Abstract
The stabilization of p53 against Mdm2-mediated degradation is an important event in DNA damage response. Initial models of p53 stabilization focused on posttranslational modification of p53 that would disrupt the p53–Mdm2 interaction. The N-terminal regions of both p53 and Mdm2 are modified in vivo in response to cellular stress, suggesting that modifications to Mdm2 also may affect the p53–Mdm2 interaction. Our NMR studies of apo-Mdm2 have found that, in addition to Mdm2 residues 25–109 that form the well ordered p53-binding domain that was observed in the p52–Mdm2 complex, Mdm2 residues 16–24 form a lid that closes over the p53-binding site. The Mdm2 lid, which is strictly conserved in mammals, may help to stabilize apo-Mdm2. It also competes weakly with peptidic and nonpeptidic antagonists. Modifications to the Mdm2 lid may disrupt p53–Mdm2 binding leading to p53 stabilization. Mdm2 and Mdm4 possess nearly identical p53-binding domains but different lids suggesting that lid modifications may select for p53 binding.
The p53 tumor suppressor protein plays an important role in maintaining genome stability and in preventing the development of cancer. In response to various stress signals, p53 activation leads to cell-cycle arrest, apoptosis, or DNA repair (1). p53 is also a transcription factor for mdm2, the product of which negatively regulates both p53 stability and activity (2, 3). Mdm2 affects p53 activity by binding the N-terminal transactivation domain, blocking transcription (4–6). Mdm2 affects p53 stability by targeting it for ubiquitin-dependent degradation (7–9). In normal cells, p53 activity is kept low by Mdm2 (10). In response to DNA damage, p53 is activated by disrupting Mdm2 association and stabilized against Mdm2-dependent degradation (11). p53 activation and stabilization likely are achieved by posttranslational modifications (12); known modifications to p53 include phosphorylation, acetylation, and ubiquitination (1, 12). In one example of p53 stabilization by posttranslational modification, the phosphorylation of p53 nuclear export signals (13–15) blocks export and leads to p53 accumulation in the nucleus. Other details, particularly concerning p53 activation, are unclear.
One model of p53 activation is to disrupt the p53–Mdm2 interaction by phosphorylation (16). p53 is heavily phosphorylated after DNA damage (17–19). Several p53 residues at the p53–Mdm2 interface have been identified as targets or potential targets for kinases that are activated in response to DNA damage (20–26). Whereas in vitro phosphorylation of some of these residues can be shown qualitatively to weaken p53–Mdm2 association, quantitative studies have determined that phosphorylation of single p53 sites (that are known substrates) has no effect on Mdm2 binding (27–29). These results are consistent with studies in which single and multiple replacement of phosphorylatable p53 residues with alanine had no affect on p53 activity (3, 30–32). These in vivo results suggest that single-site p53 phosphorylation is not critical to the disruption of the p53–Mdm2 interaction, and that alternative mechanisms should be considered.
Mdm2 also is phosphorylated after DNA-induced damage (33–35). One alternative mechanism suggests that the phosphorylation of Mdm2 can break up the p53–Mdm2 interaction (36). It is not clear, however, from the crystal structure of the p53 (15–29)–Mdm2 (17–125) complex (6) as to how Mdm2 modifications might affect p53 binding. To resolve some of the contradictory data on p53 activation, we have built a structure-based model of p53-free Mdm2 that focuses on the interaction of strictly conserved Mdm2 residues 16–24 with the well structured p53-binding domain residues 25–109. After studying the binding of apo-Mdm2 with peptides and small molecules, we were able to present a more detailed model of the p53–Mdm2 interaction.
Materials and Methods
Protein Expression and Purification.
