Abstract
Schizosaccharomyces pombe cells survive loss of telomeres by a unique pathway of chromosome circularization. Factors potentially involved in this survival mechanism include the heterodimeric Ku protein and ligase IV, both of which are involved in the repair of DNA double-strand breaks in mammalian cells. Furthermore, Ku plays a role in telomere maintenance as well as in DNA double-strand break repair in Saccharomyces cerevisiae. We have identified Ku and ligase IV homologues in S. pombe and analyzed their functions during normal growth and in cells undergoing senescence. In the absence of either a Ku subunit (pku70+) or ligase IV (lig4+), nonhomologous DNA end-joining was severely reduced. Lack of functional Ku led to shorter but stable telomeres and caused striking rearrangements of telomere-associated sequences, indicating a function for Ku in inhibiting recombinational activities near chromosome ends. In contrast to S. cerevisiae, concurrent deletion of pku70+ and the gene for the catalytic subunit of telomerase (trt1+) was not lethal, allowing for the first time the dissection of the roles of Ku during senescence. Our results support a model in which Ku protects chromosome termini from nucleolytic and recombinational activities but is not involved in the formation of chromosome end fusions during senescence. The conclusion that nonhomologous end-joining is not required for chromosome circularization was further supported by analysis of survivors in strains lacking the genes for both trt1+ and lig4+.
INTRODUCTION
Eukaryotic chromosomes end in nucleoprotein complexes known as telomeres (reviewed by Blackburn, 1991; Greider, 1996). In most species, the DNA portion consists of simple G-rich repeat sequences varying in length from 50 base pairs (bp) in hypotrichous ciliated protozoa to ∼300 bp in yeast and several kilobases in mammalian cells. Telomeric DNA is bound by a number of structural proteins that protect chromosomes from degradation and end-to-end fusion (Garvik et al., 1995; Van Steensel and de Lange, 1997; Horvath et al., 1998). Telomeres play a pivotal role in the complete replication of chromosomes, because conventional DNA polymerases fail to fully copy the ends of linear DNA molecules. In the absence of a mechanism to compensate for this “end-replication problem,” progressive telomere shortening leads to chromosome instability and cellular senescence.
Telomeric DNA is synthesized by the reverse transcriptase telomerase (Lingner et al., 1997; Nugent and Lundblad, 1998; reviewed by Bryan and Cech, 1999). This enzyme catalyzes the polymerization of telomeric repeats onto the 3′ ends of linear DNA molecules with the use of a domain in its RNA subunit as template (Greider and Blackburn, 1987; Yu et al., 1990; Singer and Gottschling, 1994). The catalytic protein subunit TERT (telomerase reverse transcriptase) is phylogenetically conserved among eukaryotes (Nakamura and Cech, 1998; O'Reilly et al., 1999). In Schizosaccharomyces pombe, TERT was identified by degenerate PCR and is encoded by the trt1+ gene (Nakamura et al., 1997). Deletion of trt1+ leads to progressive loss of telomeric sequences and causes cells to cease dividing after ∼120 generations. However, a small number of cells escape this senescence and continue proliferating without a further requirement for functional telomerase (Nakamura et al., 1998). Interestingly, many of these survivors were found to have circularized all three chromosomes. The same phenotype was observed in S. pombe strains after concurrent deletion of rad3+ and tel1+, two genes with sequence similarity to human ATM (ataxia telangiectasia mutated) (Naito et al., 1998).
Cellular senescence after the loss of telomerase function has also been reported in the budding yeast Saccharomyces cerevisiae (Lundblad and Szostak, 1989; Lendvay et al., 1996) and in mammalian cells (Bodnar et al., 1998). S. cerevisiae strains with a defective telomerase enzyme produce survivors with long and heterogeneous telomeres, which are maintained by a RAD52-dependent pathway of homologous recombination (Lundblad and Blackburn, 1993; Teng and Zakian, 1999). Immortal mammalian cell lines that lack detectable telomerase activity have also been found to contain long and heterogeneous telomeres, but it is not known whether these are formed by the same mechanism as in S. cerevisiae (Bryan et al., 1995; Reddel et al., 1997).
The Ku protein was first identified as an autoimmunoantigen in patients with polymyositis-scleroderma overlap syndrome (for review, see Featherstone and Jackson, 1999). Human Ku is a nuclear heterodimeric protein consisting of 69- and 83-kDa subunits, generally referred to as Ku70 and Ku80 (or Ku86). Ku binds tightly to overhanging or blunt ends of double-stranded DNA (Mimori and Hardin, 1986; Dynan and Yoo, 1998) and together with a 465-kDa catalytic subunit forms the DNA-dependent protein kinase (DNA-PK) (reviewed by Smith and Jackson, 1999). This enzyme plays an important role in DNA double-strand break repair and in V(D)J recombination, the rearrangement of immunoglobulin gene fragments. The S. cerevisiae Ku homologue (composed of Yku70/Hdf1 and Yku80/Hdf2) has been shown to act in the repair of DNA double-strand breaks via a pathway termed nonhomologous end-joining (NHEJ) (Boulton and Jackson, 1996a,b; Milne et al., 1996; Porter et al., 1996). This pathway restores the integrity of damaged DNA in the absence of homologous DNA sequences and is hence distinct from homologous recombination, which requires the RAD52 epistasis group genes.
Intriguingly, Ku-deficient S. cerevisiae strains also display a defect in telomere maintenance (Boulton and Jackson, 1996a; Porter et al., 1996). Telomeres are shortened and the single-stranded region at the 3′ end of the G-rich strand, normally restricted to S phase, persists throughout the cell cycle (Gravel et al., 1998; Polotnianka et al., 1998). Furthermore the telomere position effect, which confers silencing of gene expression near the telomere, is severely reduced (Boulton and Jackson, 1998; Evans et al., 1998; Laroche et al., 1998; Nugent et al., 1998) and telomeres are delocalized from the nuclear periphery (Laroche et al., 1998). A direct involvement of Ku at the telomere was confirmed by in vivo cross-linking of Ku to telomeric DNA (Gravel et al., 1998). Further genetic analysis of the telomere defect was impaired by the synthetic lethality of strains lacking functional Ku protein and either the catalytic subunit of telomerase (Est2) or a protein that binds the single-stranded portion of telomeric DNA (Cdc13) (Gravel et al., 1998; Laroche et al., 1998; Polotnianka et al., 1998).
