Abstract
Programmed cell death in prokaryotes is frequently found as postsegregational killing. It relies on antitoxin/toxin systems that secure stable inheritance of low and medium copy number plasmids during cell division and kill cells that have lost the plasmid. The broad-host-range, low-copy-number plasmid pSM19035 from Streptococcus pyogenes carries the genes encoding the antitoxin/toxin system ɛ/ζ and antibiotic resistance proteins, among others. The crystal structure of the biologically nontoxic ɛ2ζ2 protein complex at a 1.95-Å resolution and site-directed mutagenesis showed that free ζ acts as phosphotransferase by using ATP/GTP. In ɛ2ζ2, the toxin ζ is inactivated because the N-terminal helix of the antitoxin ɛ blocks the ATP/GTP-binding site. To our knowledge, this is the first prokaryotic postsegregational killing system that has been entirely structurally characterized.
Keywords: programmed cell death‖ɛ protein‖ζ protein‖toxin inactivation‖phosphotransferase
Plasmids may harbor genes that encode proteins conferring resistances to their host against antibiotics and toxic heavy atoms and are the principal players in the current crisis in antibiotic therapy. They can spread horizontally by conjugational transfer and are maintained through subsequent generations by utilizing self-encoded operons (1). The gene products of the latter act on the respective plasmids as resolvases, recombinases and, if a bacterium has lost its plasmid during cell division, as postsegregational killing (PSK) systems leading to programmed cell death (PCD). Most importantly, the PSK systems guarantee stable maintenance and inheritance of plasmids in prokaryotes with low and medium copy number (2).
The ɛ and ζ proteins studied here belong to PSK systems and are encoded as a bicistronic operon on the SegB region of the low-copy-number, broad-host-range plasmid pSM19035 that is hosted by the Gram-positive Streptococcus pyogenes (3). The system ɛ/ζ belongs to the protein PSK-type with antitoxic ɛ and toxic ζ proteins, whereas a second type features an antitoxic antisense RNA and a stable mRNA that encodes a toxic protein. In both types, bicistronic operons encode for an in vivo unstable antitoxin and a stable toxin. The products form an inactive complex in the cell cytosol. Continuous production of the antitoxin counteracts its in vivo instability and maintains a stoichiometric excess to the toxin. Loss of the entire plasmid or of the antitoxin gene results in decreasing antitoxin levels in the cytosol, so that the toxin freed from the unstable antitoxin becomes active and induces PCD. In this sense, PSK systems act as emergency buttons against ineffective inheritance of plasmids.
Several genes encoded on pSM19035, a plasmid belonging to the inc18 family (pAMβ1, pIP501), are also found on plasmids of other Gram-positive bacteria, such as Streptococci, Enterococci, and Lactococci. These bacteria are associated with the wide distribution of antibiotic resistance in food production (4) and carry genes closely related to the genes ɛ and ζ of pSM19035 (see Discussion).
The herein-reported x-ray crystal structure of the biologically nontoxic protein complex formed by ɛ and ζ is, to our knowledge, the first describing an entire antitoxin/toxin system of either Gram-positive or -negative bacteria. A search of the Protein Data Bank suggested a functional mechanism for ζ that was corroborated by site-directed mutagenesis. These studies render ɛ/ζ the first PSK system of a plasmid hosted in Gram-positive bacteria to be investigated using the tools of molecular biology. It delivers insight into the mechanism of inactivation and activation of the potent toxic protein ζ and suggests possible inactivation mechanisms of related PCD systems.
Experimental Procedures
Crystallographic Methods.
For overexpression, purification, and crystallization of wild-type and selenomethionine-derivated ɛ/ζ complex, see ref. 5. An x-ray dataset of a wild-type ɛ/ζ crystal and multiple anomalous diffraction (MAD) data of the selenomethionine derivative were measured at 100 K to a 1.95- and 3.1-Å resolution at beamlines BM14 (European Synchrotron Radiation Facility, Grenoble, France) and X31 (European Molecular Biology Laboratory/Deutsches Elektronen Synchrotron, Hamburg, Germany), respectively. For relevant characteristics describing quality and processing of the data, see ref. 5.
