Abstract
Ankyrin repeat (AR) proteins mediate innumerable protein–protein interactions in virtually all phyla. This finding suggested the use of AR proteins as designed binding molecules. Based on sequence and structural analyses, we designed a consensus AR with fixed framework and randomized interacting residues. We generated several combinatorial libraries of AR proteins consisting of defined numbers of this repeat. Randomly chosen library members are expressed in soluble form in the cytoplasm of Escherichia coli constituting up to 30% of total cellular protein and show high thermodynamic stability. We determined the crystal structure of one of those library members to 2.0-Å resolution, providing insight into the consensus AR fold. Besides the highly complementary hydrophobic repeat–repeat interfaces and the absence of structural irregularities in the consensus AR protein, the regular and extended hydrogen bond networks in the β-turn and loop regions are noteworthy. Furthermore, all residues found in the turn region of the Ramachandran plot are glycines. Many of these features also occur in natural AR proteins, but not in this rigorous and standardized fashion. We conclude that the AR domain fold is an intrinsically very stable and well-expressed scaffold, able to display randomized interacting residues. This scaffold represents an excellent basis for the design of novel binding molecules.
The importance of ankyrin repeat (AR) proteins in nature is underlined by their abundance in bacteria, fungi, plants, and animals (1). There are nuclear (e.g., IκBα), cytosolic (e.g., ankyrin), membrane-bound (e.g., notch), and secreted (e.g., black widow spider toxin) AR proteins. The function of AR domains is to mediate protein–protein interactions. They are built from tightly joined repeats of usually 33 aa. Each repeat forms a structural unit, which consists of a β-turn, followed by two antiparallel α-helices and a loop reaching the turn of the next repeat (2, 3). Up to 29 consecutive repeats can be found in a single protein (4). Yet, AR domains usually consist of four to six repeats, which stack onto each other, leading to a right-handed solenoid structure with a continuous hydrophobic core and a large solvent-accessible surface (5).
AR proteins fulfill their diverse biological functions by specific and tight binding to target polypeptides, and each repeat can contribute to target binding (3). Target affinities in the low nanomolar range have been reported (6, 7). The evolutionary success of members of this protein family originates from their ability to bind to virtually any target protein by simple adaptation of their molecular surface and by displaying highly variable residues throughout the protein. The modularity of AR domains enables surface evolution by allowing duplications and deletions of repeats and shuffling of repeats between repeat domains (5, 8). Hence, the AR domain fold is a very versatile scaffold for the evolutionary generation of protein domains displaying specific binding surfaces. These characteristics motivated us to exploit AR domains as a scaffold for the construction of libraries of novel binding molecules.
We started the library construction with the assumption that all ARs belong to a canonical ensemble, which can be described by the consensus sequence and its statistics of variation. In a first approximation, the distribution of amino acids at a given position can be described by Boltzmann's law, because this distribution reflects protein stability (9), although selection for function will introduce a bias. The collection of nearly 8,000 AR sequences in the smart database (10) facilitates such an approach. This consensus design concept has been used to generate enzymes with improved thermostability (11, 12) and to improve antibody stability (9, 13, 14). We used this concept to design a consensus AR consisting of fixed framework residues that are responsible for repeat structure maintenance (fold conservation) and of randomized interacting residues, i.e., residues, which, in a repeat domain, create a randomized surface for interaction with target proteins. Varying numbers of this repeat were assembled between capping repeats (caps), yielding combinatorial libraries of consensus AR proteins (H.K.B., M.T.S., P.F., Patrick Amstutz, and A.P., unpublished work).
Here, we show that the library design based on consensus sequences works for ARs. We analyzed the stability of six randomly chosen library members consisting of four to six repeats. Four of these proteins were screened for crystallization. With the help of the resulting 2.0-Å crystal structure of a five-repeat consensus protein, we examined differences to natural AR proteins and present structural insights into the success of the AR domain fold.
Materials and Methods
Protein Expression, Purification, and Characterization.
The detailed description of the design, expression, and purification of the consensus AR proteins will be presented elsewhere (see also patent application PCT/EP01/10454). The protein sequences have been deposited in GenBank and are listed in Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org. Throughout this article, subscript indices will be used to describe consensus repeat positions (e.g., Gly27) whereas normal indices denote residues in the protein sequence (e.g., Met-34). In addition, X will denote any amino acid except C, G, or P, whereas Z will indicate any of the amino acids H, N, or Y. The limitations to the indicated amino acids were possible by using mixed trinucleotides as a building block for oligonucleotide synthesis (15). Briefly, AR protein libraries were assembled joining an N-terminal cap, two to four designed internal ARs (consensus repeat), and a C-terminal cap (N2C, N3C and N4C libraries). The N-terminal cap sequence is (including a His tag): MRGSHHHHHH GSDLGKKLLE AARAGQDDEV RILMANGADV NAX, where Asp-13 is the first residue of the cap and where the last X corresponds to a consensus repeat position 33. The internal repeat sequences are based on the following designed consensus sequence: D1X2X3G4X5T6P7L8H9L10 A11A12X13X14G15H16L17E18I19V20 E21V22L23L24K25Z26G27A28D29V30 N31A32X33. The C-terminal cap sequence is: QDKFGKTAFD ISIDNGNEDL AEILQ, where the first Gln corresponds to consensus repeat position X33 of the preceding repeat.