Together, these results suggest that, at least in the budding yeast, Ku protein is involved directly in telomere maintenance as well as in double-strand break repair. This is an apparent contradiction, because the two mechanisms serve opposing goals. Binding of Ku to the ends of DNA double-strand breaks leads directly or indirectly to their repair. In contrast, at the natural ends of chromosomes, end-to-end fusions are specifically repressed.
The identification of a S. pombe Ku70 homologue enabled us to investigate its functions in telomere maintenance in an organism that is evolutionarily as distant from S. cerevisiae as it is from humans. We report that telomeres are shortened in Ku-deficient S. pombe and that telomere-proximal sequences undergo frequent rearrangements. Unlike in S. cerevisiae, double mutants of telomerase and pku70 were viable and displayed accelerated senescence, which led exclusively to survivors with three circular chromosomes. Ku-dependent NHEJ, therefore, is not required for chromosome circularization in S. pombe but plays a role in limiting the access of nucleolytic and recombinational activities to chromosome ends.
MATERIALS AND METHODS
S. pombe Strain Construction
The diploid strain heterozygous for deletions of trt1+ and pku70+ (PP29) was generated by transformation of CF248 (h+/h− leu1-32/leu1-32 ura4-D18/ura4-D18 his3-D1/his3-D1 ade6-M210/ade6-M216 trt1+/trt1−::his3+) with a linear DNA fragment containing the KanMX4 gene (Wach et al., 1994) flanked by approximately 800 bp of S. pombe DNA corresponding to sequences upstream and downstream of the large central exon in the pku70+ gene (accession number O94395). Transformations were performed with the use of the lithium acetate method as described by Alfa et al. (1993). Transformants were selected on YEA plates supplemented with geneticin disulfate (100 μg/ml). After restreaking twice on selective medium, correct insertion of the KanMX4 gene was confirmed by PCR. The diploid strain heterozygous for deletions of trt1+ and lig4+ (PP28) was generated in a similar way except that 800-bp fragments from upstream and downstream of the lig4+ gene (accession number O74833) were used to flank the KanMX4 marker.
Culture Media
Strains were propagated on YEA plates (0.5% yeast extract [Difco, Detroit, MI], 3% glucose, 2% agar, 0.01% leucine, 0.01% uracil, 0.01% histidine-HCl, 0.001% adenine). Diploid strains were sporulated on ME plates (3% malt extract [BIO101, Vista, CA], 2% agar). Genotypes of haploid strains were identified by streaking on YEA plates supplemented with 100 μg/ml geneticin disulfate (Sigma Chemical, St. Louis, MO) and pombe minimal glutamate (PMG) supplemented with leucine, uracil, and adenine (Alfa et al., 1993). YES (0.5% yeast extract [Difco], 3% glucose, 0.01% leucine, 0.01% uracil, 0.01% histidine-HCl, 0.01% adenine) was used for growth in liquid before transformation and during the recording of growth curves.
In Vivo DNA Repair Assay
The plasmid pBG1 (Burke and Gould, 1994) was linearized with XhoI and gel purified. S. pombe strains were transformed with 1 μg of linear or supercoiled pBG1 with the use of the lithium acetate method described by Alfa et al. (1993). Cells were plated on minimal medium, and colonies were counted after incubation at 32°C for 5 d.
Monitored Growth in Liquid Culture
Diploid strains PP28 and PP29 were sporulated at 30°C, and the resulting tetrads were dissected. Colonies grown from each spore at 32°C were transferred into 3 ml of YES and incubated in a shaker for 6 h at 32°C. Cells were then counted with the use of a hemacytometer, and 20 ml of YES was inoculated at a cell density of 2.5 × 104 cells/ml. Cultures were grown under vigorous shaking (250 rpm) at 32°C for 24 h, at which point the cell density was determined by counting, and cells were diluted into 20 ml of fresh YES at a density of 2.5 × 104 cells/ml. The remaining cells were collected by centrifugation, washed twice in SP1 buffer (1.2 M d-sorbitol, 50 mM sodium citrate, 50 mM Na2HPO4·7H2O, 40 mM EDTA), frozen in liquid nitrogen, and stored at −80°C for later preparation of genomic DNA. These procedures were repeated every 24 h for 26 d. To obtain sufficient cells from strains undergoing senescence, three identical 20-ml cultures were maintained. Relative growth rates were determined by comparing cell densities after growth for 24 h.
Genomic DNA Preparation
S. pombe genomic DNA was prepared with the use of an adaptation of the method described by Alfa et al. (1993). Frozen S. pombe cell pellets (∼2 × 108 cells) were thawed and resuspended in 1 ml of SP1 buffer containing Zymolyase-100T at 0.5 mg/ml. The cell suspension was incubated at 37°C for 30 min. Cells were collected by centrifugation at 10,000 × g in a tabletop centrifuge and resuspended in 1 ml of 5 × TE (50 mM Tris-HCl, pH 8.0, 10 mM EDTA) followed by the addition of 75 μl of SDS (20% [wt/vol] in H2O) and incubation at 65°C for 10 min. Potassium acetate (328 μl; 5 M) was added, and samples were incubated on ice for 15 min, followed by centrifugation at 10,000 × g for 10 min. The clarified supernatant was mixed with 1 volume of isopropanol, and nucleic acids were precipitated on ice for 20 min, collected by centrifugation, and resuspended in 500 μl of 5 × TE containing DNase-free RNase A (60 μg/ml). After incubation at 37°C for 1 h, organic extraction, and ethanol precipitation, the concentration of genomic DNA was determined by UV spectroscopy and by comparing 1-μl samples on ethidium bromide–stained agarose gels.
Southern Hybridization
Genomic DNA (15 μg) was digested for 6–8 h with EcoRI or NsiI in the buffers supplied by the manufacturer. Restriction fragments were loaded directly onto agarose gels and run in 0.5 × TBE (45 mM Tris, 45 mM borate, 1 mM EDTA, pH 8.3) at 2 V/cm for 16 h. To confirm equal loading, gels were stained for 30 min in ethidium bromide (1 μg/ml), and DNA was visualized under UV light. DNA was denatured by sodium hydroxide treatment and transferred onto a nylon membrane (Hybond NX, Amersham, Arlington Heights, IL) in 10 × SSC (1.5 M NaCl, 0.15 M sodium citrate) with the use of a PosiBlot 30-30 pressure blotter (Stratagene, La Jolla, CA).
