ZFP-expressing plasmid. For control transfections, the C63-4-1 and/or the ZFP plasmid were replaced by pcDNA3.1(−)/His/LacZ (Invitrogen). The electroporated samples were cultured for a further 24 h in 8 ml of medium followed by harvesting and resuspension in PBS. Samples were removed for normalization of cell numbers, and the remaining cells were lysed in Reporter Lysis buffer (Promega). CAT activity was assayed by using the Quan-T-CAT assay system (Amersham Pharmacia). Samples were resuspended in liquid scintillation mixture (Beckman) and levels of 3H were measured for 5 min in a scintillation counter.
Transient Assays Using the Luciferase Reporter Vector.
The Jurkat human T cell line was cultured at 37°C under a 7% CO2/93% air atmosphere in RPMI 1640 medium containing penicillin (100 units/ml) and streptomycin (100 μg/ml), supplemented with 10% FCS. Cells were transfected with transfection mix by using Effectene (Qiagen) in six-well plates, according to the manufacturer's instructions. Initial experiments were performed to determine the optimal amount of phorbol 12-myristate 13-acetate (PMA) required to stimulate the maximal level of basal HIV transcription from the LTR and the optimal concentration of Tat required for full activation of the LTR in T cells. These were determined to be 50 ng/ml for PMA and 25 ng for C63-4-1 (Tat-expressing vector) (data not shown).
In the experiments, each transfection mix contained 600 ng of LTR-FF, 150 ng of pRL-TK (plasmid containing the Renilla luciferase gene under the control of the thymidine kinase promoter, Promega), 25 ng of C63-4-1, and 150–300 ng of either a ZFP plasmid or pcDNA3.1(−). Concentrations of DNA were kept constant throughout by the addition of pUC18. DNA was mixed in a total volume of 150 μl of EC buffer, and 8 μl of Enhancer was added for every μg of DNA. The transfection mixes were vortexed and incubated at room temperature for 5 min, and then 10 μl of Effectene was added per μg of DNA. These were incubated for a further 5 min at room temperature, then 0.5 ml of normal growth medium was added to each. The total mix was then added to 2 ml of cells, resuspended at 2.5 × 105 cells per ml in fresh medium. The cells were incubated at 37°C for 2 h, and then 2.5 ml of normal growth medium was added to each. Cells were activated 24 h after transfection by the addition of phytohemagglutinin (PHA) (Sigma) and PMA (Sigma) to a final concentration of 10 μg/ml and 50 ng/ml, respectively. Cells were then harvested after a further 24 h, washed once in PBS, and then lysed in 150 μl of 1× passive lysis buffer (Promega) for 30 min at room temperature. Samples (10 μl) of the lysates were assayed by using 50 μl of Luciferase Assay Reagent II reagent and 50 μl of Stop and Glo reagent from the dual luciferase assay system kit (Promega). Firefly luciferase and Renilla luciferase activities were measured sequentially by using a microplate luminometer with an injection unit (Berthold Detection Systems, Pforzheim, Germany).
Toxicity assays were performed in parallel with luciferase assays by transferring 100 μl of transfected cell mix into a well of a 96-well plate. Normal growth medium (100 μl) was added 2 h after transfection. These cells were treated in parallel with PMA and PHA on day 2, and cell proliferation was measured on day 3 by the addition of 40 μl of CellTiter 96 aqueous one solution cell proliferation assay reagent (Promega). Cells were then incubated at 37°C for 2–4 h, and the level of colored product produced was determined by measuring the absorbance at 490 nm.
The pRL-TK plasmid was included in these experiments as a control for transfection efficiency. This plasmid expresses the Renilla luciferase gene under the control of the herpes simplex virus thymidine kinase promoter. Toxicity assays were performed in parallel to account for toxic effects of PMA and PHA and to detect any possible toxicity of the ZFP-expressing plasmids. All results were corrected for toxicity, and the HIV-1 5′ LTR firefly luciferase results were then adjusted for transfection efficiency.
Transfection of DNA Constructs and Challenge with HIV-1.