Selenium sites were determined with SOLVE V.1.17 (6) based on MAD data to a 4-Å resolution. When using a noncrystallographic twofold axis (NCS), phase extension to 3.1 Å by using the program DM (7) improved the electron-density map, and the polypeptide chains were traced with the program o (8). Ill defined parts of the polypeptide chains were traced as polyalanine. Rigid body refinement of this model with the wild-type protein dataset, using the cns program package (9), enabled phase extension to 1.95 Å. Structure refinement was supported by manual model building and simulated annealing, and NCS restraints were deleted. Near convergence of refinement, solvent molecules and sulfate ions were added in the Fo − Fc map. All refinement steps used the cns package and were monitored with the free R factor based on 5.1% of the x-ray data; for refinement statistics, see Table 1. The stereo-chemical quality of the final model was assessed by WHAT_CHECK (10), and the figures were drawn with MOLSCRIPT V.2.1.2 (11) and RASTER3D (12). Surface potentials were calculated using delphi (13) and mapped on surfaces generated with msms (14). Sequence alignments by CLUSTALW V.1.74 (15) were plotted using alscript (16).
Table 1.
Crystallographic data
| Space group | P212121 |
| Lattice parameters, Å | a = 59.54 |
| b = 79.85 | |
| c = 191.44 | |
| Number of ɛ2ζ2 heterotetramers per AU | 1 |
| Resolution limit, Å | 20.00–1.95 |
| Rfree | 0.235 |
| Rwork | 0.199 |
| rmsd bonds, Å | 0.005 |
| rmsd angles, ° | 1.11 |
| Number of amino acids per AU | 712 |
| Number of identified water molecules per AU | 695 |
| Number of bound [SO4]2− ions per AU | 2 |
| Number of atoms per AU | 6,482 |
AU, asymmetric unit; rmsd, rms deviation.
Mutagenesis Studies.
DNA purification, PCR, cloning, and sequencing followed standard procedures. Mutations were introduced by the two-stage overlap extension method, using the pBT288-borne wild-type ζ gene as template. After NcoI (5′ restriction site) and BamHI (3′ restriction site) restriction digest, the fragment was ligated in pTrc99A (Stratagene) and transformed into Escherichia coli ER2566 (New England Biolabs). Plasmid DNA of positive transformants was isolated and sequenced. The construct ζ(V234Stop) was amplified in one step from pBT288 plasmid DNA by using a reverse primer that generates a stop codon downstream from the codon for Val-234ζ. Cloning was performed as described above.
Results
Model Building and Refinement.
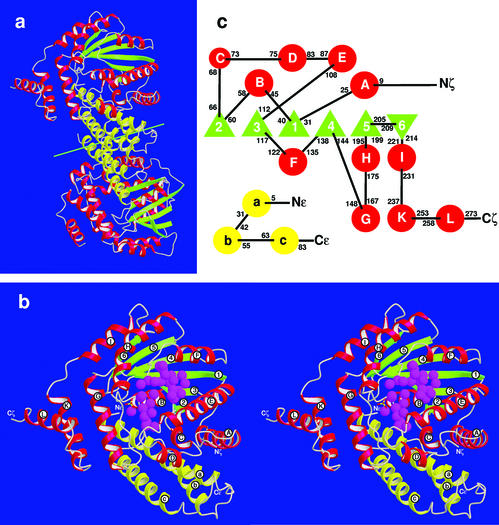
The copurified proteins ɛ and ζ crystallized as a heterotetrameric, dumbbell-shaped ɛ2ζ2 complex with dimensions 41 × 47 × 97 Å3 (Fig. 1a). The NCS axis cuts the handgrip that is formed by the ɛ2 dimer and stabilized mainly by hydrophobic interactions; the ζ proteins forming the outer parts of the dumbbell interact with the ɛ2 dimer but not with each other (Fig. 1a). In agreement with matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) mass spectrometry (data not shown), ɛ and ζ proteins do not contain N-terminal methionines. There are some differences in electron densities of the halves ɛ,ζ I and ɛ,ζ II. No electron density could be attributed to Ala-2ɛ and Ala-90ɛ in ɛ,ζ I or to the segment Glu-87ɛ to Ala-90ɛ in ɛ,ζ II (for amino acid sequences, see Fig. 5). In the C-terminal regions of both ɛ polypeptide chains, an increase of temperature factors indicates partial disorder. Because of poorly defined electron density, the solvent-exposed, proline-rich C termini of the ζ polypeptide chains could not be fitted so that they terminate with Pro-273ζ in ɛ,ζ I and Leu-271ζ in ɛ,ζ II. Structure refinement converged at Rwork of 19.9% and Rfree of 23.5%. Structural differences between the ɛ,ζ halves I and II in the ɛ2ζ2 complex of 0.909 Å rmsd on Cα atoms can be attributed to contacts with crystal-symmetry-related molecules. Atomic parameters were deposited in the Protein Data Bank under PDB ID code 1gvn, and chain identifiers A,B and C,D correspond to ɛ,ζ I and ɛ,ζ II, respectively.