In the present study, six randomly chosen full-length library members, two each from the N2C (E2_5 and E2_17), N3C (E3_5 and E3_19), and N4C (E4_2 and E4_8) libraries, were expressed and purified by immobilized metal affinity chromatography to virtual homogeneity. Proteins were used at 2 mg/ml in 50 mM Tris⋅HCl, pH 8.0, 50 mM NaCl for dynamic light scattering measurements by using a ProteinSolutions DynaPro MS/X instrument at 20°C with the software DYNAMICS 4.0. Equilibrium unfolding of 10 μM protein in 50 mM Tris⋅HCl, pH 7.4, 150 mM NaCl, and variable concentrations of guanidinium chloride (GdmCl) was followed at 222 nm by CD with a Jasco (Tokyo) J-715 instrument. Samples were equilibrated overnight and measured at 23°C in duplicates. Data were evaluated by fitting to a two-state unfolding model where appropriate (16). The assumption of two-state unfolding is, however, preliminary and fluorescence measurements as a complementary method to monitor the equilibrium unfolding were not conclusive because of the lack of buried fluorophors.
Crystallization and Data Collection.
Proteins were rebuffered in 50 mM Tris⋅HCl, pH 8.0, 150 mM NaCl and concentrated to ≈9 mg/ml. E3_5 was crystallized by using the hanging or sitting drop vapor diffusion method at 20°C, in 24-well crystallization plates. The drops contained 2 μl of protein and 2 μl of reservoir solution (16–22% PEG 4000, 0.2 M Li2SO4, and 0.1 M NH4OAc, pH 4.6–5.0), with 0.5 ml of reservoir buffer in each well. The crystals grew in ≈1–2 weeks from precipitated protein. For cryoprotection, crystals were soaked for 30 sec in reservoir solution containing additional PEG 4000 (final concentration 40%), before flash-freezing them at 110 K for data collection.
X-ray diffraction analysis was performed by using CuKα radiation generated by a Nonius FR 591 rotating anode generator (Nonius, Delft, The Netherlands) equipped with a double-focusing mirror system (XRM-216; Prophysics, Zurich). Data were recorded on an imaging plate detector (300 mm; Mar Research, Norderstedt, Germany) with a detector to crystal distance of 90 mm. Under these conditions crystals were stable and diffracted x-rays to 2.0-Å resolution. A data set from a single crystal (85 × 85 × 850 μm) was collected and processed with the denzo and scalepack (17) crystallographic data reduction package. The crystal belonged to space group P21212 and the Matthews coefficient of VM = 2.3 Å3/Da was calculated by using the molecular mass of 17.7 kDa, which corresponds to an estimated water content of 46.1%. Statistics on data collection are given in Table 1. In addition, crystals of two different space groups (indexed P21 and R3) were found and data were collected. However, higher diffraction limits suggested the use of the P21212 data set for the structure determination.
Table 1.
Statistics for data collection and refinement
| Data collection | |
| Space group | P21212 |
| Cell dimensions, Å | a = 73.864, b = 47.360, C = 47.003, α = β = γ = 90.00° |
| Resolution limits, Å | 20.0–2.03 |
| Observed reflections | total: 173,985; unique: 11,291 |
| Completeness, % | 98.4 (83.8) |
| Redundancy | 15.4 |
| Rsym (% on I) | 10.0 (33.0) |
| Refinement | |
| Resolution range, Å | 20.0–2.03 |
| Rfactor/Rfree, % | 18.4/23.0 |
| Ordered water molecules | 178 |
| rmsd from ideal geometry | |
| Bond lengths, Å | 0.022 |
| Bond angles, ° | 1.926 |
| Average B factor, Å2 | 17.98 |
Numbers in parentheses refer to the highest-resolution shell.
Molecular Replacement, Model Building, and Refinement.
The crystal structure was determined by molecular replacement by using the program amore (18), with the structure of the GA-binding protein β1 (GABPβ1) (Protein Data Bank ID code 1AWC; ref. 19) as a search model. All nonidentical residues in the search model were replaced by Ala. A conventional amore protocol (rotation, translation, rigid body refinement) was applied and yielded a single clear solution. Model building was carried out by using the program o (20). The structure was refined in cns (21), followed by refmac (22), resulting in a final model with an R factor of 18.4% and an Rfree factor of 23.0%. Water molecules were picked by using arpwarp (23) in the solvent building mode. Crystallographic data are given in Table 1. The N-terminal His tag cannot be seen in the electron density. Clear density starts at residue 11 and extends throughout the entire molecule, with the exception of the side chains of residues 12, 16, 45, and 68. Additional electron density was interpreted as 178 water molecules, two sulfate ions, and one Tris(hydroxymethyl)aminomethane molecule.
Analysis and Bioinformatics.