Probes specific for the pol1+ gene and the chromosomal C, I, K, L, and M fragments were generated by random-primed labeling of gel-purified PCR products with the use of [α-32P]dCTP and high-prime mix (Boehringer Mannheim, Indianapolis, IN). Probes specific for the telomeric and telomere-associated sequences were created by the same method with the use of gel-purified fragments of pNSU70 (Sugawara, 1988). Hybridizations were carried out in Church-Gilbert buffer at 65°C (Church and Gilbert, 1984). To allow sequential hybridization of the same membrane, probes were removed by incubation at 65°C for 12 min in 0.4 N NaOH, 0.2% SDS. Membranes were then washed extensively in water before rehybridization in Church-Gilbert buffer.
Pulsed-Field Gel Electrophoresis
Cells were thawed and resuspended at a density of 5.5 × 108 cells/ml in SP1 buffer. For each four agarose plugs, 108 cells were treated with Zymolyase-100T (0.375 mg/ml) at 37°C for 1.5–2 h. Cells were collected by centrifugation and resuspended in 140 μl of TSE buffer (10 mM Tris-HCl, pH 7.5, 0.9 M sorbitol, 45 mM EDTA). A total of 220 μl of low-melt preparative-grade agarose (Bio-Rad, Richmond, CA) was added from a 1% solution in TSE equilibrated at 43°C, and the cell suspension was transferred into four plug molds. Solidified plaques were washed in PW1 (50 mM Tris-HCl, pH 7.5, 0.25 M EDTA, 1% SDS) at 50°C for 2–6 h, transferred into PW2 (10 mM Tris-HCl, pH 9.0, 0.5 M EDTA, 1% [wt/vol] N-lauroyl sarcosine, 1 mg/ml proteinase K), and incubated at 50°C for 24 h. Plugs were incubated for another 24 h at 50°C in fresh PW2 and subsequently washed extensively in T10xE (10 mM Tris-HCl, pH 7.5, 10 mM EDTA).
At this stage, plugs were either stored in T10xE at 4°C or loaded directly onto 0.8% agarose gels in 1 × TAE (40 mM Tris-acetate, 2 mM EDTA). Electrophoresis was performed in a CHEF DR III pulsed-field electrophoresis system (Bio-Rad) with the use of the settings suggested by the manufacturer. For NotI digests, plugs were washed twice for 15 min in TE (10 mM Tris-HCl, pH 7.5, 1 mM EDTA) and then incubated in NotI buffer (50 mM Tris-HCl, pH 7.9, 10 mM MgCl2, 100 mM NaCl, 1 mM DTT, 100 μg/ml BSA) for 2–3 h. Plugs were transferred into fresh NotI buffer containing 80 U NotI (New England Biolabs, Beverly, MA) and incubated at 37°C for 6 h. Plugs were then washed in T10xE, equilibrated in 0.5 × TBE for at least 30 min, and loaded onto 1% agarose gels. Gels were run in 0.5 × TBE with buffer circulation at 14°C. The run time was 24 h at 6 V/cm with a 60- to 120-s switch time ramp at an included angle of 120 degrees.
DNA was visualized by staining with ethidium bromide (1 μg/ml) for 30 min. Gels were then irradiated with 120 mJ/cm2 in a UV Stratalinker1800 (Stratagene) to nick the DNA. After sodium hydroxide treatment, DNA was transferred onto Hybond NX membrane (Amersham) and hybridized as described above.
RESULTS
S. pombe Homologues of Ku70 and Ligase IV Are Involved in DNA Repair
To investigate whether factors involved in DNA double-strand break repair play a role in telomere maintenance in fission yeast, we searched the S. pombe genome for homologues of the mammalian genes for the Ku and ligase IV proteins. A BLAST search identified the S. pombe gene SPCC126.02c (hereafter referred to as pku70+) as encoding a protein with 48% similarity (27% identity) to human Ku70. A comparison of the inferred pku70+ gene product with the S. cerevisiae Ku70 protein revealed a slightly lower homology, with 41% similarity (23% identity). The pku70+ gene contains five putative introns and encodes a 69-kDa protein.
A search of the S. pombe genome for proteins homologous to human ligase IV revealed the ORF SPCC1183.05c encoding a protein with 53% similarity (32% identity). The S. pombe gene contains nine putative introns and codes for a protein of 107 kDa. The sequence similarity with the S. cerevisiae ligase IV homologue LIG2 is 48% (28% identity). We hereafter refer to the corresponding S. pombe gene as lig4+.
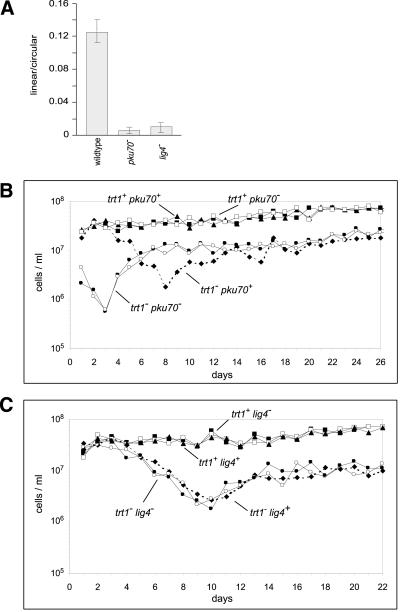
To test whether the putative pku70+ and lig4+ gene products were in fact involved in double-strand break repair, the plasmid pBG1 (Burke and Gould, 1994) was linearized within the his3+ sequence and transformed into strains containing the his3-D1 deletion. To account for differences in transformation efficiency between different strains and between experiments, each strain was transformed in parallel with the supercoiled form of pBG1. In the absence of a chromosomal copy of the his3+ gene, growth on minimal medium requires circularization of the plasmid via NHEJ. The number of transformants obtained with the linear substrate, normalized to the circular control, reflects a strain's ability to accurately repair double-strand breaks via NHEJ. It was found that deletion of pku70+ or lig4+ caused an ∼10-fold decrease in the number of transformants obtained with the linear substrate (Figure 1A). Therefore, we conclude that pku70+ and lig4+ are indeed involved in DNA double-strand break repair, as indicated by their sequence homology with the respective mammalian and S. cerevisiae proteins.
Figure 1.