Figure 1.
(a) Structure of the ɛ2ζ2 heterotetramer. The green line near the center shows the NCS axis. α-Helices of proteins ɛ and ζ are yellow and red, respectively, and β-strands of protein ζ are green. (b) Heterodimeric ɛ,ζ-half in stereo; coloring as in a and labeling of secondary structure elements as in c. The well ordered water molecules in the ATP site (the large crevice near the center of the complex) and the substrate site (closer to the viewer and located between α-helices E and F) are indicated by magenta spheres. (c) Topography of secondary structure elements; α-helices are shown as circles labeled in lower- and uppercase letters in proteins ɛ and ζ, respectively, and β-strands in ζ as triangles and numbered. Black numbers give the respective N- and C-terminal residues within the amino acid sequences.
Figure 5.
Sequence alignment of gene products from Enterococci, Lactococci, and Streptococci that are related to proteins ɛ and ζ. Secondary structure elements of proteins ɛ and ζ are indicated by cylinders (helices) and arrows (β-strands) on top of the alignments. Coloring and labeling are as in Fig. 1 b and c. Amino acids of protein ɛ that are shown in Fig. 4 to interfere with binding of ATP to the ɛ2ζ2 complex and catalytically important amino acids in protein ζ are red. Parts of the amino acid sequence that are conserved in four or more sequences are beige, and those conserved in only three sequences are blue.
Protein Folding.
Protein ɛ (10.7 kDa) is folded into a three-helix bundle and protein ζ (32.4 kDa) features an α/β structure (Fig. 1b), the central twisted β-sheet comprising six β-strands in the order 2/3/1/4/5/6 (Fig. 1 b and c). β-strands 1–5 are parallel but β-strand 6 is antiparallel and connected by a short loop to β-strand 5. α-Helices are inserted between and flank the β-strands (Fig. 1c). The two C-terminal helices K and L are only weakly bound to the bulk of the complex and form a solvent-exposed helix-loop-helix appendage. Helices A, B, C, and G of ζ confine a crevice containing a well ordered network of water molecules (Fig. 1b). The top of this crevice is closed by the N-terminal half of helix a of the adjacent protein ɛ.
The Toxin ζ Is Folded Like a Phosphotransferase.
A PROSITE (17) search showed that ζ features a Walker A motif (18), also known as P-loop (19) and typical for ATP/GTP-binding proteins. This motif is formed by 40GXXGXGKT47 as a loop between β-strand 1 and α-helix B and binds the β-phosphate of ATP/GTP or inorganic phosphate (20, 21). In agreement with the positive electrostatic potential of this segment, the electron density of the ɛ2ζ2 heterotetramer showed two tetrahedral-shaped molecules, each bound by residues 43GSGK46 of the Walker A motifs. These molecules were identified as sulfate ions because the protein was exposed to 1.3 M ammonium sulfate during purification (5).