The model was evaluated by using the program procheck (23). A thorough analysis of the consensus AR protein structure and several other AR protein structures, [Protein Data Bank ID codes: 1A5E (24), 1AWC (19), 1BD8 (25), 1DCQ (26), 1IHB (27), 1K1A (28), 1MYO (29), 1NFI (30), 1SW6 (31), and 1YCS (2)], was carried out by using the following programs: hbplus (32) and ligplot (33) for analysis of the H-bonding networks and the hydrophobic contacts; grasp (34) for surface calculations and cavity search; swiss pdb-viewer (35) and insight ii (Accelrys, Cambridge, U.K.) for model dissection, superposition and rms deviation (rmsd) calculations; the pride server (36) for determining the evolutionary relation, and treeview (37) for visualizing this relation.
For the analysis, the different AR protein structures were split into single repeats. Splitting was done on a visual basis at the start of the β-turn, which corresponds to position D1 of the consensus sequence proposed by Sedgwick and Smerdon (3).
Results and Discussion
We designed a consensus AR consisting of fixed framework residues and randomized interacting residues based on sequence and structure analyses. Varying numbers of this AR were cloned between N- and C-terminal caps, i.e., ARs that shield the hydrophobic core of the AR domain from the solvent. This process resulted in designed AR protein libraries of distinct repeat numbers (N2C, N3C, and N4C representing four-, five-, and six-repeat proteins, respectively; N and C denote the caps and digits denote the number of consensus repeats). All randomly chosen full-length library members analyzed so far (>20) could be expressed in soluble form in the cytoplasm of Escherichia coli to ≈10–30% of total cellular protein (up to 200 mg protein per liter shake flask culture) and did not aggregate in standard buffers (20–50 mM Tris⋅Cl, pH 7–8.5/50–500 mM NaCl).
Equilibrium Unfolding.
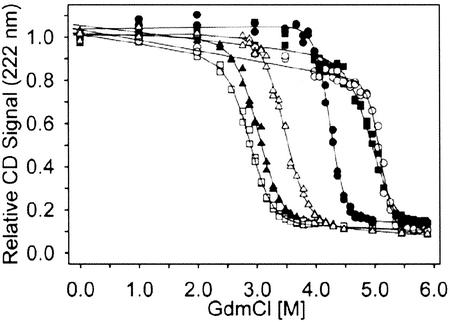
Two consensus AR proteins each from the N2C (E2_5, E2_17), N3C (E3_5, E3_19), and N4C (E4_2, E4_8) AR protein libraries were analyzed in more detail. The GdmCl equilibrium unfolding of these four-, five-, and six-repeat proteins was measured by CD (Fig. 1). These designed AR proteins show cooperative, reversible unfolding with midpoints of unfolding between 2.9 and 5 M GdmCl and possess values for the free energy of unfolding (ΔG) between 9.5 and 21 kcal/mol, assuming two-state unfolding (16) (Table 2). Even though all proteins are very stable, their variable residues (10–14% of total residues) do have an influence on stability when comparing proteins of the same length. Therefore, a definite statement relating the number of ARs to biophysical properties has to await AR proteins with identical repeats. To date, the equilibrium unfolding data of four natural AR proteins have been reported. These are myotrophin (N2C), with ΔG = 5.1 kcal/mol (38); the INK4 family members p16 (N2C), with ΔG = 3.1 kcal/mol (39) and p19 (N3C), where no two-state unfolding was observed (40) and notch (N5C), with ΔG = 8 kcal/mol (41). The thermodynamic stability of our consensus repeat proteins is thus clearly higher than that of the reported natural AR proteins. This finding underlines the success of the consensus design strategy and demonstrates the intrinsic high stability of the AR domain fold. In addition, our consensus AR domains tolerate many variable surface residues, emphasizing their potential as novel binding molecules.
Figure 1.
Equilibrium unfolding of six randomly chosen full-length members of consensus AR protein libraries. The equilibrium denaturation was followed by CD spectroscopy (see Materials and Methods). The CD signal is displayed as fraction of the CD value of each sample at 0 M GdmCl. Note that this representation makes no assumption about the pretransition or posttransition baseline. Solid lines correspond to the two-state fits (16), which are evaluated in Table 2. ▵, E2_5; ▴, E2_17; ■, E3_5; □, E3_19; ○, E4_2; and ●, E4_8.
Table 2.
GdmCl equilibrium unfolding of unselected N2C, N3C, and N4C consensus AR protein library members
| Protein | ΔG, kcal/mol | m, kcal/(mol⋅M) | Dm, M |
|---|---|---|---|
| E2_5 | 11.40 ± 0.70 | 3.29 ± 0.19 | 3.46 ± 0.01 |
| E2_17 | 9.53 ± 0.59 | 3.16 ± 0.18 | 3.02 ± 0.01 |
| E3_5 | 14.84 ± 1.98 | 3.00 ± 0.37 | 4.95 ± 0.06 |
| E3_19 | 9.59 ± 0.51 | 3.33 ± 0.17 | 2.88 ± 0.01 |
| E4_2* | — | — | 5.11 ± 0.02 |
| E4_8 | 21.13 ± 1.30 | 4.98 ± 0.30 | 4.24 ± 0.01 |
ΔG, m value, and the midpoint of denaturation (Dm) of each measurement have been determined under the assumption of a two-state unfolding equilibrium (16).
E4_2 is oligomeric; the other proteins are monomers.
Crystallization.