An NP2 cell line that stably expressed CD4 (NP2/CD4) was cultured at 37°C under 5% CO2 in DMEM containing penicillin (100 units/ml), streptomycin (100 μg/ml), and G418 (500 μg/ml), supplemented with 5% FCS (36). Cells were seeded at 105 cells per well of a six-well plate. The following day the cells were transfected with transfection mix by using Lipofectin (GIBCO). Transfection mix contained 0.4 μg of pcDNA3.1-CXCR4 with either 2 μg of the required pcDNA3.1(−) vector (empty or containing HIVBA′, HIVBA′-KOX, or TFZ-KOX, a control ZFP). At 24 h after transfection, the samples were reseeded at 2.5 × 104 cells per well into a 48-well plate and grown for a further 24 h. The transfected cells were then challenged with 10-fold serial dilutions of the HXB2 strain of HIV-1. Virus supernatant (100 μl) was added to each well and incubated for 3 h, and then 0.5 ml of growth medium was added. The samples were grown for a further 72 h and then washed in PBS and fixed in cold (−40°C) methanol/acetone (1:1) for 10 min. The samples were further washed with PBS and PBS/1% FCS and immunostained by using an anti-p24 mouse primary mAb with anti-mouse IgG β-galactosidase-conjugated secondary Ab, before incubation with substrate (37). Foci of infection, which were stained blue, were estimated by light microscopy and the number of focus-forming units per milliliter was calculated.
Replication of the HIV-1 HXB2 strain was assessed by measuring supernatant reverse transcriptase by using an ELISA kit (Cavidi Tech, Uppsala), according to the manufacturer's instructions.
Results
Repression of Transcription from the HIV-1 LTR in a Model System.
Seven three-finger proteins had previously been constructed for binding to HIV-1 LTR DNA sequences and had been shown to bind correctly in ELISA and electrophoretic mobility-shift assay (12). However, to give more coverage of the HIV-1 promoter an eighth protein, HIVA′, was produced to bind between the second and third Sp1 sites (Fig. 1). This protein was selected against the target site 5′-GCC TGG GCG from a small library of Zif268 variants containing mutations at positions −1, 1, and 3 of finger 3. Position 3 of finger 1 was also mutated from Glu to Val to accommodate a C to T variant in the HIV-1 sequence (27). The binding affinity and specificity of HIVA′ were verified by ELISA, and its apparent Kd was measured to be 4.9 nM.
Figure 1.
DNA target sites and the corresponding ZFPs engineered to bind the HIV-1 promoter. (a) DNA sequence from the HIV-1 (HXB2 strain) 5′ LTR (GenBank accession no. K03455). Nucleotides are numbered relative to the transcription start site (+1). The sites targeted by the ZFPs HIVA–HIVG are underlined. Binding sites for NF-κB and Sp1 are also highlighted (adapted from refs. 27 and 38). (b) DNA recognition by the three-finger peptides in this study. Potential protein–DNA contacts are indicated by dotted arrows. The products shown are derived from two combinatorial libraries whose randomized amino acid positions are shown as circles (numbered relative to their helical positions). Each library binds a complementary DNA sequence, “HIJKLM” or “NOPQ,” so that when these subdomains are recombined they make a full-length protein that potentially binds all 10 bp of the DNA, “HIJKLMNOPQ,” as shown (12). (c) Selection of DNA-binding domains to recognize the HIV-1 promoter. (i) Nucleotide sequences from HIV-1 of the form 3′-HIJKLMNOPQ-5′ (see above), as recognized by clones HIVA to HIVG. (ii) Amino acid sequences of the helical regions from recombinant ZFPs that recognize HIV-1 sequences. The origin of the amino acids is indicated by shading Lib12 and Lib23 residues. HIVA and HIVA′ contain some WT Zif268 residues (underlined). (iii) Apparent Kd for the interaction of the ZFPs for their cognate sequences as measured by phage ELISA (7, 12).