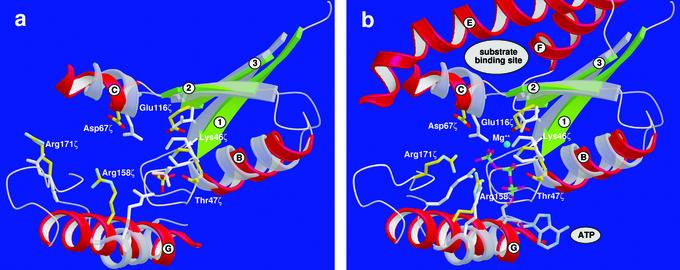
A DALI (22) superposition of the tertiary structure of ζ to other structures in the Protein Data Bank showed that the folding of ζ is similar to adenylate kinase (AK) from Bacillus stearothermophilus (23) with a 3.2-Å rmsd on Cα-atom positions and 15% sequence identity and to chloramphenicol phosphotransferase (Cmp) from Streptomyces venezuelae (24) with a 3.4-Å rmsd on Cα-atom positions and 8% sequence identity. The next five highest scores all belong to nucleoside monophosphate (NMP) kinases. These α/β proteins feature a central parallel β-sheet with the strand sequence 2/3/1/4/5 (19). Although the sequence identity to ζ is low, the folding of the active site of NMP kinases and Cmp is similar to that of ζ, and amino acids that are catalytically important in AK and Cmp are found at corresponding positions in ζ. Fig. 2a shows a superposition of ζ with the active site of the free Cmp in the open conformation (24). Amino acids involved in Cmp ATP binding are: the entire P-loop (Gly-10 to Ser-17), Arg-133, and Arg-136. In free Cmp, Arg-136 points away from the active site but the side chain is rotated if ATP is bound, its guanidinium group being moved by 9 Å. For ζ, the corresponding residues are Gly-40ζ to Thr-47ζ (the P-loop) and amino acids Arg-158ζ and Arg-171ζ. In accordance with Arg-136 of Cmp, Arg-171ζ is located within a flexible loop segment distinguished by high temperature factors. In Cmp, Asp-92 binds a Mg2+ ion to promote ATP hydrolysis, whereas in ζ, electron density at this particular position is attributed to Glu-116ζ. Additionally, Asp-37 in Cmp deprotonates the chloramphenicol (C-3) hydroxyl group and conducts the nucleophilic attack. This amino acid is conserved in ζ with Asp-67ζ.
Figure 2.
Superposition of the ATP-binding site of Cmp with the large crevice in protein ζ. α-Helices of the latter are red, β-strands are green, relevant side chains are yellow and labeled, and side chains in Cmp are gray and not labeled. (a) The open conformation of Cmp with inorganic phosphate (gray; sulfate ion in protein ζ are yellow/red) bound to the P-loop (Lys-46ζ in protein ζ). (b) Superposition as in a, but both enzymes with bound ATP and of Mg2+ according to the Cmp crystal structure; the binding site for the yet unknown substrate of protein ζ is indicated. Conformations of Arg-158ζ and Arg-171ζ (with the appending loop region) are modeled according to the analogous amino acids in the closed conformation of Cmp and in NMP kinases.
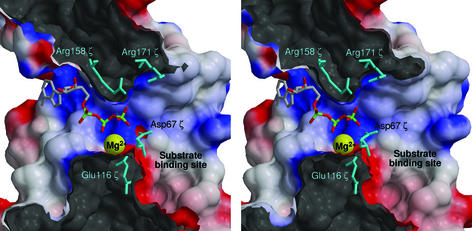
The substrate-binding site of NMP kinases and of the related Cmp is located around β-strand 3 and the adjacent region (25). In the tertiary structure of ζ, the equivalent helix E, β-strand 3, and helix F form a pocket that is 7 Å deep, 15 Å long, and 10 Å wide and filled by well ordered water molecules (Fig. 1b). The pocket is connected with the large crevice of ζ where ATP binds (Fig. 3).
Figure 3.
Cut through protein ζ (stereo view) showing the substrate-binding site (right) forming a hydrophilic entrance to the large crevice of ζ to which ATP is modeled as in Fig. 2b. Positive electrostatic potential is blue and negative red; maximum values are ±10 kT/e. Catalytically active amino acids and Mg2+ (yellow) are labeled. The two Arg and Mg2+ stabilize the negative charge of ATP, Glu-116ζ stabilizes Mg2+, and Asp-67ζ is essential for deprotonation of the substrate (not shown) that creates the functionally essential nucleophile.
Mutagenesis Studies on ζ.