A clone was obtained (Y. Wang, Schering-Plough Research Institute) and expressed as a thioredoxin-Mdm2 fusion protein in BL21 (DE3) Escherichia coli. To produce labeled protein, the cells were grown in M9 minimal medium using 15NH4 Cl as the nitrogen source and 13C glucose as the carbon source. The fusion protein was isolated on a Nickel column, eluted, and pooled. Thrombin cleavage yielded Mdm2 (16–125) with seven nonnative N-terminal residues. The protein was applied to a gel filtration column (Superdex 75) and isolated. The protein sequence identity was confirmed by mass spectroscopy. The protein stability was optimized by varying pH, salt, and glycerol concentration. The final sample conditions were 100–400 μM Mdm2 in a 75 mM potassium phosphate buffer, pH 6.5, with 5 mM DTT. The sample was stable (>1 week) at 20°C. An Mdm2 (17–125) protein without any additional N-terminal amino acids also was produced by the incorporation of a factor Xa cleavage site into the clone for the thioredoxin-Mdm2 fusion protein. Dynamic light scattering on a 1.2 mg/ml, 20-μl apo-Mdm2 (16–125) sample found a hydrodynamic radius of 1.749 nm and a calculated molecular mass of 12.4 kDa with no polydispersion, indicating that the protein is monodisperse and monomeric.
NMR Spectroscopy.
Data were acquired on 500- and 600-MHz spectrometers. Standard triple resonance NMR experiments (37) were used to assign a 13C15N-labeled MDM2 (+7, 16–125) sample. Binding studies were performed on 15N-labeled Mdm2 (+7, 16–125) with 100 μM protein and typical ligand concentrations of 100 μM.
Results
Mdm2 N Terminus Is Structured.
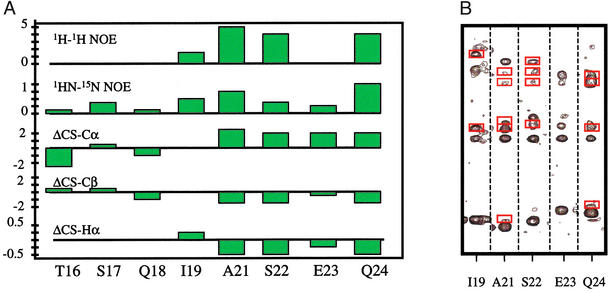
Residues 16–125 of Mdm2 were expressed as a thioredoxin-fusion protein, the cleavage of which yielded a seven-residue peptide (GSHMLEG) N-terminal to Mdm2 (16–125). Standard triple resonance NMR experiments (37) were used to assign 109 of 110 native Mdm2 residues and 5 of 7 nonnative residues. Restraints from nuclear Overhauser enhancement (NOE) measurements were used to fold the protein. In the x-ray crystal structure of the p53 (15–29)–Mdm2 (17–125) complex, Mdm2 (25–109) and p53 (15–28) were well ordered, whereas no electron density was observed for Mdm2 (17–24) or Mdm2 (110–125). In the absence of p53, our NMR data (1H-1H and 1HN-15N NOEs) confirmed that the fold and tertiary structure of Mdm2 (25–109) are consistent with the x-ray structure and, unexpectedly, determined that residues I19–Q24 also are structured. There are a number of interresidue 1H-1H NOEs for this region (Fig. 1) and the Cα, Cβ, and Hα chemical shift index (38) indicates that A21–Q24 are helical. Furthermore, residues T16–Q24 have positive 1HN-15N NOEs, indicating no rapid internal motion. In contrast, Mdm2 residues E114–N125 have no 1H-1H NOEs, random coil chemical shifts and large negative 1HN-15N NOEs, indicating that they are very flexible.
Figure 1.
(A) The presence of secondary structure for residues S21–Q24 is indicated by a number of interresidue 1H-1H NOEs (1–5), positive heteronuclear 1HN-15N NOEs, and chemical shift index values for Cα, Cβ, and Hα atoms [parts per million (ppm)] that are consistent with a helix. (B) 2D planes from a 15N-edited 3D-NOESY-heteronuclear single quantum coherence (HSQC) show interresidue 1H-1H NOEs (red boxes) throughout this region.