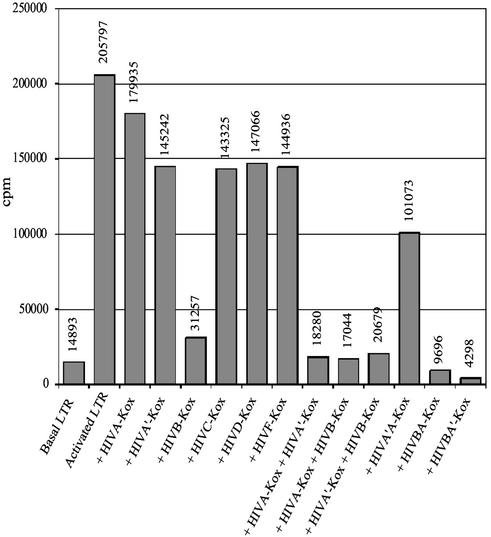
To generate functional transcription factors, the ZFPs listed in Fig. 1 were fused to the KRAB repressor domain from the human KOX1 protein. This domain was chosen because it had been shown to be an effective repressor of a chimeric HIV-1 when fused to TetR (26). The six tightest-binding three-finger ZFPs (HIVA-, HIVA′-, HIVB-, HIVC-, HIVD-, and HIVF-KOX) were subsequently tested for in vivo activity by using the HIV-1 LTR in a CAT reporter construct. HIV Tat was coexpressed and caused a 14-fold activation of basal CAT expression from the HIV-1 5′ LTR promoter (Fig. 2). Of the six three-finger ZFPs, HIVB-KOX was the most potent, repressing the activated LTR by 85%. All other proteins repressed the LTR by ≈30%.
Figure 2.
CAT assays for HIV-1 5′ LTR activity in the presence of ZFP-KOX transcription factors. Controls were basal LTR activity (without Tat) and activated LTR activity (0.1 μg of Tat plasmid added). ZFP repressors were coexpressed with the LTR-CAT reporter plasmid (10 μg) and HIV-1 Tat. Results are shown for three-finger repressor proteins, combinations of two three-finger ZFPs, and six-finger proteins.
The six-finger transcription factors were constructed from the three-finger peptides described above, on the basis that increasing the affinity and stability of the protein–DNA complex may improve their repressive function. Previous data from our lab and others (9, 10, 15, 24, 28, 29) have demonstrated that six-finger proteins have increased affinity for DNA relative to three-finger proteins, and therefore, in this instance, activity alone was tested. The six-finger protein ZFP HIVA′A-KOX was created by fusion of the two ZFPs HIVA′ and HIVA with a canonical-type linker containing two Gly-Ser-Gly (-GSG-) insertions. The resultant linker, TG(GSG)2ERP, was able to span a gap of 2 bp between the two 9-bp-binding sites (M.M., unpublished data). ZFPs HIVB and HIVA′ were fused with the linker peptide TGGSGERP, which can bridge a 1-bp gap, to create the six-finger peptide HIVBA′. Another six-finger ZFP, HIVBA, was constructed by joining HIVB and HIVA with the 26-residue linker LRQKDG(GSG)5GERP. This linker probably spans the minor groove of the DNA double helix, allowing the HIVB and HIVA domains to bind their respective binding sites some 12 bp apart. In the functional assays, all six-finger ZFPs repressed the activated HIV-1 5′-LTR. HIVBA-KOX and HIVBA′-KOX repressed the activated LTR to below basal level (95% and 98% repression, respectively), which was significantly greater than the best three-finger protein, HIVB-KOX (Fig. 2). Perhaps surprisingly, HIVA′A-KOX reduced the activity of the activated LTR by only 50%, which was similar to the values obtained for the other six-finger ZFPs HIVFE-KOX and HIVGF-KOX, and the nine-finger protein, HIVGFE-KOX (data not shown).
The three-finger ZFPs HIVA-, HIVA′-, and HIVB-KOX were also tested in pairs without covalent linkers. The combinations HIVA-KOX + HIVA′-KOX, HIVA-KOX + HIVB-KOX, and HIVA′-KOX + HIVB-KOX were assayed and found to repress CAT expression from the activated 5′-LTR to almost basal levels (91%, 92%, and 90%, respectively) (Fig. 2). The use of multiple proteins may mimic more closely the activity of natural transcription factors, which form a multicomponent complex on a promoter. That said, the six-finger ZFPs HIVBA-KOX and HIVBA′-KOX were the better repressors.