Because ζ is toxic for E. coli, the wild-type gene ζ could not be cloned and expressed without coexpression of gene ɛ. The importance of functional amino acids conserved among phosphotransferases and ζ was studied by site-directed mutagenesis. Changes Lys46ζAla in the Walker A motif and Asp67ζThr enabled cloning and expression of the genes ζ(K46A) and ζ(D67T) in E. coli without coexpression of the antagonistic gene ɛ. Construct ζ(R158A) could be cloned into E. coli but not overexpressed. Among the obtained nonlethal constructs for ζ(R171S), four were chosen for sequencing and they all showed a second point mutation. However, a construct for ζ(R158A+R171S) could be cloned and expressed. Another ζ variant truncated after Val-234 was obtained that lacks the helix K–loop–helix L appendage.
Toxin–Antitoxin Interactions.
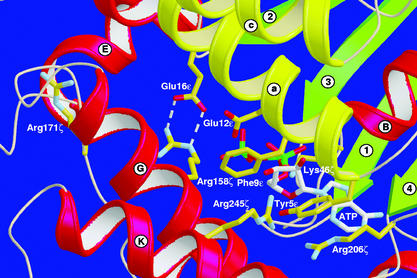
The predominant contacts between antitoxin ɛ and toxin ζ involve helix a of ɛ. While the C-terminal part of helix a binds to ζ, the N-terminal part closes the large crevice of ζ like a lid (Fig. 4), with side chains of Tyr-5ɛ and Phe-9ɛ pointing into the crevice and impeding binding of ATP/GTP. In addition, in both halves of ɛ2ζ2 the side chains of Glu-12ɛ point toward ζ; the carboxylate groups of Glu-16ɛ hydrogen bond to Arg-158ζ (with short donor–acceptor distances in the 2.76- to 2.84-Å range) and fix their orientation away from the crevice. In ɛ,ζ I, Arg-158ζ hydrogen bonds to the backbone C⩵O group of Arg-171ζ (3.26 Å), whereas in ɛ,ζ II, this interaction is mediated by a water molecule.
Figure 4.
Steric hindrance of ATP binding to ɛ2ζ2. The putative position of ATP is modeled according to Cmp and NMP kinases. α-Helices οf proteins ɛ and ζ are yellow and red, respectively, and β-strands are green and labeled as in Fig. 1c. The hydrogen bonds between Arg-158ζ and Glu-16ɛ are indicated by dashed lines. α-Helix a of ɛ covers the large crevice in ζ, its side chains filling the crevice and interfering with ATP binding. The substrate binding pocket is located at the bottom of the crevice.
The Operon ɛ/ζ Is Frequently Found.
A blast search (26) indicated several genes encoding proteins closely related to ɛ and ζ. The amino acid sequence alignment in Fig. 5 shows the similarity of several Enterococcus, Streptococcus, and Lactococcus proteins to the ɛ/ζ system. In Enterococcus faecalis plasmid pRE25, two ɛ/ζ homologous operons (orf18/orf19 and orf49/orf50) could be identified (27). High sequence identities are found for proteins pRE25orf18 (78% to ɛ) and pRE25orf19 (91% to ζ), as well as for pRE25orf49 (80% identity to ɛ) and pRE25orf50 (81% identity to ζ). Although the N-terminal 70 aa in pRE25orf50 are highly conserved, it seems to be truncated and restricted to only 85 residues. In Enterococcus faecium, a plasmid-borne, truncated ζ-homologous protein corresponding to ζ amino acids 101–287 (97% sequence identity) was identified and designated ζ (28). The published sequence (GenBank accession no. AAL02178.1) is artificially truncated and the authors confirmed the presence of the entire ɛ/ζ operon but did not publish the sequence. Streptococcus agalactiae plasmid pIP501 encodes for protein pIP501orfX (87% identity to ζ; ref. 29) but lacks amino acids 214–269 of ζ (Fig. 5), and a hypothetical protein sharing 77% sequence identity with ɛ (pIP501ɛ) was identified (27). The plasmid-borne gene product orfA of Enterococcus hirae (AAK96239.1) shares 96% sequence identity with ζ, but the existence of the ɛ-related gene in this particular strain remained unidentified. Finally, an ORF of Lactococcus lactis plasmid pCRL291.1 (AAK59285.1) shows 55% sequence identity with ɛ (pCRL291.1ɛ). Recently, the chromosomal gene SP1051 from Streptococcus pneumoniae TIGR4 was described sharing 45% sequence identity and 65% sequence homology with gene ζ (not shown in Fig. 5). SP1051 is encoded in a bicistronic operon together with upstream gene SP1050 that encodes a transcriptional regulator protein homologous to Cro that bears no resemblance with ɛ (30). In preliminary investigations we found that proteins encoded by SP1050 and SP1051 form a stable complex in vitro and behave in E. coli like a PCD system (data not shown).