(A) Double-strand break repair defect in pku70− and lig4− strains. For each strain, the efficiency of repair is expressed as the number of transformants obtained with linear plasmid divided by the number of transformants obtained with circular plasmid. Numbers of colonies for a typical experiment were as follows: 1012 wild-type circular; 127 wild-type linear; 1160 pku70− circular; 12 pku70− linear; 1560 lig4− circular; and 22 lig4− linear. (B) Senescence and generation of survivors in the absence of telomerase and pku70+. A diploid strain heterozygous for deletions of pku70+ and trt1+ was sporulated, and the tetrads were dissected and germinated on YES plates. The resulting colonies were used to inoculate precultures in YES liquid medium. Growth curves were recorded as described in MATERIALS AND METHODS. Day 1 on the x axis corresponds to ∼30 generations after germination. The cell density on each day is plotted for wild type (▴), pku70− trt1+ (▪ and □), pku70+ trt1− (♦), and pku70− trt1− (● and ○). (C) As in B except that the diploid starter strain was heterozygous for deletions of lig4+ and trt1+. The cell density on each day is plotted for wild type (▴), lig4− trt1+ (▪ and □), lig4+ trt1− (♦), and lig4− trt1− (● and ○).
Lack of pku70+ but not lig4+ Accelerates Senescence in Telomerase-deficient S. pombe Strains
To analyze the functions of the Ku and ligase IV proteins in telomere maintenance, diploid strains were constructed in which one copy of either the pku70+ or lig4+ gene was replaced by the kanamycin resistance marker. Both strains were also heterozygous for a deletion of the trt1+ gene, which encodes the catalytic subunit of telomerase. Heterozygous diploids were then sporulated, and the resulting tetrads were dissected. Haploid sister strains were propagated by successive restreaks on plates. Wild-type and pku70− strains formed round pin-sized colonies within 2 d on each of eight restreaks. As reported previously, trt1− cells grew well on the first and second restreak but formed fewer and smaller colonies on the third restreak (Nakamura et al., 1997). Cells taken from this plate were viable and formed pin-sized colonies within 3 d on subsequent restreaks, indicating that these were telomerase-independent survivors. The pku70− trt1− spores formed normal-sized colonies after germination, but on the first restreak few colonies appeared, and these were small and most of them had ragged edges. On subsequent restreaks, most colonies appeared normal but required 3 d to reach pin size.
To analyze the effect of the pku70− and trt1− mutations under competitive growth conditions, the products of two complete tetrads were used to assay growth in liquid culture. During 26 d, the growth of haploid strains that were trt1+ but pku70− was indistinguishable from that of wild-type controls (Figure 1B). Similarly, the absence of ligase IV in a telomerase-proficient strain did not cause any growth defects (Figure 1C). The growth rate of trt1− cultures was similar to that of wild-type controls for the first 2–3 d and then decreased gradually as the number of severely elongated, branched, and dead cells increased. After 8–10 d, the growth rate increased again as a result of the generation of survivors that lack a requirement for functional telomerase. For most trt1− cultures, the growth rate then stabilized at a level fourfold to sixfold below the growth rate of telomerase-positive cultures (Figure 1, B and C).
Consistent with the growth on plates, the growth rate of pku70− trt1− strains declined much faster than that of trt1− strains, with cultures reaching a point of lowest viability after only 3 d (Figure 1B). At this time, the generation time of pku70− trt1− cells was ∼5.4 h, compared with ∼3.7 h for trt1− cells at their low point, indicating that the double mutant undergoes a more severe crisis. Similar to trt1−, the generation time of pku70− trt1− cultures then decreased and plateaued at ∼2.6 h.
The accelerated and more severe senescence seen in the pku70− trt1− strains could be attributable to a role of pku70p in telomere maintenance. In addition, pku70p might be involved in the circularization of chromosomes via a nonhomologous end-joining pathway, such that pku70− strains are impaired in generating survivors. In the latter case, deletion of another component involved in the same double-strand break repair pathway should also lead to a more severe crisis, because survivors would be generated at a lower frequency. Although deletion of lig4+ reduced NHEJ by ∼10-fold, it had no effect on senescence, and growth rates deteriorated and then recovered with the same kinetics in trt1− lig4− strains as in isogenic trt1− controls (Figure 1C). Thus, pku70p is implicated as being important for telomere maintenance rather than for chromosome circularization.
Absence of pku70p Leads to Telomere Shortening and Rearrangements of Telomere-associated Sequences
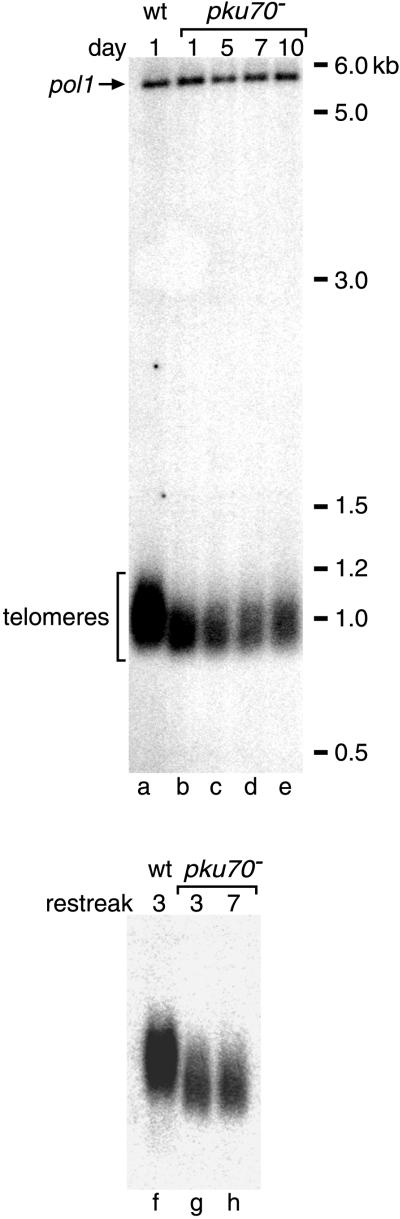
We next investigated whether the absence of pku70p or lig4p would cause a telomere phenotype even in a telomerase-positive strain. Genomic DNA was prepared from pku70− and lig4− cells as well as from isogenic controls grown in liquid culture for various times. DNA samples were digested with EcoRI, which cuts ∼1 kilobase (kb) from the ends of wild-type S. pombe chromosomes. Southern blot analysis with a telomeric probe revealed that the terminal restriction fragments were not shortened in lig4− strains. In contrast, in pku70− strains, telomeres were ∼100 bp shorter than in wild-type controls (Figure 2). This reduced telomere length was reached in less than 30 generations from the time of sporulation and was stably maintained for more than 100 generations in liquid culture and on plates (Figure 2). Reduction in telomere length could perhaps be due to the inability of telomerase to compensate for increased telomere degradation in the absence of Ku. However, although overexpression of trt1+ increased telomerase activity approximately fivefold as measured by an in vitro assay, it did not lead to an increase in telomere length in wild-type or pku70− strains (our unpublished results).