Repression of Transcription from the HIV-1 LTR in a Physiological System.
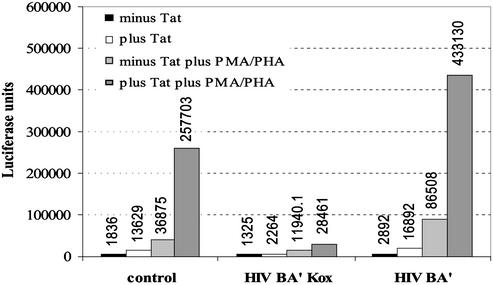
Experiments were performed to determine whether the ZFPs described could inhibit HIV-1 LTR-driven transcription of the luciferase gene under more physiological conditions within human T cells, the natural targets of HIV. Initial experiments used the plasmid pHIVBA′-KOX, which expresses the most effective six-finger ZFP, HIVBA′-KOX. This construct was tested at various concentrations in the absence and presence of HIV-1 Tat and the mitogens PMA and PHA. In the control experiment, the activity of the basal and stimulated LTR was maximal from 40 ng of the Tat-expressing plasmid (data not shown). However, upon coexpression of HIVBA′-KOX, both basal and Tat-activated transcription in the absence and presence of mitogens was repressed. In fact, the level of transcription detected in mitogen-stimulated cells in the presence of Tat was reduced by 90% by using 150 ng of pHIVBA′-KOX. Doubling the amount of the pHIVBA′-KOX plasmid did not result in further inhibition (data not shown). Because HIVBA′-KOX was able to inhibit transcription efficiently in the presence of PMA and PHA, it is clear that the binding of NF-κB to its upstream binding sites in activated T cells cannot overcome the inhibitory action of the ZFP repressor, which binds over downstream Sp1 sites. This result is consistent with the observation that a cooperative interaction between NF-κB and Sp1 is required for HIV enhancer activity (39). Similar results were obtained for HIVBA-KOX (data not shown).
Further experiments were performed to determine whether the binding of HIVBA′ to the HIV-1 5′ LTR was sufficient to inhibit transcription in the absence of the KOX1 domain. For this study, HIVBA′ was created to contain a nuclear localization sequence but no KOX1 domain. In agreement with previous data, the expression of HIVBA′-KOX inhibited HIV transcription by 89%; however, HIVBA′ had a slight stimulatory effect on transcription, particularly in the presence of PMA and PHA (Fig. 3).
Figure 3.
Luciferase assays for HIV-1 5′ LTR activity in human T cells in the presence or absence of Tat, PMA/PHA, and control vector pcDNA3.1(−), HIVBA′-KOX, or HIVBA′.
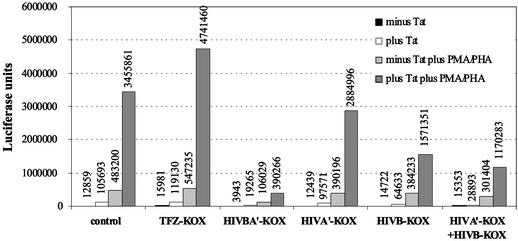
In an experiment similar to the CAT assay described above, the six-finger ZFP HIVBA′-KOX and the three-finger ZFPs HIVB-KOX and HIVA′-KOX were assayed separately, and in concert, to compare the extent of inhibition that could be achieved by combinations of ZFPs in T cells. As an additional control, ZFP TFIIIAZIF-KOX (TFZ-KOX) (28), which does not target the HIV-1 5′ LTR but binds a G-rich sequence, was used (Fig. 4). The results demonstrated that expression of TFZ-KOX in these cells had no effect on HIV-1 5′ LTR transcription, as expected, and provided an important control for any possible trans effects from the KOX1 domain. Again three-finger proteins were less effective at inhibiting HIV transcription than the six-finger protein. ZFPs HIVB-KOX and HIVA′-KOX alone reduced the level of activated transcription in the presence of Tat, PMA, and PHA by 55% and 17%, respectively, whereas in combination a reduction of 66% was observed. In contrast, HIVBA′-KOX reduced the level of activated transcription in the presence of Tat by 89%.
Figure 4.