Discussion
In all known protein systems responsible for PCD in prokaryotes, the stoichiometric relationship of antitoxin and toxin, the lability of the antitoxin, and the stability of the toxin play the key regulatory role. Although proteases responsible for continuous degradation of protein ɛ are still unidentified, protein ζ was shown to be more stable than protein ɛ in the cell cytosols of Bacillus subtilis and E. coli (31). The toxicity of ζ could be described, but its functional mechanism and target substrate are still unknown (2).
Binding of ATP to the P-loop in the large crevice of ζ was homology modeled on the structures of NMP kinases and Cmp (Fig. 2b). As in the related structures, ATP binding to ζ may involve hydrogen bonding from peptide N-H groups of the P-loop to the β-phosphate and of Lys-46ζ in the Walker A motif to one oxygen atom each of the β- and γ-phosphates. Additionally, Arg-158ζ and Arg-171ζ may form hydrogen bonds to the phosphate groups. As in Cmp, the adenine would be located in the tertiary structure of ζ at a position that is occupied by side chains Tyr-5ɛ and Phe-9ɛ of helix a of ɛ in the ɛ2ζ2-complex (Fig. 4), which impede ATP binding by steric hindrance. Furthermore, Glu-16ɛ of helix a binds tightly to the side chain of Arg-158ζ, thereby stabilizing it in a conformation pointing away from the ATP-binding site. An association of ATP binding with the toxic function of free ζ has been verified by changes Lys46ζAla, Arg158ζAla, and Arg171ζSer. Substitution of only one of the two arginines Arg-158ζ and Arg-171ζ was not sufficient to entirely destroy the toxic effect of ζ, but exchanging both arginines in one construct resulted in a variant that was no longer toxic and could be expressed in high levels in E. coli. Similarly, a Lys46ζAla replacement in the Walker A motif abolished the toxic effect of ζ.
We can assign toxin ζ as phosphotransferase because the P-loop is located between β-strand 1 and the adjacent helix B, similarly found in protein kinases and bacterial phosphotransferases. Additionally, two strictly invariant aspartates are of fundamental importance in phosphotransferases (32–35). In Cmp, Asp-92 coordinates the catalytically essential Mg2+, and Asp-37 deprotonates the C-3 hydroxyl of the substrate chloramphenicol, enabling the latter to conduct nucleophilic attack on the γ-phosphate of ATP (24). In ζ, the carboxylate groups of Glu-116ζ and of Asp-67ζ are in positions to perform these tasks. The change Asp67ζThr resulted in a nontoxic variant of ζ, indicating that the toxicity of ζ is associated with a phosphorylation reaction because in Cmp, the related Asp-37 interacts solely with the substrate and not with ATP.
Asp-67ζ lies beneath the large crevice, between the ATP- and substrate-binding sites (Figs. 2 and 3). This is comparable to the NMP family (25), where the substrate-binding site is formed by β-strand 3 and the flanking N- and C-terminal helices, analogous to the proposed substrate-binding site of ζ (α-helix E–β-strand 3–α-helix F). Consequently, we propose the pocket that is connected with the large crevice to be the substrate-binding site (Fig. 3). Assuming that no dramatic conformational changes occur on substrate binding, the substrate for ζ can only be a small molecule or a solvent-exposed segment of a protein or nucleic acid.
We have derived a possible functional mechanism of ζ by analogy with the structurally related Cmp and adenylate kinase (AK), suggesting that Asp-67ζ deprotonates the substrate. The resultant nucleophile then attacks the γ-phosphate of ATP to yield a pentacovalent transition state that is stabilized by Arg-171ζ and Arg-158ζ through hydrogen bonding to the β- and γ-phosphate groups. Because the side chain of Arg-171ζ points away from the ATP-binding site in the ɛ2ζ2 complex, it has to rotate toward the binding site in free ζ when ATP is coordinated, as similarly described for Arg-136 in Cmp (24). This assumption is supported by high B factors of the segment containing Arg-171ζ, which indicates flexibility. Although a Mg2+ ion could not be identified in the ɛ2ζ2 electron density, we assume that Glu-116ζ binds this catalytically important ion to favor product formation through neutralization of the developing negative charge and that the Walker A motif stabilizes the transferred phosphoryl group. Collapse of the pentacovalent transition state yields the phosphorylated substrate and ADP.