Figure 2.
Telomere length in wild-type and pku70− strains. Genomic DNA of pku70+ trt1+ and pku70− trt1+ strains was prepared on the indicated days of monitored growth in liquid culture (lanes a–e) or after three and seven restreaks on plates (lanes f–h). A total of 15 μg of DNA from each sample was digested with EcoRI and subjected to agarose gel electrophoresis and Southern transfer onto a nylon membrane. The blot was subsequently probed with a 32P-labeled telomeric fragment. As a loading control, a 32P-labeled fragment of the single-copy pol1+ gene was included in the hybridization mix. Size markers were 100-bp and 1-kb ladders from New England Biolabs.
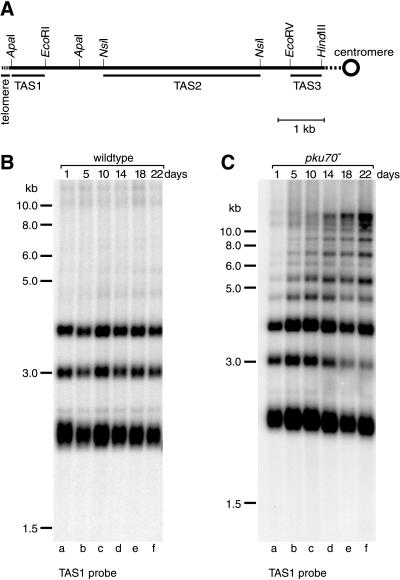
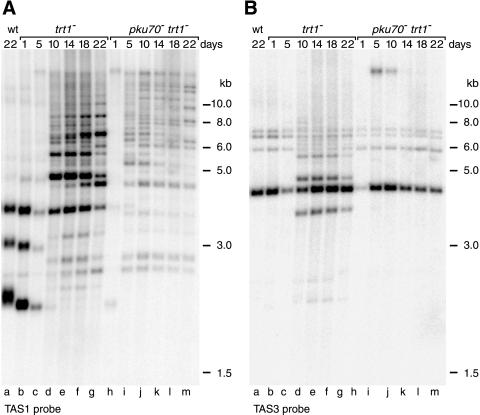
In S. pombe, the telomeric repeat sequences are internally flanked by at least 19 kb of repetitive telomere-associated sequence (TAS) (Sugawara, 1988). Depending on the strain background, TAS are found on four, five, or all six chromosome ends (Sugawara, 1988; Nakamura et al., 1998). To analyze the restriction pattern of subtelomeric sequences, three probes were used that hybridize to distinct regions of TAS (referred to as TAS1, TAS2, and TAS3 in Figure 3A). In the strains used in this study, digestion of total DNA with NsiI generated terminal fragments of ∼2.3, 3, and 3.8 kb. These fragments can be visualized by Southern blotting with the use of a TAS1 probe (Figure 3B). In a wild-type strain, the restriction pattern remained constant for 22 d of growth in liquid culture (Figure 3B, compare lanes a and f).
Figure 3.
Stability of TAS. (A) Restriction enzyme sites in the telomeric and telomere-associated sequences of one chromosome arm cloned in the plasmid pNSU70 (Sugawara, 1988). Locations of the probes used for Southern blotting are indicated by the bottom bars. Because of extensive homology between the TAS on different chromosomes, these probes hybridize to subtelomeric fragments on all six chromosome arms. (B and C) Genomic DNA (15 μg) was prepared from cells grown in liquid culture for the indicated number of days (see Figure 1), digested with NsiI, and fractionated on agarose gels. DNA was transferred onto a nylon membrane and hybridized to a 32P-labeled TAS1 probe. Size markers are as in Figure 2.
In contrast, frequent rearrangements of TAS were observed in the absence of pku70p (Figure 3C). This was indicated by the appearance of DNA fragments of ∼4.7 and 5.5 kb that hybridized to the TAS1 probe (lanes a–c). During extended growth in liquid culture, further rearrangements occurred, as shown by numerous higher-molecular-mass DNA fragments containing TAS1 sequences (lanes d–f). In contrast to the telomere-proximal TAS1 sequence, very few rearrangements were observed when the same blot was hybridized with the TAS2 or TAS3 probe, indicating that the effects of deleting pku70+ are limited to sequences near the ends of chromosomes (our unpublished results). Rearrangements of TAS were not observed in the absence of pku70+ and rad22+ (the homologue of S. cerevisiae RAD52), suggesting that these events are due to homologous recombination between TAS (P.B., T.M. Nakamura, and T.R.C., unpublished data). Together, our results indicate that pku70p has at least two functions in telomere maintenance. First, rapid shortening of telomeres after deletion of pku70+ indicates an involvement in telomere length regulation. Second, after extended growth of pku70− strains, a role in the protection of terminal sequences from recombinational activities becomes apparent.
pku70p Delays Telomere Erosion in the Absence of Telomerase
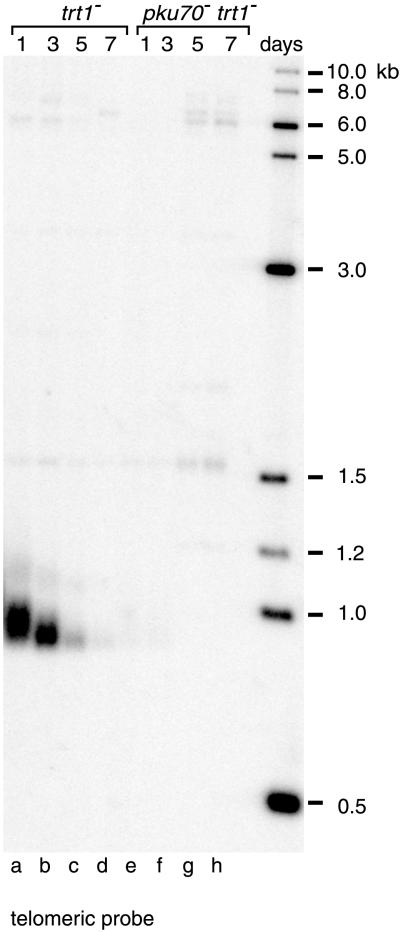
Despite the changes in telomere length and subtelomeric organization observed in pku70− strains, growth rate and cell viability were not affected. In contrast, deletion of trt1+ led to a gradual increase in generation time and loss of viability (Figure 1, B and C). To examine changes in the length of telomeric repeats as cells undergo senescence, total DNA was prepared from cells grown for 1, 3, 5, and 7 d in liquid culture. For trt1− strains, the length of the terminal EcoRI fragments as well as the hybridization intensity decreased gradually, indicating progressive loss of telomeric sequences (Figure 4, lanes a–d). These results are consistent with a previous study analyzing trt1− cells from successive restreaks on plates (Nakamura et al., 1997).