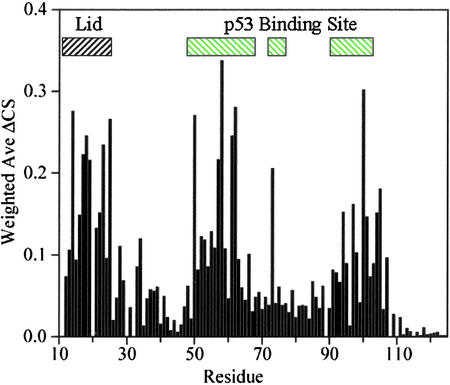
In addition to the region A21–Q24 being structured, ligand binding studies (Fig. 2) revealed that residues T16–Q24 are in close contact with the p53-binding site of Mdm2. Residues in the p53-binding site of Mdm2 are strongly affected by binding of a compound with Kd ≈ 20 μM; residues 16–24 are also strongly affected. Similar results are obtained for a wide variety of peptides and small organic molecules. In all cases of competitive binding, perturbation of the p53-binding pocket is accompanied by perturbation of residues 16–24. An Mdm2 construct for residues 17–125 showed the same perturbation patterns and NOEs.
Figure 2.
Binding of Mdm2 antagonists to p53-free Mdm2 is detected in heteronuclear single quantum coherence spectra by monitoring changes in 1HN-15N chemical shifts as a function of ligand concentration. We used 100 μM of 15N-labeled Mdm2 and 100 μM of the ligand. The Kd of this compound was ≈20 μM. Residues in the p53-binding site of Mdm2 are strongly perturbed upon ligand binding, as are residues 16–24. Nonnative residues 13 and 14 also are perturbed. Peptides and small organic molecules that bind into the p53-binding site perturb the Mdm2 lid as well.
Structure-Based Model of the Mdm2 Lid.
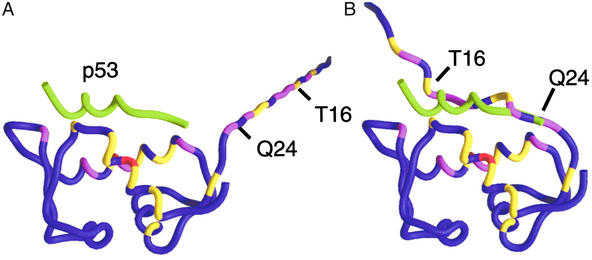
Fig. 3 maps chemical shift perturbations (from Fig. 2) to the backbone of Mdm2. A model for p53-free Mdm2 (16–125) where residues 16–24 are extended away from the p53-binding site (Fig. 3A) is clearly not compatible with the data in Figs. 1 and 2. Fig. 3B satisfies the experimental data by folding residues A21–Q24 into a coil and placing residues T16–Q24 into close contact with the p53-binding site. In this model, residues 16–24 form a “flexible lid” that stabilizes Mdm2 residues 25–109 in the absence of p53. Mdm2 residues 1–15 also may bind weakly. The lid helps to bury part of the largely hydrophobic 1,498-Å2 p53–Mdm2 interaction site stabilizing Mdm2 when p53 is not bound. The Mdm2 lid competes weakly with p53 but is easily displaced by WT-p53, high-affinity peptides, and small molecules.
Figure 3.
Chemical shift perturbations (ΔCS) from Fig. 2 are mapped onto the Mdm2 backbone. Coloring indicates the strength of the perturbations: red, ΔCS > 0.3 ppm; magenta, 0.2 ppm < ΔCS < 0.3 ppm; and yellow, 0.1 ppm < ΔCS < 0.2 ppm. The protein used in the binding studies contained nonnative residues 9–15. Residues 9 and 10 were not assigned and were excluded. Nonnative residues 11–15 and native Mdm2 residues 16–24 were appended to the crystal structure of the p53–Mdm2 complex. (A) The chemical shift perturbations are inconsistent with a flexible, extended N terminus because, in this model, many strong perturbations occur 20–40 Å from the p53-binding site. (B) Docking the flexible Mdm2 N terminus places perturbed residues 16–24 within 5 Å of perturbed Mdm2 atoms in the p53-binding site. The Mdm2 residues 16–24 form a lid that competes weakly with p53. Perturbations to the Mdm2 residues in the p53-binding site are primarily ligand-induced; they change sign and intensity as a function of ligand composition, whereas perturbations to the lid are caused by its nonspecific displacement and are independent of the chemical nature of the ligand. The position of p53 (green tube) is included as a point of reference.