Luciferase assays for HIV-1 5′ LTR activity in human T cells in the presence or absence of Tat, PMA/PHA, and various ZFP repressor constructs (150 ng of plasmid). The control contained pcDNA3.1(−) in place of the ZFP expression plasmid.
It is clear from these experiments that the inhibitory function of HIVBA′-KOX is mediated by the repression domain and is not solely a result of occlusion of Sp1 or RNA polymerase II from the LTR. However, given the absence of inhibition from TFZ-KOX, we conclude that binding to the HIV-1 LTR is required for inhibition. The stimulatory effect of HIVBA′ may result from the opening up of the DNA structure around the promoter, allowing easier access for transcription factors such as NF-κB.
Inhibition of HIV-1 HXB2 Strain.
In the assays described above, HIVBA′-KOX was shown to inhibit transcription of reporter genes from the HIV-1 5′ LTR. Whereas one assay was purely a model to determine activity against the LTR, the other assay was performed in host T cells under stimulation to mimic more physiological conditions. However, we wished to test HIVBA′-KOX against a more challenging and relevant target, the HIV virus. In this study, pHIVBA′-KOX was cotransfected with a vector that expressed CXCR4 into NP2 cells that had been stably transfected with human CD4. NP2 is a human glioma cell line that does not express the common HIV and SIV (simian immunodeficiency virus) coreceptors (37) and does not become infected in the absence of expression of both CD4 and CXCR4. When challenged with HIV, only cells that express CD4 and the coreceptor, CXCR4, would become susceptible to infection. In this experiment, it was assumed that cells that had been transfected and expressed CXCR4 would also coexpress the ZFP.
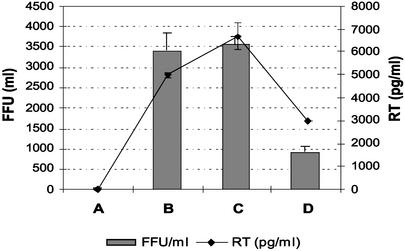
The results from the HIV-1 infection were analyzed in two ways: first, cells were stained with an anti-p24 Ab to quantify foci of infection; second, supernatants were analyzed for virus particles by using an ELISA for reverse transcriptase. The results of the first assay demonstrated that the six-finger HIVBA′-KOX inhibited HIV-1 (HXB2 strain) replication by 75%, as judged by the reduction in the number of foci (Fig. 5). Meanwhile, the level of reverse transcriptase in the supernatant was reduced 3-fold. The control construct, pTFZ-KOX, did not inhibit HIV-1 replication. This result demonstrates that the inhibitory effect of HIVBA′-KOX on viral replication cannot be attributed to nonspecific inhibition from the KOX domain or from nonspecific DNA binding. Replication in this control was slightly enhanced compared with the CXCR4 control.
Figure 5.
Assays to demonstrate inhibition of HIV-1 replication. The bars show HIV-1 focus-forming units (FFU)/ml, and the line shows the level of HIV-1 reverse transcriptase (RT) in the culture supernatant. The negative control contains pcDNA3.1 alone (A), whereas positive controls contain pcDNA3.1 + CXCR4 (B) or TFZ-KOX + CXCR4 (C). HIVBA′-KOX was tested and showed a reduction in the number of foci and the levels of viral RT (D).
Discussion
ZFPs have been designed to specifically target sequences within the HIV-1 5′ LTR. Eight three-finger repressors were made that covered a region of the promoter including the three Sp1 sites, the TATA box, and the 5′ untranslated region. All three-finger ZFPs had Kd values in the nanomolar range, which were comparable to those of the natural proteins, Sp1 and Zif268. However, HIVB-KOX, which bound across two Sp1 sites, demonstrated the greatest effect on gene transcription. This finding suggests that in vivo activity may be determined by position of binding and DNA accessibility, as well as affinity for the target site.