Figure 4.
Telomere-shortening phenotype of trt1− and pku70− trt1− strains. Sister strains obtained by tetrad dissection of a trt1+/trt1− pku70+/pku70− strain were grown in liquid culture. Genomic DNA was prepared on the indicated days, and 20 μg of each sample was digested with EcoRI. DNA was transferred onto a nylon membrane and hybridized with a 32P-labeled telomeric fragment.
Intriguingly, in pku70− trt1− strains, the telomeric repeats were already absent on d 1 of growth in liquid culture, corresponding to ∼30 generations after germination (Figure 4, lane e). This rapid loss of terminal sequence in the double mutant is consistent with the accelerated senescence observed in Figure 1B. At least in the absence of telomerase, pku70p seems to protect the ends of chromosomes from rapid degradation, possibly by binding to the ends and preventing the access of nucleases.
TAS Amplification and Chromosome Circularization in Survivors of Senescence
The rapid disappearance of telomeric sequences in pku70− trt1− strains suggested that TAS might also be affected more severely in the double mutant. Therefore, we analyzed genomic DNA isolated from trt1− and pku70− trt1− cultures by Southern hybridization with the three subtelomeric probes depicted in Figure 3A. In a trt1− strain, initial shortening of the terminal NsiI fragments and a reduction in hybridization to the TAS1 probe were observed (Figure 5A, lanes b and c). Reduced hybridization was also observed with the use of a TAS3 probe, suggesting that after 5 d of growth many cells had lost more than 5 kb of terminal sequence (Figure 5B, lane c). The survivors of senescence, however, had retained TAS1 sequences, which were strikingly rearranged and amplified (Figure 5A, lanes d–g). Similarly, although to a lesser extent, TAS2 and TAS3 sequences were found to be rearranged in these cells (Figure 5B, lanes d to g; our unpublished results).
Figure 5.
Dynamic rearrangements of TAS in survivors of senescence. (A) Genomic DNA (15 μg) was prepared from trt1− and pku70− trt1− cultures on the indicated days, digested with NsiI, and fractionated on agarose gels. DNA was transferred onto a nylon membrane and hybridized to a 32P-labeled TAS1 probe. Size markers are as in Figure 2. (B) The blot shown in A was stripped as described in MATERIALS AND METHODS and hybridized to a TAS3 probe.
As expected for a pku70− trt1− culture, TAS were lost more rapidly (Figure 5A, compare lanes b and h). In fact, on d 1, the majority of cells had already lost more than 5 kb of terminal sequence (Figure 5B, lane h). The emerging survivors, however, contained telomere-proximal sequences that were undergoing frequent rearrangements (Figure 5A, lanes i–m). As in trt1− strains, fewer rearrangements were observed with the use of the more distal TAS2 and TAS3 probes (Figure 5B, lanes i–m; our unpublished results).
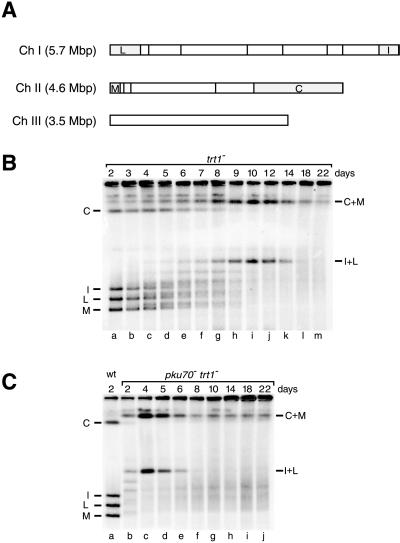
Telomerase-deficient S. pombe cells have been shown to escape senescence through two distinct pathways: 1) loss of 4–5 kb of terminal sequence followed by circularization of all three chromosomes, and 2) amplification of telomeric and telomere-associated sequences (Nakamura et al., 1998). Survivors of the latter category were thought to maintain linear chromosomes through homologous recombination based on the resemblance of this phenotype to that observed in telomerase-deficient S. cerevisiae and mammalian cells. To test this notion, genomic DNA from the same cultures used above was digested with NotI and analyzed by pulsed-field gel electrophoresis. (A NotI restriction map of S. pombe chromosomes is shown in Figure 6A.) Southern blots were then hybridized simultaneously with four probes that visualize the terminal fragments of chromosomes I and II in presenescent trt1− cells (Figure 6B, lane a). When DNA samples from later times were analyzed, these terminal fragments were no longer detected. Instead, a number of less well-defined bands suggested the occurrence of chromosome fusions and rearrangements (lanes c–h). As survivors emerged, two bands were predominantly visualized by the probes (lanes h–k). Sequential probing identified these bands as C+M and I+L, the products of chromosome circularization.
Figure 6.
Chromosome circularization in trt1− and pku70− trt1− strains. (A) Scheme of NotI restriction sites in S. pombe chromosomes. The terminal fragments on chromosomes I and II are shown in gray. Chromosome III lacks a NotI restriction site. (B and C) Pulsed-field gel analysis of NotI-digested genomic DNA from the same trt1− and pku70− trt1− strains used in Figure 5. The gel was run and processed as described in MATERIALS AND METHODS. DNA was transferred onto a nylon membrane and hybridized to internal probes on the I, L, M, and C fragments. The terminal fragments of linear chromosomes I and II are indicated on the left, and the fragments resulting from chromosome circularization (I+L and C+M) are indicated on the right.