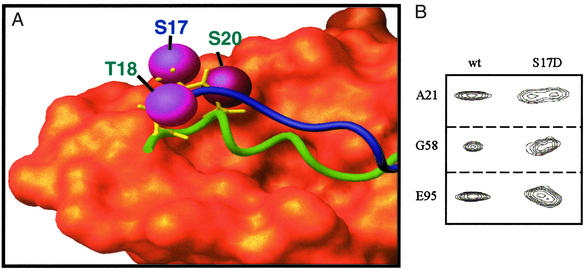
A critical function of the Mdm2 lid is suggested by the superimposition of the structure-based model (Fig. 3B) with the x-ray crystal structure of the p53–Mdm2 complex. The side chains of p53 residues Thr-18 and Ser-20 and Mdm2 residue Ser-17 are highlighted. Fig. 4A suggests that key phosphorylatable residues in the p53 and Mdm2 N termini are in close proximity. Simultaneous phosphorylation of Mdm2 and p53 residues would prevent p53 binding, because it would require the close proximity of negatively charged residues from both molecules. The phosphorylation of Mdm2 residue Ser-17 alone also may affect the affinity of nonphosphorylated p53 by formation of a salt bridge with His-73 or Lys-94, which would stabilize the Mdm2 lid. It is likely that simultaneous modifications to both p53 and Mdm2 affect their interaction. Note that the residues in the Mdm2 lid are strictly conserved in mammals (Fig. 5), suggesting their functional importance.
Figure 4.
(A) Superimposition of the x-ray crystal structure of p53–Mdm2 complex with a structure-based model of p53-free Mdm2. The Van der Waals surface for Mdm2 residues 25–109 from the crystal structure is orange. Green and blue tubes show the p53 peptide (from the crystal structure) and p53-free Mdm2 residues 16–24, respectively. In the crystal structure of the p53–Mdm2 complex, Mdm2 residues 17–24 are displaced by p53; there is no density for these residues. Magenta spheres indicate phosphorylatable residues on both p53 and Mdm2. Phosphate groups added to these residues have been shown to disrupt the p53–Mdm2 interaction. Note the close proximity of phosphorylatable p53 and Mdm2 residues. Rapid phosphorylation of p53-free Mdm2 followed by p53 phosphorylation of the p53 N terminus would make the displacement of the Mdm2 lid difficult due to the close proximity of several phosphate groups. (B) Selected peaks from heteronuclear single quantum coherence spectra demonstrate that the lid regions of WT-MDM2 and Mdm2–S17D have different affinities for the p53-binding site. Doubling of the peaks indicates two distinct conformations that interconvert on a long, slow time scale.
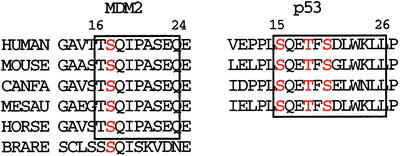
Figure 5.
Sequence alignment for flexible N-terminal residues of Mdm2 are strictly conserved in mammals. Boxed regions roughly indicate p53 and Mdm2 residues that compete for the p53-binding site; both regions are highly conserved. Phosphorylatable residues are red.
Discussion
Helical protein lids and flexible flaps covering parts of active protein active sites are well known. For example, the lipase lid is a six-residue helix that covers a largely hydrophobic active site in the absence of substrate (39). The lipase lid moves upon substrate binding, allowing access. Whereas the Mdm2 lid also buries a hydrophobic active site, phosphorylatable Mdm2 residues in the lid occupy a region on the Mdm2 surface that places them in close spatial proximity to the binding locations of p53 residues Thr-18 and Ser-20. The Mdm2 lid is therefore well positioned to disrupt p53 binding. This model is supported by binding assays that demonstrate that the phosphorylation of Mdm2 Ser-17 by DNA-PK disrupts the p53–Mdm2 interaction (36). There is also cell-based evidence that a S17A mutation decreases p53 activity (36). Less direct evidence comes from ATM (ataxia-telangiectasia-mutated), a critical component of cell-cycle arrest and DNA repair (18). In response to DNA damage from ionizing radiation, ATM first phosphorylates Mdm2 and then p53 (34). Although there is no data regarding the in vivo ATM-dependent phosphorylation of the Mdm2 N terminus, ATM phosphorylates Mdm2 in vitro at multiple sites, including one site between residues 1 and 115 (33). In this region only Ser-17 fits the SQ-motif that is required for DNA-PK/ATM/ATR (ATM- and Rad3-related) activity and is therefore the likely in vitro target of ATM. We examined the role of Ser-17 phosphorylation by making an S17D mutant protein. NMR spectra of Mdm2–S17D (Fig. 4B) found that resonances for lid residues 21–24 as well as residues 19, 58, 59, 93, 95, 96, and 98 in the p53-binding site showed two distinct conformations that interconvert on a slow time scale. This result confirms that the lid of the S17D mutant protein has a higher affinity for Mdm2 than the wild-type protein. Binding assays were performed to verify this model. The results for phosphorylated peptides binding to WT-Mdm2 are consistent with previous studies (28, 29). The results for phosphorylated peptides binding to Mdm2–S17D are consistent with previous results that found a reduction of the p53–Mdm2 interaction due to DNA-PK phosphorylation of Mdm2 residue Ser-17, rather than phosphorylation of p53 residue Ser-15.