We performed crystallization trials with four immobilized metal affinity chromatography-purified consensus AR proteins (E2_5, E3_5, E3_19, and E4_2). Dynamic light scattering experiments showed monodisperse behavior for E2_5, E3_5, and E3_19, which is advantageous for crystallization (42). E4_2 was polydisperse, confirming gel filtration data that indicated a monomer–oligomer mixture (data not shown). Of the four proteins used for crystallization screens, the two five-repeat proteins gave crystals. The crystallization of E3_5 was further refined.
Overall Structure.
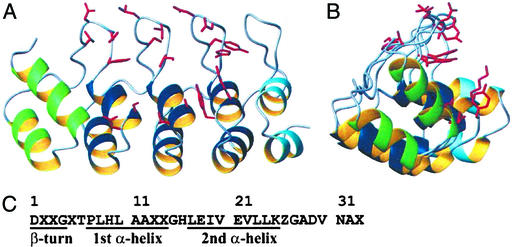
The crystals of the consensus AR protein E3_5 were analyzed, a complete data set was collected, and the structure was determined as described in Materials and Methods. A 2.0-Å resolution structure of E3_5 was obtained. The B factors are generally low and do not vary much, which can be taken as an indication for very little thermal movement and therefore a rigid structure of the protein in the crystal. E3_5 shows a very regular and ordered AR domain fold, and the structure is highly homologous (rmsdCα = 0.9 Å) to mouse GABPβ1 [69.7% sequence identity (19)].
Evaluation of the Designed Consensus Repeats.
The designed consensus repeats show essentially no backbone deviation between each other (rmsdCα 2–3: 0.52 Å; 2–4: 0.37 Å; 3–4: 0.49 Å). Most consensus amino acids fulfill their assigned functions, and residues of the hydrophobic core and some exposed residues are identical throughout the molecule on the rotamer level (consensus positions: Asp1, Thr6, Pro7, Leu8, His9, Leu10, His16, Leu17, Ile19, Val20, Glu21, Leu23, Leu24, Asp29, Val30, and Asn31; see Materials and Methods and Fig. 2C for consensus sequence and numbering). These are framework residues forming the repeat scaffold. All glycines and alanines in framework positions are also superimposable. Other residues are less similar on the rotamer level, such as all randomized positions (X2, X3, X5, X13, X14, and X33) but also the framework positions Glu18, Val22, and Lys25. This finding indicates that other residues may also be allowed in these framework positions. The residues at framework position Z26 have a hydrophobic stem and a polar end with rotamers showing similar orientations. Interestingly, very similar overall findings are obtained for the internal repeats of GABPβ1 (19). However, in this molecule not only side chains corresponding to the consensus positions 18, 22, and 25 adopt variable rotamers, but also those corresponding to positions 10, 17, and 21. This may be because of the presence of different amino acids at these positions. The amino acids in positions corresponding to consensus positions 19, 20, 26, 29, 30, and 31 vary in GABPβ1, but still adopt similar conformations as in E3_5. H-bonding is very regular in E3_5. In GABPβ1, however, the second repeat contains fewer H-bonds (27 H-bonds) than the consensus repeat (31 H-bonds; Fig. 3).
Figure 2.
Structure of the consensus AR protein E3_5. (A and B) Perpendicular views of E3_5 prepared with molmol (53). Ribbon representation of E3_5 showing the helices of the N-terminal, internal (consensus), and C-terminal repeats in green, dark blue, and light blue, respectively. The side chains of amino acids at randomized positions are highlighted in red. (C) The consensus AR sequence. X, any amino acid but C, G, or P; Z, any of the amino acids H, N, Y.
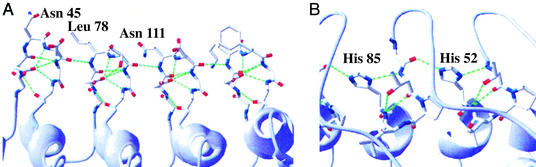
Figure 3.
H-bonding networks in the consensus AR protein E3_5. H-bonds are shown in green. (A) β-Turn H-bond network spanning the entire molecule. Each β-turn is formed by the sequence X33Asp1X2X3Gly4X5 (Xi being randomized positions). The X2 positions in the first three β-turns are labeled for orientation (Asn-45, Leu-78, and Asn-111). (B) TPLH motifs of repeats 2 and 3. The side chains of His-52 and His-85 form H-bonds to the β-turn of the position X5 of the third or fourth repeat, respectively.
Caps.
We used the terminal repeats of GABPβ1 (19) as the starting point for the engineering of our caps, because GABPβ1 was most homologous to our consensus repeats. We adapted the caps of GABPβ1 in the loop (N-terminal cap) and the β-turn (C-terminal cap) regions to fit the consensus repeats and we replaced cysteines to prevent dimerization on oxidation. The designed caps of E3_5 are therefore highly homologous (≈90% sequence identity) to the terminal repeats of GABPβ1. Nevertheless, the designed loop region of the N-terminal cap shows, as expected, significant differences to GABPβ1, affecting the entire cap structure. The implemented consensus loop (GADVNA) is more densely packed (causing Met-34 to rotate to fill the altered hydrophobic core), contains more H-bonds and is more polar than the corresponding loop (GAPFT) of GABPβ1. The C-terminal cap of E3_5 is slightly more compact compared with GABPβ1. The change of the β-turn from SKFC (GABPβ1) to DKFG (E3_5) had no impact on the structure and H-bonds. In GABPβ1, an atypical AR H-bond of Asn-149 to Asn-115 is observed, whereas in E3_5, Asn-158 makes a “consensus-like” H-bond to Ala-121. Overall, the designed caps shield the continuous hydrophobic core formed by the assembled consensus repeats as anticipated.