There is much known about the chromatin organization, and the occupancy of transcription factors at the HIV-1 5′ LTR, that may help us to understand the activity of the HIVB-KOX and HIVBA′-KOX proteins. Under basal conditions the transcription factor sites for Sp1 and NF-κB lie between two nucleosomes positioned at −415 to −255 (nuc-0) and −3 to 141 (nuc-1) in a region that contains two DNase I-hypersensitive sites, DHS2 (−232 to −130) and DHS3 (−65 to −6) (40, 41). The Sp1 and NF-κB sites have been shown to be essential for proviral activity in vivo, and other transcription factors such as LEF1 and ETS-1 require Sp1 for activity in vitro (42, 43). It is thought that only Sp1 and NF-κB occupy sites in vivo within DHS2 and DHS3 and can generate increased nucleosome remodeling without displacement of underlying histones (44). Owing to their accessibility, such DHSs have been the preferred targets of engineered ZFP transcription factors for endogenous gene regulation experiments (22, 23). With this in mind, the greater activity of HIVB-KOX may be due, at least in part, to its having easier access to its target DNA site in comparison to other three-finger proteins. However, it should also be noted that the initial assays were performed in reporter systems in which the organization of proteins on the LTR may be different from the integrated proviral LTR. Nevertheless, it is interesting that both HIVA- and HIVA′-KOX also bind across Sp1 sites but do not have the same effect as HIVB-KOX. ZFPs HIVC- and HIVD-KOX bind either side of the TATA element and it is assumed that this site would also be accessible to transcription factors. In contrast, HIVF-KOX binds in the region protected by nucleosome nuc-1. nuc-1 becomes displaced or disrupted under stimulation with cytokines or phorbol esters which, in turn, leads to HIV-1 gene expression (41, 45).
The six-finger proteins, HIVBA-KOX and HIVBA′-KOX, were constructed in an attempt to create a more potent inhibitor than the three-finger HIVB-KOX. These proteins reduced transcription from the Tat-activated LTR to below basal level. They were also more effective than combinations of HIVB-KOX, HIVA-KOX, and HIVA′-KOX, demonstrating that increased affinity through the covalent linkage of two three-finger units leads to greater repression of gene activity. This is probably because the half-life of a six-finger ZFP/DNA complex is generally far longer than that of a three-finger complex (15), so the six-finger KOX repressor can be active for longer periods. Such high-affinity artificial transcription factors may also benefit from being better able to exclude endogenous transcription factors (such as Sp1 and NF-κB) from the target promoter. Interestingly, the same was not true for HIVA′A-KOX, but the reasons for this are unclear.
The possibility that the ZFP repressors were effective only by occluding Sp1 and other factors from their sites in the HIV-1 5′ LTR was investigated. Removal of the KOX1 domain from HIVBA′-KOX greatly reduced its repression effect, indicating that this domain was essential for full activity. It was surprising that only ZFP-KOX fusions targeted to the Sp1 sites showed significant repression of the HIV-1 LTR. This is in contrast with a previous study in which a TetR-KRAB fusion protein caused an 80% reduction in viral titers when two or seven TetR-binding sites were introduced 6 kb from the promoter (26).
The repressors described here were also tested in a system that mimicked stimulated physiological conditions, in which binding of NF-κB and the recruitment of its coactivator complex to the LTR occur during T cell activation. Under these conditions, the six-finger ZFP HIVBA′-KOX was still able to repress expression from the LTR. It is possible that the protein abrogates any stimulatory effects from the binding of NF-κB to the HIV-1 5′ LTR through the recruitment of heterochromatin silencing proteins that are associated with the KRAB domain-binding protein, KAP-1 (46).
When HIVBA′-KOX was assayed with a more clinically relevant target, the HIV-1 HXB2 strain, it behaved essentially as seen in the earlier reporter assays, reducing viral replication and viral infectivity 75%. The effect of HIVBA′-KOX was shown to be specific, as a control protein, TFZ-KOX, that contained functional DNA-binding zinc fingers had a slightly stimulatory effect on viral production.
In this work we have demonstrated that customized ZFPs can inhibit HIV-1 replication by repression of the 5′ LTR promoter. It is too early to suggest that this would represent an alternative therapy for HIV infection, as there are many issues still to overcome, such as transgene delivery and possible immunogenicity. However, these problems are associated with other gene therapies and are the focus of much research; lentiviral vectors, for example, are constantly under development (47). These considerations notwithstanding, ZFP transcription factors have been shown to be able to reduce the infection of cells with HIV-1, and in the accompanying paper (17) we demonstrate that this method is applicable to other viral diseases.