Our model also suggests a mechanism for Mdm4 to compete with Mdm2 for p53. Mdm4 has significant sequence similarity with Mdm2 (29). Mdm4, however, lacks ubiquitin-ligase activity; it stabilizes p53 by preventing Mdm2-dependent degradation. Mdm4 and Mdm2 bind p53 with similar affinity (29), so it is difficult to rationalize how Mdm4 can successfully compete with Mdm2 for p53. The residues that comprise the Mdm2 lid are not conserved in Mdm4. However, 10 of the first 20 residues of the Mdm4 N terminus are serine or threonine, and the only strictly conserved residues between human Mdm2 and Mdm4 in this region are all phosphorylatable: Thr-4, Ser-7, Ser-17, and Ser-22. Specificity of different kinases for substrates on the N termini of Mdm2 and Mdm4 may therefore select for binding to p53.
In this work, restraints from NMR data were used to dock flexible Mdm2 residues to the surface of the Mdm2 crystal structure. NMR is well suited to study flexible protein regions; it is also an excellent tool for quantifying weak interactions (40). The existence of the Mdm2 lid demonstrates the connection between flexibility and functionality. The flexibility is likely needed for the lid to be accessible to kinases. The timing of modifications to the lid is also likely to be important. We believe that this model is compatible with much of the current data that show p53 phosphorylation as an important event in response to cellular stress. It is our view that knowledge of modifications to both p53 and the Mdm2 N terminus may be necessary to understand the mechanism of the regulation of p53 activity.
Acknowledgments
We thank Mr. Paul Reichert for performing the dynamic light scattering experiments.
Abbreviations
- NOE
nuclear Overhauser enhancement
- ATM
ataxia-telangiectasia-mutated
- ppm
parts per million
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Momand J, Zambetti G P, Olson D C, George D, Levine A J. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 3.Wu Z, Earle J, Saito S, Anderson C W, Appella E, Xu Y. Mol Cell Biol. 2002;22:2441–2449. doi: 10.1128/MCB.22.8.2441-2449.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J, Marechal V, Levine A J. J Virol. 1993;65:6001–6007. [Google Scholar]
- 5.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 6.Kussie P H, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine A J, Pavletich N P. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 7.Haupt Y, Maya R, Kazaz A, Oren M. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 8.Honda R, Tanaka H, Yasuda H. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 9.Kubbutat M H, Jones S N, Vousden K H. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 10.Fuchs S Y, Adler V, Buschmann T, Wu X, Ronai Z. Oncogene. 1998;17:2543–2547. doi: 10.1038/sj.onc.1202200. [DOI] [PubMed] [Google Scholar]
- 11.Shieh S Y, Ikeda M, Taya Y, Prives C. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 12.Oren M. J Biol Chem. 1999;274:36031–36034. doi: 10.1074/jbc.274.51.36031. [DOI] [PubMed] [Google Scholar]
- 13.Freedman D A, Levine A J. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Xiong Y. Science. 2001;292:1910–1915. doi: 10.1126/science.1058637. [DOI] [PubMed] [Google Scholar]