β-Turn.
Fig. 3A shows the β-turns with their hydrogen bonding network. Five intrarepeat H-bonds per β-turn involve (i) three side-chain/main-chain interactions from the carboxyl Asp1O to X3, Gly4, and X5, (ii) a Asp1NH-Gly4O backbone–backbone H-bond, and (iii) a Asp1NH-X5O backbone–backbone H-bond. One interrepeat H-bond is formed from X3O to X2NH of the neighboring repeat. This arrangement leads to a continuous, regular array of the β-turns including five intrarepeat and one interrepeat H-bonds per β-turn (Fig. 3A). Natural AR proteins with known structures do not possess such a regular arrangement of the β-turns, which is also manifested in a lower number of H-bonds present (24, 29). An increased number of H-bonds is one of the few factors that seems to correlate with increased thermostability in all protein families (43).
TPLH Motif.
The first α-helix of ARs is capped at the N terminus by a highly conserved TPLH motif (3) (Fig. 3B). Pro7 initiates the helix and Leu8 forms hydrophobic contacts to the inner side of the antiparallel helix of the same repeat. Thr6 forms three H-bonds with the His9 residue: (i) the backbone CO and (ii) the side-chain OH form H-bonds to the backbone NH of His9. (iii) The backbone NH forms an H-bond to the imidazole Nδ of the His9 side chain. Thereby, the His9 side chain is in a conformation that allows the formation of one or two H-bonds toward X33 of the same or X5 of the next repeat. The high conservation of this motif in natural AR proteins underlines the importance of this interrepeat H-bond network, propagating over the entire molecule.
Loops.
Natural ARs sometimes carry insertions in the loop regions (2, 28, 30, 44). Even an additional helix may be present. Such irregularities were eliminated by our consensus design. The GADVNAX loop connecting helix 2 with the β-turn of the next repeat is very regular in E3_5 (rmsdCα <0.25 Å). Gly27 breaks out of helix 2. Ala28 and especially Val30 are hydrophobic anchors of the loop, whereas Asp29 and Asn31 are involved in H-bonding to the neighboring repeat loops. The GHLE loop connecting the two helices in a repeat is structurally conserved in E3_5 (rmsdCα <0.2 Å). It establishes several main-chain/main-chain H-bonds with helix 1 of the same repeat. The design further anticipated a side-chain/main-chain H-bond from His16 to the X13 of the previous repeat. This H-bond is seen in repeats two and four, but not in repeat three, which may be caused by the reorientation of the His-92 side-chain to form a H-bond with Thr-90. Again, the strict conservation of the loops may be stabilizing the consensus repeats compared with less conserved natural repeats, as the H-bonding pattern is more pronounced.
Bioinformatics.
To further understand the high in vitro stability of consensus AR proteins, we dissected published repeat structures and analyzed them concerning irregularities, interrepeat and intrarepeat H-bonds, hydrophobic contacts, buried surface area, cavities, and rmsdcα. In addition, we subjected all suitable repeats (minimal length 30 aa) to a pride cross comparison (36).
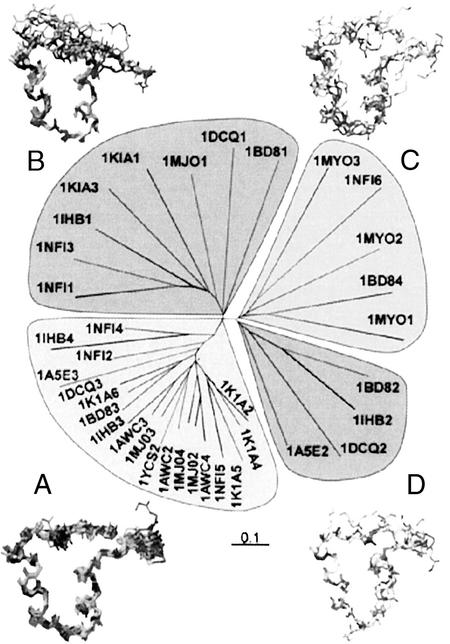
The pride server divides the ARs mainly into four groups (Fig. 4). Group A is by far the largest and most conserved group and encloses the regular ARs. In this group all internal repeats of E3_5 as well as the internal repeats of 1AWC, 1K1A, 1IHB, 1YCS, and 1NFI can be found. A typical repeat in this group was found to have ≈30 H-bonds, ranging from 17 (1A5E3) to 34 (1DCQ3). The internal repeats of E3_5 are found to have on average ≈31 H-bonds. The rmsdCα in this group is well below 1.0 Å (usually 0.5 Å). A second group (B) includes mostly N-terminal caps, which are structurally more distant to the consensus repeat, lacking the β-turn or having irregular loop regions. In a third group (C), irregular repeats and the repeats of 1MYO were grouped, which can be explained by their locally different architecture in the β-turn and in the repeat-connecting loop region, leading to a high rmsdCα, >3 Å. The fourth group (D) consists only of the second repeat of INK4 family proteins, which have a shortened first α-helix, leading to an elongated loop between the helices (24–27). These irregular repeats form only 22 H-bonds and destabilize the overall structure of the INK4 proteins (45). Nevertheless, they are of biological importance, as they are part of the INK4/CDK interaction interface.