Acknowledgments
We acknowledge Aine McKnight and David Marchent of the Wohl Virion Centre, London, for their assistance with the HIV assays.
Abbreviations
- ZFP
zinc-finger protein
- PMA
phorbol 12-myristate 13-acetate
- PHA
phytohemagglutinin
- CAT
chloramphenicol acetyltransferase
- NF
nuclear factor
- KRAB
Krüppel-associated box
References
- 1.Desjarlais J R, Berg J M. Proc Natl Acad Sci USA. 1992;89:7345–7349. doi: 10.1073/pnas.89.16.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desjarlais J R, Berg J M. Proc Natl Acad Sci USA. 1993;90:2256–2260. doi: 10.1073/pnas.90.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jamieson A C, Kim S H, Wells J A. Biochemistry. 1994;33:5689–5695. doi: 10.1021/bi00185a004. [DOI] [PubMed] [Google Scholar]
- 4.Rebar E J, Pabo C O. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 5.Choo Y, Klug A. Proc Natl Acad Sci USA. 1994;91:11163–11167. doi: 10.1073/pnas.91.23.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choo Y, Klug A. Proc Natl Acad Sci USA. 1994;91:11168–11172. doi: 10.1073/pnas.91.23.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isalan M, Choo Y. Methods Enzymol. 2001;340:593–609. doi: 10.1016/s0076-6879(01)40444-7. [DOI] [PubMed] [Google Scholar]
- 8.Choo Y, Isalan M. Curr Opin Struct Biol. 2000;10:411–416. doi: 10.1016/s0959-440x(00)00107-x. [DOI] [PubMed] [Google Scholar]
- 9.Pabo C O, Peisach E, Grant R A. Annu Rev Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 10.Beerli R R, Barbas C F., III Nat Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 11.Greisman H A, Pabo C O. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 12.Isalan M, Klug A, Choo Y. Nat Biotechnol. 2001;19:656–660. doi: 10.1038/90264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreier B, Beerli R R, Segal D J, Flippin J D, Barbas C F., III J Biol Chem. 2001;276:29466–29478. doi: 10.1074/jbc.M102604200. [DOI] [PubMed] [Google Scholar]
- 14.Liu Q, Segal D J, Ghiara J B, Barbas C F., III Proc Natl Acad Sci USA. 1997;94:5525–5530. doi: 10.1073/pnas.94.11.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J-S, Pabo C O. Proc Natl Acad Sci USA. 1998;95:2812–2817. doi: 10.1073/pnas.95.6.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H, Yang W P, Barbas C F., III Proc Natl Acad Sci USA. 1995;92:344–348. doi: 10.1073/pnas.92.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papworth M, Moore M, Isalan M, Minczuk M, Choo Y, Klug A. Proc Natl Acad Sci USA. 2003;100:1621–1626. doi: 10.1073/pnas.252773399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen J. Science. 2002;296:2320–2324. doi: 10.1126/science.296.5577.2320. [DOI] [PubMed] [Google Scholar]
- 19.Cohen J. Science. 2002;296:2325–2326. doi: 10.1126/science.296.5577.2325. [DOI] [PubMed] [Google Scholar]
- 20.Friesen W J, Darby M K. J Biol Chem. 2001;276:1968–1973. doi: 10.1074/jbc.M008927200. [DOI] [PubMed] [Google Scholar]
- 21.McColl D J, Honchell C D, Frankel A D. Proc Natl Acad Sci USA. 1999;96:9521–9526. doi: 10.1073/pnas.96.17.9521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, Spratt S K, Liu Q, Johnstone B, Qi H, Raschke E E, Jamieson A C, Rebar E J, Wolffe A P, Case C C. J Biol Chem. 2000;275:33850–33860. doi: 10.1074/jbc.M005341200. [DOI] [PubMed] [Google Scholar]
- 23.Liu P Q, Rebar E J, Zhang L, Liu Q, Jamieson A C, Liang Y, Qi H, Li P X, Chen B, Mendel M C, et al. J Biol Chem. 2001;276:11323–11334. doi: 10.1074/jbc.M011172200. [DOI] [PubMed] [Google Scholar]