- 16.Shieh S-Y, Taya Y, Prives C. EMBO J. 1999;18:1815–1823. doi: 10.1093/emboj/18.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canman C E, Lim D S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 18.Banin S, Moyal L, Shieh S, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, Ziv Y. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 19.Jimenez G S, Khan S H, Stommel J M, Wahl G M. Oncogene. 1999;18:7656–7665. doi: 10.1038/sj.onc.1203013. [DOI] [PubMed] [Google Scholar]
- 20.Dumaz N, Milne D M, Jardine L J, Meek D W. Biochem J. 2001;359:459–464. doi: 10.1042/0264-6021:3590459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bean L J H, Stark G R. J Biol Chem. 2002;277:1864–1871. doi: 10.1074/jbc.M108881200. [DOI] [PubMed] [Google Scholar]
- 22.Hirao A, Kong Y Y, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge S J, Mak T W. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 23.Chehab N H, Malikzay A, Stavridi E S, Halazonetis T D. Proc Natl Acad Sci USA. 1999;96:13777–13782. doi: 10.1073/pnas.96.24.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chehab N H, Malikzay A, Appel M, Halazonetis T D. Genes Dev. 2000;14:278–288. [PMC free article] [PubMed] [Google Scholar]
- 25.Unger T, Juven-Gershon T, Moallem E, Berger M, Siononv R V, Lozano G, Oren M, Haupt Y. EMBO J. 1999;18:1805–1814. doi: 10.1093/emboj/18.7.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Unger T, Sionov R V, Moallem E, Yee C Y, Howley P M, Oren M, Haupt Y. Oncogene. 1999;18:3205–3212. doi: 10.1038/sj.onc.1202656. [DOI] [PubMed] [Google Scholar]
- 27.Kane S A, Fleener C A, Zhang Y S, Davis L J, Musselman A L, Huang P S. Anal Biochem. 2000;278:29–38. doi: 10.1006/abio.1999.4413. [DOI] [PubMed] [Google Scholar]
- 28.Lai Z, Auger K R, Manubay C M, Copeland R A. Arch Biochem Biophys. 2000;381:278–284. doi: 10.1006/abbi.2000.1998. [DOI] [PubMed] [Google Scholar]
- 29.Bottger V, Bottger A, Garcia-Echeverria C, Ramos Y F, Van der Eb A J, Jochemsen A G, Lane D P. Oncogene. 1999;18:189–199. doi: 10.1038/sj.onc.1202281. [DOI] [PubMed] [Google Scholar]
- 30.Ashcroft M, Kubbutat M H, Vousden K H. Mol Cell Biol. 1999;19:1751–1758. doi: 10.1128/mcb.19.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blattner C, Tobiasch E, Litfen M, Rahmsdorf H J, Herrlich P. Oncogene. 1999;18:1723–1732. doi: 10.1038/sj.onc.1202480. [DOI] [PubMed] [Google Scholar]
- 32.Abraham J, Spaner D, Benchimol S. Oncogene. 1999;18:1521–1527. doi: 10.1038/sj.onc.1202454. [DOI] [PubMed] [Google Scholar]
- 33.Khosravi R, Maya R, Gottlieb T, Oren M, Shiloh Y, Shkedy D. Proc Natl Acad Sci USA. 1999;96:14973–14977. doi: 10.1073/pnas.96.26.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Toledo S M, Azzam E I, Dahlberg W K, Gooding T B, Little J B. Oncogene. 2000;19:6185–6193. doi: 10.1038/sj.onc.1204020. [DOI] [PubMed] [Google Scholar]
- 35.Hay T J, Meek D W. FEBS Lett. 2000;478:183–186. doi: 10.1016/s0014-5793(00)01850-0. [DOI] [PubMed] [Google Scholar]
- 36.Mayo L D, Turchi J J, Berberich S J. Cancer Res. 1997;57:5013–5016. [PubMed] [Google Scholar]
- 37.Bax A, Grzesiek S. Acc Chem Res. 1993;26:131–138. [Google Scholar]
- 38.Wishart D S, Sykes B D. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 39.Brzozowski A M, Derewenda U, Derewenda Z S, Dodson G G, Lawson D M, Turkenburg J P, Bjorkling F, Huge-Jensen B, Patkar S A, Thim L. Nature. 1991;351:444–445. doi: 10.1038/351491a0. [DOI] [PubMed] [Google Scholar]
- 40.Shuker S B, Hajduk P J, Meadows R P, Fesik S W. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]