Consistent with the accelerated senescence in pku70− trt1− cells, chromosome fusions were already apparent on d 2 (Figure 6C, lane b). By d 4, bands C+M and I+L indicative of circular chromosomes were the predominant species (lane c). Intriguingly, hybridization to these junction fragments diminished on subsequent days (lanes d and e) and was then largely replaced by a diffuse smear (f–j). A similar situation occurred later in trt1− cultures: survivors with circular chromosomes emerged, the culture returned to a stable growth rate, and then on d 18–22 the bands indicative of circular chromosomes diminished (Figure 6B, lanes l and m).
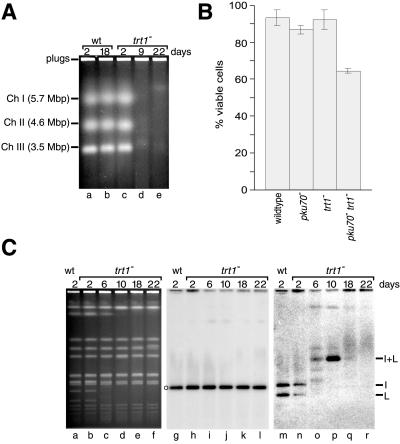
These results raised the question of whether loss or diminution of the fusion fragments indicated that at least some of the chromosomes had reverted to a linear form. Whereas intact linear S. pombe chromosomes can be separated by pulsed-field gel electrophoresis, circular chromosomes fail to enter a pulsed-field gel (Fan et al., 1992). In DNA preparations from a wild-type strain, the three linear chromosomes were separated by pulsed-field gel electrophoresis (Figure 7A, lanes a and b). The same result was obtained with trt1− cells before senescence (lane c). However, neither in young survivors (lane d) nor in cells that had been cultured for 22 d (lane e) did we observe linear chromosomes of discrete size.
Figure 7.
Chromosome dynamics in survivors. (A) Intact S. pombe chromosomal DNA was prepared from wild-type and trt1− strains and fractionated by pulsed-field gel electrophoresis. DNA was stained with ethidium bromide. (B) Viability of survivors. Cells were grown in liquid culture and counted with the use of a hemacytometer, and the equivalent of 500 cells were plated in triplicate for each strain. Colonies were counted after 2.5 d. (C) NotI-digested chromosomal DNA was fractionated by pulsed-field gel electrophoresis and stained with ethidium bromide (lanes a–f). The DNA was then transferred onto a nylon membrane that was sequentially hybridized with a probe specific for the K fragment on chromosome I (lanes g–l) and probes specific for the I and L fragments on chromosome I (lanes m–r). The K fragment is an internal restriction fragment on chromosome I and is indicated by an open circle next to lane g.
The fact that the growth rate of trt1− cultures was fourfold to sixfold below that of wild type could be due to an increase in the duration of the cell cycle. Alternatively, progression through the cell cycle might be normal but might frequently lead to inviable daughter cells. It was conceivable, therefore, that our cell samples contained >80% dead cells, possibly with partially degraded chromosomal DNA. To test these possibilities, plating assays were performed with cells after growth in liquid culture for 26 d. Strikingly, trt1− cells showed the same viability as wild-type controls, and only a moderate reduction was observed in pku70− trt1− cells (Figure 7B). Furthermore, when NotI-digested genomic DNA was visualized by ethidium bromide staining, the overall banding pattern was unchanged between early and late survivors (Figure 7C, lanes d–f), and hybridization to a chromosome-internal DNA restriction fragment was observed in samples in which I and L fragments could not be detected (Figure 7C, compare lanes k and l and lanes q and r). Together, these results suggest that failure to detect the terminal fragments of chromosomes I and II is not due to general DNA degradation. Therefore, it appears that chromosomes are mostly circular, with heterogeneity in the fusion fragments caused by extensive rearrangements of TAS. Alternatively, or in addition, the high frequency of rearrangements observed among TAS sequences indicates that many chromosomes may be undergoing recombination at any given time, resulting in recombination intermediates that either will fail to enter a pulsed-field gel or will run as a heterogeneous population of DNA molecules.
DISCUSSION
The results described here establish a function for the fission yeast Ku protein in maintaining the integrity of chromosome ends. In pku70− cells, telomeres are ∼100 bp shorter than in wild-type strains and telomere-adjacent sequences undergo frequent rearrangements. The latter observation provides direct evidence that Ku prevents recombinational activities from acting near the termini of chromosomes. Combining pku70+ and trt1+ deletions leads to accelerated senescence resulting from rapid loss of telomeric and subtelomeric sequences. However, despite the majority of cells losing in excess of 5 kb of terminal sequence, survivors with circular chromosomes are generated. In these, frequent rearrangements of subtelomeric sequences lead to great heterogeneity of the joining fragments. The occurrence of chromosome circularization in the absence of the Ku protein or ligase IV suggests that chromosome end fusions are not mediated by nonhomologous end-joining.
Ku Protects Chromosome Ends from Degradation and Recombination
Mutations in a number of genes in S. pombe and S. cerevisiae have been shown to cause altered but stable telomere length. Telomere length regulation in these strains may be affected in several different ways: shortened telomeres could result from a down-regulation of telomerase activity as well as from overactivation of an exonucleolytic activity that trims chromosome ends. The opposite can be imagined for factors that cause telomere elongation. In addition, telomere length can be modified in a number of more indirect ways, such as an imbalance between replication of the genome and telomere replication or a defect in the synthesis of the opposite strand, which appears to be tightly coupled to telomerase activity (Price, 1997; Diede and Gottschling, 1999; Adams Martin et al., 2000). The shortened but stable telomeres that were observed in pku70− strains are consistent with a role for Ku in mediating the equilibrium between synthesis and degradation of telomeric DNA. Deletion of pku70+ also led to substantial rearrangements of TAS, demonstrating a role for Ku in preventing recombinational activities from acting near the chromosome ends.
Protection from nucleolytic as well as recombinational activities appears to be conserved between S. pombe and S. cerevisiae. When an elongated telomere is introduced into S. cerevisiae, wild-type telomere tract length is restored by one of two mechanisms: slow, continual shortening or a single-step deletion of the artificially long telomere (Li and Lustig, 1996). The latter pathway has been shown to involve homologous recombination. In strains that are lacking one of the Ku subunits, gradual shortening of the elongated telomere is accelerated and single-step deletion events occur 50 times more frequently than in wild-type strains (Polotnianka et al., 1998).