Fig. 5 shows that ɛ and ζ occur as highly homologous proteins in plasmids from various pathogenic bacteria, among which several, such as Streptococci, Enterococci, and Lactococci, are found in food production (4). Less conserved amino acid sequences are located in loop segments of ɛ and in helix A of ζ, which are not involved in ɛ2ζ2 complex formation. In the L. lactis plasmid pCRL291.1ɛ gene product, only N- and C-terminal amino acids of α-helix a that are responsible for inactivation of toxin ζ by complex formation are conserved. We suggest that the proteins shown in Fig. 5 are PSK systems that are structurally and functionally homologous to ɛ/ζ. In S. agalactiae, pIP501orfX lacks the C-terminal three helices I, K, and L, but the last 17 aa, which are rich in prolines, are strictly conserved. Because ζ truncated after V234ζ is not toxic, this proline-rich C-terminal segment region might also be engaged in substrate binding.
Among themselves, PCD systems show significant sequence homology. For example, the PSK system kis/kid shows 34% sequence identity and 69% sequence homology to the chromosomal mazE/F operon, and 37% sequence identity and 77% sequence homology to the chpBI and chpBK genes, both of which are involved in PCD in E. coli (36, 37). Systems similar to ɛ/ζ (either chromosomal or plasmid-encoded) offer a specific, unique opportunity to combat bacteria hosting these operons. Because the antitoxin is steadily degraded in vivo, a specific strategy in medicinal applications would inhibit transcription or translation of the PCD system. This could be achieved by preventing the dissociation of regulatory proteins from the promoter region of the PCD operon. More sophisticated would be a stable antisense RNA inhibiting translation of the antitoxin gene. On the other hand, toxin inhibition of a PSK system would deregulate plasmid inheritance and result in the loss of a plasmid(s) that harbors the genes responsible for antibiotic resistance. Because the substrate- and ATP-binding sites of the toxin ζ are now known, new possibilities are opened for structure-based drug design. Using an inhibitor for a PSK system as comedication with conventional antibiotic therapy would offer novel approaches, because the present plasmid-encoded resistances of host bacteria would get lost.
Acknowledgments
We thank Paul Tucker of the European Molecular Biology Laboratory outstation at Deutsches Elektronen Synchrotron and Vivian Stojanoff of the European Synchrotron Radiation Facility for assistance during synchrotron experiments, Claudia Alings for support in protein crystallization, and Ana Camacho for some collaboration. This work was supported by Deutsche Forschungsgemeinschaft Grant Sa196/38-2 (to W.S.), European Union Grant QLK3-CT-2001-00277 (to W.S. and J.C.A.), and Fonds der Chemischen Industrie (W.S.).
Abbreviations
- Cmp
chloramphenicol phosphotransferase
- NMP
nucleoside monophosphate
- PCD
programmed cell death
- PSK
postsegregational killing
- rmsd
rms deviation
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1gvn).