Figure 4.
pride (36) analysis of single AR. The analysis divides the repeats into four branches, which are shown as groups. The repeats, including terminal repeats, were numbered according to their occurrence in the full-length proteins. This number is given directly after the appropriate Protein Data Bank identifier. The respective superpositions of the repeats are shown next to the four groups.
In AR domains the two helices of a single repeat do not pack closely together to form a single tight hydrophobic core. Using a cut-off of 3.9 Å for hydrophobic contacts, hbplus showed often none or only a few hydrophobic contacts within a single AR. Most of the hydrophobic contacts between the two antiparallel helices range from 4.0 to 4.5 Å. These values are rather high but are still in agreement with the general CH3–CH3 group van der Waals distances, known for proteins (46). In contrast to the weak intrarepeat hydrophobic contacts the interrepeat contacts are in the normal range (<3.9 Å). This results in two rows of packed helices perpendicular to the orientation of the repeats. The main hydrophobic contacts are thus formed in an interrepeat rather than in an intrarepeat manner (3). A careful comparison of the hydrophobic core packaging of E3_5 with that of natural AR proteins of known structures did not reveal any significant differences. In general, cavities are present in AR proteins including E3_5, but are rather small (<30 Å3).
The interrepeat contacts in E3_5 and known natural AR protein structures are mostly of hydrophobic nature. A typical contact area between two repeats is formed by four to five H-bonds, ≈80–100 atoms, which potentially contribute to the hydrophobic contact (cut-off 3.9 Å), and comprises 1,100–1,700 Å2 of buried surface. The contact area is a substantial part of the hydrophobic core, but shows some analogies to typical protein–protein interaction surfaces, which are characterized by a size-equivalent surface but more H-bonds [protein–protein interaction: 1,600 ± 400 Å2, 9 ± 5 H-bonds (47)]. The covalent linkage between two repeats certainly increases the interface stability. Still, the few interrepeat H-bonds (e.g., the H-bonds in the TPLH motif, between the β-turns and in the loop regions), which are conserved throughout the structures and are part of our consensus sequence, may help in the proper orientation of the repeats during assembly and may also be a stabilizing factor (2).
Cooperativity.
The crystal structure of E3_5 enabled us to rationalize the basis of its cooperative unfolding (Fig. 1; Table 2). The hydrophobic core and the H-bond networks in the β-turn region, the TPLHLAA motif, and the GADVNA loop motif are features that extend throughout the entire molecule. Thus, these features lead to mutual stabilization of stacked repeats and thus to cooperative unfolding of the assembly. The weak hydrophobic intrarepeat contacts suggest that a single AR is not stable (2). In contrast, interrepeat contacts are stronger, suggesting that individual repeats are stabilized by their neighbors and thus stability and cooperativity increase as the number of repeats increases. Such a behavior has been reported for p16, where the minimal folding unit was determined to consist of two repeats (45). The increased stability and cooperativity of E4_8 (Table 2) also supports this reasoning. Nevertheless, this observation needs more detailed analysis with perfectly repetitive AR proteins.
Ramachandran Plot and Protein Surface.
In the so-called turn region (left-handed α-helical region) of the Ramachandran plot of, e.g., GABPβ1 (19) mostly Gly but also other amino acids such as Asp-71, Lys-104, Asn-115, and Cys-137 are found. In E3_5, however, all amino acids found in the turn region are Gly. Gly to Ala mutations, where both residues retain the conformation and are in the turn region, can destabilize a protein by as much as 1.9 kcal/mol (48). Mutations from Gly to Ala not constrained to the turn region are in general stabilizing (≈1 kcal/mol) because of an increase in the entropy of unfolding (49). The replacement of non-Gly residues found in the turn region by Gly may thus be a stabilizing feature of the consensus AR proteins. At neutral pH, E3_5 has many conserved surface exposed charges, which are well separated and occur as “belts” on the surface formed by several repeats. Hydrophobic surface patches are less pronounced in comparison to known structures of natural AR proteins. In turn, this might render E3_5 more soluble and less prone to aggregation.
Summary and Conclusions.
The canonical sequence approximation has previously been used to create molecules with improved stabilities (9, 11–14). We have successfully applied this consensus design to ARs. We showed that consensus AR proteins are well expressed and possess a high thermodynamic stability. This high stability of the AR protein framework may rationalize the widespread occurrence of AR proteins. With the help of a 2.0-Å crystal structure of such a consensus AR protein, we were able to pinpoint stability-determining aspects. The absence of irregularities leading to refined intrarepeat and interrepeat interactions seems to be important. Especially the improved mutual stabilization of neighboring repeats seems to be beneficial (extended H-bonding networks, continuous hydrophobic core). Furthermore, only glycines are found in the turn region of the Ramachandran plot. Although many of these features occur in natural AR proteins as well, they are not present in this rigorous and standardized fashion as implemented in the designed molecules.