- 24.Beerli R R, Dreier B, Barbas C F., III Proc Natl Acad Sci USA. 2000;97:1495–1500. doi: 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren D, Collingwood T N, Rebar E J, Wolffe A P, Camp H S. Genes Dev. 2002;16:27–32. doi: 10.1101/gad.953802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herchenroder O, Hahne J C, Meyer W K, Thiesen H J, Schneider J. Biochim Biophys Acta. 1999;1445:216–223. doi: 10.1016/s0167-4781(99)00046-9. [DOI] [PubMed] [Google Scholar]
- 27.Kirchhoff F, Greenough T C, Hamacher M, Sullivan J L, Desrosiers R C. Virology. 1997;232:319–331. doi: 10.1006/viro.1997.8586. [DOI] [PubMed] [Google Scholar]
- 28.Moore M, Choo Y, Klug A. Proc Natl Acad Sci USA. 2001;98:1432–1436. doi: 10.1073/pnas.98.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore M, Klug A, Choo Y. Proc Natl Acad Sci USA. 2001;98:1437–1441. doi: 10.1073/pnas.98.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalderon D, Roberts B L, Richardson W D, Smith A E. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 31.Margolin J F, Friedman J R, Meyer W K, Vissing H, Thiesen H J, Rauscher F J., III Proc Natl Acad Sci USA. 1994;91:4509–4513. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evan G I, Lewis G K, Ramsay G, Bishop J M. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dingwall C, Ernberg I, Gait M J, Green S M, Heaphy S, Karn J, Lowe A D, Singh M, Skinner M A. EMBO J. 1990;9:4145–4153. doi: 10.1002/j.1460-2075.1990.tb07637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.West M J, Lowe A D, Karn J. J Virol. 2001;75:8524–8537. doi: 10.1128/JVI.75.18.8524-8537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dingwall C, Ernberg I, Gait M J, Green S M, Heaphy S, Karn J, Lowe A D, Singh M, Skinner M A, Valerio R. Proc Natl Acad Sci USA. 1989;86:6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soda Y, Shimizu N, Jinno A, Liu H Y, Kanbe K, Kitamura T, Hoshino H. Biochem Biophys Res Commun. 1999;258:313–321. doi: 10.1006/bbrc.1999.0633. [DOI] [PubMed] [Google Scholar]
- 37.McKnight A, Weiss R A, Shotton C, Takeuchi Y, Hoshino H, Clapham P R. J Virol. 1995;69:3167–3170. doi: 10.1128/jvi.69.5.3167-3170.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones K A, Peterlin B M. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 39.Perkins N D, Edwards N L, Duckett C S, Agranoff A B, Schmid R M, Nabel G J. EMBO J. 1993;12:3551–3558. doi: 10.1002/j.1460-2075.1993.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verdin E. J Virol. 1991;65:6790–6799. doi: 10.1128/jvi.65.12.6790-6799.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verdin E, Paras P J, Van Lint C. EMBO J. 1993;12:3249–3259. doi: 10.1002/j.1460-2075.1993.tb05994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J Y H, Gonzales-Scarano F, Zeichner S L, Alwine J C. J Virol. 1993;67:1658–1662. doi: 10.1128/jvi.67.3.1658-1662.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheridan P L, Sheline C T, Cannon K, Voz M L, Pazin M J, Kadonaga J T, Jones K A. Genes Dev. 1995;9:2090–2104. doi: 10.1101/gad.9.17.2090. [DOI] [PubMed] [Google Scholar]
- 44.Steger D J, Workman J L. EMBO J. 1997;16:2463–2472. doi: 10.1093/emboj/16.9.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Lint C, Emiliani S, Ott M, Verdin E. EMBO J. 1996;15:1112–1120. [PMC free article] [PubMed] [Google Scholar]
- 46.Ryan R F, Schultz D C, Ayyanathan K, Singh P B, Friedman J R, Fredericks W J, Rauscher F J., III Mol Cell Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lois C, Hong E J, Pease S, Brown E J, Baltimore D. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]