References
- 1.Lilley A, Young P, Bailey A. In: The Horizontal Gene Pool. Thomas C M, editor. London: Harwood; 2000. pp. 287–300. [Google Scholar]
- 2.Gerdes K, Ayora S, Canosa I, Ceglowski P, Diáz-Orejas R, Franch T, Gultyaev A P, Bugge Jensen R, Kobayashi I, Macpherson C, et al. In: The Horizontal Gene Pool. Thomas C M, editor. London: Harwood; 2000. pp. 49–86. [Google Scholar]
- 3.Ceglowski P, Boitsov A, Chai S, Alonso J C. Gene. 1993;136:1–12. doi: 10.1016/0378-1119(93)90441-5. [DOI] [PubMed] [Google Scholar]
- 4.Perreten V, Schwarz F, Cresta L, Boeglin M, Dasen G, Teuber M. Nature. 1997;389:801–802. doi: 10.1038/39767. [DOI] [PubMed] [Google Scholar]
- 5.Meinhart A, Alings C, Sträter N, Camacho A G, Alonso J C, Saenger W. Acta Crystallogr D. 2001;57:745–747. doi: 10.1107/s0907444901004176. [DOI] [PubMed] [Google Scholar]
- 6.Terwilliger T C, Berendzen J. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowtan K. Joint CCP4 ESF-EACBM Newslett Protein Crystallogr. 1994;31:34–38. [Google Scholar]
- 8.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 9.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 10.Hooft R W W, Vriend C, Sander C, Abola E E. Nature. 1996;381:272–272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 11.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 12.Meritt E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 13.Honig B, Sharp K, Yang A-S. J Phys Chem. 1993;97:1101–1109. [Google Scholar]
- 14.Sanner M F, Spehner J-C, Olson A J. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 15.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barton G J. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- 17.Bairoch A, Bucher P, Hofmann K. Nucleic Acids Res. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker J R, Saraste M, Runswick M J, Gay M N. EMBO J. 1992;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murzin A G, Brenner S E, Hubbard T, Chothia C. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 20.Saraste M, Sibbald P R, Wittinghofer A. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 21.Smith C A, Rayment I. Biophys J. 1996;70:1590–1602. doi: 10.1016/S0006-3495(96)79745-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 23.Berry M B, Phillips G N., Jr Proteins. 1998;32:276–288. doi: 10.1002/(sici)1097-0134(19980815)32:3<276::aid-prot3>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 24.Izard T, Ellis J. EMBO J. 2000;11:2690–2700. doi: 10.1093/emboj/19.11.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Branden C, Tooze J. Introduction to Protein Structure. New York: Garland; 1991. pp. 99–110. [Google Scholar]
- 26.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwarz F V, Perreten V, Teuber M. Plasmid. 2001;46:170–187. doi: 10.1006/plas.2001.1544. [DOI] [PubMed] [Google Scholar]
- 28.Mann P A, Xiong L, Mankin A S, Chau A S, Mendrick C A, Najarian D J, Cramer C A, Loebenberg D, Coates E, Murgolo N J, et al. Mol Microbiol. 2001;41:1349–1356. doi: 10.1046/j.1365-2958.2001.02602.x. [DOI] [PubMed] [Google Scholar]
- 29.Langella P, Le Loir Y, Ehrlich S D, Gruss A. J Bacteriol. 1993;175:5806–5813. doi: 10.1128/jb.175.18.5806-5813.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tettelin H, Nelson K E, Paulsen I T, Eisen J A, Read T D, Peterson S, Heidelberg J, DeBoy R T, Haft D H, Dodson R J, et al. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 31.Camacho A G, Misselwitz R, Behlke R, Ayora S, Welfle K, Meinhart A, Lara B, Saenger W, Welfle H, Alonso J C. Biol Chem. 2002;383:1701–1713. doi: 10.1515/BC.2002.191. [DOI] [PubMed] [Google Scholar]
- 32.Cox S, Radzio-Andzelm E, Taylor S S. Curr Opin Struct Biol. 1994;4:893–901. doi: 10.1016/0959-440x(94)90272-0. [DOI] [PubMed] [Google Scholar]
- 33.Goldsmith E J, Cobb M H. Curr Opin Struct Biol. 1994;4:833–840. doi: 10.1016/0959-440x(94)90264-x. [DOI] [PubMed] [Google Scholar]
- 34.Bossemeyer D. FEBS Lett. 1995;369:57–61. doi: 10.1016/0014-5793(95)00580-3. [DOI] [PubMed] [Google Scholar]
- 35.Brenner S. Nature. 1987;329:21. doi: 10.1038/329021a0. (lett.). [DOI] [PubMed] [Google Scholar]
- 36.Top E M, Moënne-Loccoz Y, Pembroke T, Thomas C M. In: The Horizontal Gene Pool. Thomas C M, editor. London: Harwood; 2000. pp. 249–286. [Google Scholar]
- 37.Engelberg-Kulka H, Glaser G. Annu Rev Microbiol. 1998;53:43–70. doi: 10.1146/annurev.micro.53.1.43. [DOI] [PubMed] [Google Scholar]