As we have well-behaved AR proteins in hand consisting of different repeat numbers with randomized surfaces, we can probe these molecules for binding against target proteins. Indeed, libraries of N2C and N3C AR proteins were selected by using ribosome display (50) against several globular proteins, and specific binding molecules with affinities in the low nanomolar range were isolated (H.K.B., P. Amstutz, M.T.S., P.F., and A.P., unpublished work). The intrinsic stability, the polar surface, and the selectable binding properties of our designed AR proteins suggest that these molecules could also assist crystallization of difficult targets in analogy to antibody cocrystallization (51).
Supplementary Material
Acknowledgments
We thank Dr. Guido Capitani, Christophe Briand, Daniel Frey, and Patrick Amstutz for valuable discussions and the referees for valuable comments. H.K.B. was the recipient of a predoctoral fellowship from the Roche Research Foundation. M.T.S. was the recipient of a Kekulé Stipendium of the Fonds der Chemischen Industrie, with financial participation of the Bundesministerium für Bildung und Forschung. This project was supported by the National Center of Competence in Research Structural Biology.
Abbreviations
- AR
ankyrin repeat
- cap
capping repeat
- GdmCl
guanidinium chloride
- GABPβ1
GA-binding protein β1
- rmsd
rms deviation
Note:
While we revised the present manuscript, Mosavi et al. (52) published the crystal structures of two AR proteins consisting of three or four full consensus repeats. Although the two approaches have differences (protein libraries vs. full consensus proteins, different consensus sequences, caps in our molecules, different solution behavior, different number of repeats) the structural and stability findings are similar. Thus the two studies complement each other.
Footnotes
References
- 1.Bork P. Proteins. 1993;17:363–374. doi: 10.1002/prot.340170405. [DOI] [PubMed] [Google Scholar]
- 2.Gorina S, Pavletich N P. Science. 1996;274:1001–1005. doi: 10.1126/science.274.5289.1001. [DOI] [PubMed] [Google Scholar]
- 3.Sedgwick S G, Smerdon S J. Trends Biochem Sci. 1999;24:311–316. doi: 10.1016/s0968-0004(99)01426-7. [DOI] [PubMed] [Google Scholar]
- 4.Walker R G, Willingham A T, Zuker C S. Science. 2000;287:2229–2234. doi: 10.1126/science.287.5461.2229. [DOI] [PubMed] [Google Scholar]
- 5.Kobe B, Kajava A V. Trends Biochem Sci. 2000;25:509–515. doi: 10.1016/s0968-0004(00)01667-4. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki F, Goto M, Sawa C, Ito S, Watanabe H, Sawada J, Handa H. J Biol Chem. 1998;273:29302–29308. doi: 10.1074/jbc.273.45.29302. [DOI] [PubMed] [Google Scholar]
- 7.Malek S, Huxford T, Ghosh G. J Biol Chem. 1998;273:25427–25435. doi: 10.1074/jbc.273.39.25427. [DOI] [PubMed] [Google Scholar]
- 8.Marcotte E M, Pellegrini M, Yeates T O, Eisenberg D. J Mol Biol. 1999;293:151–160. doi: 10.1006/jmbi.1999.3136. [DOI] [PubMed] [Google Scholar]
- 9.Steipe B, Schiller B, Plückthun A, Steinbacher S. J Mol Biol. 1994;240:188–192. doi: 10.1006/jmbi.1994.1434. [DOI] [PubMed] [Google Scholar]
- 10.Letunic I, Goodstadt L, Dickens N J, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley R R, Ponting C P, Bork P. Nucleic Acids Res. 2002;30:242–244. doi: 10.1093/nar/30.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehmann M, Kostrewa D, Wyss M, Brugger R, D'Arcy A, Pasamontes L, van Loon A P. Protein Eng. 2000;13:49–57. doi: 10.1093/protein/13.1.49. [DOI] [PubMed] [Google Scholar]
- 12.Lehmann M, Loch C, Middendorf A, Studer D, Lassen S F, Pasamontes L, van Loon A P, Wyss M. Protein Eng. 2002;15:403–411. doi: 10.1093/protein/15.5.403. [DOI] [PubMed] [Google Scholar]
- 13.Ohage E, Steipe B. J Mol Biol. 1999;291:1119–1128. doi: 10.1006/jmbi.1999.3019. [DOI] [PubMed] [Google Scholar]
- 14.Knappik A, Ge L, Honegger A, Pack P, Fischer M, Wellnhofer G, Hoess A, Wölle J, Plückthun A, Virnekäs B. J Mol Biol. 2000;296:57–86. doi: 10.1006/jmbi.1999.3444. [DOI] [PubMed] [Google Scholar]
- 15.Virnekäs B, Ge L, Plückthun A, Schneider K C, Wellnhofer G, Moroney S E. Nucleic Acids Res. 1994;22:5600–5607. doi: 10.1093/nar/22.25.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pace C N, Scholtz J M. In: Protein Structure: A Practical Approach. Creighton T E, editor. London: Oxford Univ. Press; 1997. pp. 299–321. [Google Scholar]
- 17.Otwinowski Z, Minor W. In: Methods in Enzymology. Carter C W Jr, Sweet R M, editors. Vol. 276. San Diego: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 18.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 19.Batchelor A H, Piper D E, de la Brousse F C, McKnight S L, Wolberger C. Science. 1998;279:1037–1041. doi: 10.1126/science.279.5353.1037. [DOI] [PubMed] [Google Scholar]
- 20.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 21.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Murshudov G N, Vagin A A, Lebedev A, Wilson K S, Dodson E J. Acta Crystallogr D. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 23.Collaborative Computational Project. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 24.Byeon I-J, Li J, Ericson K, Selby T L, Tevelev A, Kim H J, O'Maille P, Tsai M D. Mol Cell. 1998;1:421–431. doi: 10.1016/s1097-2765(00)80042-8. [DOI] [PubMed] [Google Scholar]
- 25.Baumgartner R, Fernandez-Catalan C, Winoto A, Huber R, Engh R A, Holak T A. Structure (London) 1998;6:1279–1290. doi: 10.1016/s0969-2126(98)00128-2. [DOI] [PubMed] [Google Scholar]
- 26.Mandiyan V, Andreev J, Schlessinger J, Hubbard S R. EMBO J. 1999;18:6890–6898. doi: 10.1093/emboj/18.24.6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venkataramani R, Swaminathan K, Marmorstein R. Nat Struct Biol. 1998;5:74–81. doi: 10.1038/nsb0198-74. [DOI] [PubMed] [Google Scholar]
- 28.Michel F, Soler-Lopez M, Petosa C, Cramer P, Siebenlist U, Müller C W. EMBO J. 2001;20:6180–6190. doi: 10.1093/emboj/20.22.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y, Nanduri S, Sen S, Qin J. Structure (London) 1998;6:619–626. doi: 10.1016/s0969-2126(98)00063-x. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs M D, Harrison S C. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 31.Foord R, Taylor I A, Sedgwick S G, Smerdon S J. Nat Struct Biol. 1999;6:157–165. doi: 10.1038/5845. [DOI] [PubMed] [Google Scholar]
- 32.McDonald I K, Thornton J M. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 33.Wallace A C, Laskowski R A, Thornton J M. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 34.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 35.Guex N, Peitsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 36.Carugo O, Pongor S. J Mol Biol. 2002;315:887–898. doi: 10.1006/jmbi.2001.5250. [DOI] [PubMed] [Google Scholar]
- 37.Page R D M. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 38.Mosavi L K, Williams S, Peng Z. J Mol Biol. 2002;320:165–170. doi: 10.1016/S0022-2836(02)00441-2. [DOI] [PubMed] [Google Scholar]
- 39.Tang K S, Guralnick B J, Wang W K, Fersht A R, Itzhaki L S. J Mol Biol. 1999;285:1869–1886. doi: 10.1006/jmbi.1998.2420. [DOI] [PubMed] [Google Scholar]
- 40.Zeeb M, Rosner H, Zeslawski W, Canet D, Holak T A, Balbach J. J Mol Biol. 2002;315:447–457. doi: 10.1006/jmbi.2001.5242. [DOI] [PubMed] [Google Scholar]
- 41.Zweifel M E, Barrick D. Biochemistry. 2001;40:14357–14367. doi: 10.1021/bi011436+. [DOI] [PubMed] [Google Scholar]
- 42.Zulauf M, D'Arcy A. J Crystallogr Growth. 1992;122:102–106. [Google Scholar]
- 43.Kumar S, Tsai C-J, Nussimov R. Protein Eng. 2000;13:179–191. doi: 10.1093/protein/13.3.179. [DOI] [PubMed] [Google Scholar]
- 44.Huxford T, Huang D B, Malek S, Ghosh G. Cell. 1998;95:759–770. doi: 10.1016/s0092-8674(00)81699-2. [DOI] [PubMed] [Google Scholar]
- 45.Zhang B, Peng Z. J Mol Biol. 2000;299:1121–1132. doi: 10.1006/jmbi.2000.3803. [DOI] [PubMed] [Google Scholar]
- 46.Li A J, Nussinov R. Proteins. 1998;32:111–127. [PubMed] [Google Scholar]
- 47.Lo Conte L, Chothia C, Janin J. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 48.Masumoto K, Ueda T, Motoshima H, Imoto T. Protein Eng. 2000;13:691–695. doi: 10.1093/protein/13.10.691. [DOI] [PubMed] [Google Scholar]
- 49.Matthews B W, Nicholson H, Becktel W J. Proc Natl Acad Sci USA. 1987;84:6663–6667. doi: 10.1073/pnas.84.19.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanes J, Plückthun A. Proc Natl Acad Sci USA. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ostermeier C, Michel H. Curr Opin Struct Biol. 1997;7:697–701. doi: 10.1016/s0959-440x(97)80080-2. [DOI] [PubMed] [Google Scholar]
- 52.Mosavi L K, Minor D L, Peng Z-Y. Proc Natl Acad Sci USA. 2002;99:16029–16034. doi: 10.1073/pnas.252537899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koradi R, Billeter M, Wüthrich K. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.