S. pombe Ku Mutants Lack Temperature Sensitivity and Generate Survivors in the Absence of Telomerase
Despite many similarities between Ku-deficient S. cerevisiae and S. pombe strains, we also observed some notable differences. S. cerevisiae lacking either Ku subunit grow well at 25°C but display severe growth defects at 37°C (Feldmann and Winnacker, 1993; Boulton and Jackson, 1996a). When cells are shifted to the restrictive temperature, viability remains high for a few generations but decreases to 20% within 24 h (Barnes and Rio, 1997). A recent study suggested that the temperature-induced lethality is due to a defect in telomere metabolism, because the few cells that form colonies at 37°C show considerable amplification of subtelomeric Y′ DNA (Fellerhoff et al., 2000). In contrast to these observations, the growth rate and plating efficiency of S. pombe pku70− cells was indistinguishable from that of wild-type controls even after prolonged incubation at 37°C (our unpublished data). Therefore, it appears that the temperature-sensitive component of telomere maintenance is not conserved between S. cerevisiae and S. pombe.
The most striking difference between Ku mutants in the two yeasts concerns the generation of telomerase-independent survivors of senescence. In S. cerevisiae, concurrent deletion of the catalytic subunit of telomerase (EST2) and YKU70 or YKU80 causes a synthetic near lethality, which is characterized by double mutants undergoing a few cell divisions before losing viability (Gravel et al., 1998; Nugent et al., 1998). In contrast, pku70− trt1− fission yeast cells go through accelerated senescence but efficiently generate survivors that display a stable growth rate and high viability.
It is possible that Ku has additional functions in S. cerevisiae that are responsible for the lethality in the double mutant. However, the differences in the pathways by which the two yeast species escape senescence provide an alternative explanation. In S. cerevisiae lacking Est2, survivors have linear chromosomes with long and heterogeneous telomeres that undergo gradual shortening and continuous structural rearrangements (Lundblad and Blackburn, 1993; Teng and Zakian, 1999). In contrast, the majority of trt1− survivors and all pku70− trt1− survivors that we examined had circularized all three chromosomes. In light of the rapid degradation of terminal sequences observed in pku70− trt1− S. pombe, one can speculate that maintenance of chromosome ends by recombinational activities may be insufficient to counteract the loss of terminal sequences in S. cerevisiae.
Chromosome Circularization and TAS Rearrangements
Comparison of survivors generated on plates and in liquid culture revealed some intriguing differences. When trt1− or pku70− trt1− cells went through senescence in liquid culture, chromosome circularization as well as rearrangement of TAS was observed. In fact, even after circularization allowed cells to escape from senescence, dynamic rearrangements continued and led to great heterogeneity and amplification of TAS1 sequences. Curiously, when trt1− or pku70− trt1− cells were propagated by successive restreaks on plates, we consistently found that survivors had circular chromosomes that lacked TAS1 and TAS2 sequences but retained TAS3 sequences (Nakamura et al., 1998; P.B., unpublished data). Therefore, it appears that chromosome circularization but not dynamic rearrangements are essential for the generation of survivors in S. pombe. Indeed, in the absence of trt1+ and rad22+, the S. pombe homologue of RAD52, survivors generated in liquid culture are identical to those isolated from plates. They contain circular chromosomes but lack rearranged and amplified TAS1 sequences (our unpublished data). Although the different environments of plates versus liquid culture provide numerous reasons for the distinct phenotypes, one possibility is that continuous recombination provides a growth advantage in liquid culture. On plates, in contrast, individual survivors are not in competition, because they each form individual colonies.
Involvement of Ku in Telomere Maintenance Is Evolutionarily Conserved
Our results are consistent with a function for the S. pombe Ku protein in the protection of chromosome ends from nucleolytic degradation and recombinational activities. Similar conclusions were reached from experiments in the budding yeast. Considering the large evolutionary distance between these two yeast species, an involvement of the Ku protein is likely to be more widely conserved among eukaryotes. Indeed, recent reports indicate that rodent and human Ku can associate with telomeric DNA in vitro and in vivo (Bianchi and de Lange, 1999; Hsu et al., 1999), and increased chromosome fusions were observed in mouse cells lacking Ku70, Ku80, or DNA-PKcs (Bailey et al., 1999). It will now be very interesting to elucidate how cells distinguish between DNA double-strand breaks and natural ends of chromosomes, because both seem to be bound by at least some of the same factors. During senescence, a transition between the two forms occurs and chromosome ends start to be perceived as DNA double-strand breaks. The fact that S. pombe, unlike any other known organism, can survive this transition through the formation of circular chromosomes provides an important tool for further studies.
ACKNOWLEDGMENTS
We thank Diana Baumann, Julia Cooper, Robert West, and the members of the Cech laboratory for their comments and suggestions. This work was supported by a grant from the National Institutes of Health (GM28039) and by the Howard Hughes Medical Institute. P.B. is supported by a Wellcome Prize Traveling Research Fellowship (grant reference 054549/Z/98/Z).
REFERENCES
- Adams Martin A, Dionne I, Wellinger RJ, Holm C. The function of DNA polymerase alpha at telomeric G tails is important for telomere homeostasis. Mol Cell Biol. 2000;20:786–796. doi: 10.1128/mcb.20.3.786-796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with Fission Yeast. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1993. [Google Scholar]
- Bailey SM, Meyne J, Chen DJ, Kurimasa A, Li GC, Lehnert BE, Goodwin EH. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc Natl Acad Sci USA. 1999;96:14899–14904. doi: 10.1073/pnas.96.26.14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes G, Rio D. DNA double-strand-break sensitivity, DNA replication, and cell cycle arrest phenotypes of Ku-deficient Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:867–872. doi: 10.1073/pnas.94.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, de Lange T. Ku binds telomeric DNA in vitro. J Biol Chem. 1999;274:21223–21227. doi: 10.1074/jbc.274.30.21223. [DOI] [PubMed] [Google Scholar]
- Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–572. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Quellette M, Frolkis M, Holt SE, Chiu C-P, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996a;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996b;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan TM, Cech TR. Telomerase and the maintenance of chromosome ends. Curr Opin Cell Biol. 1999;11:318–324. doi: 10.1016/S0955-0674(99)80043-X. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JD, Gould KL. Molecular cloning and characterization of the Schizosaccharomyces pombe his3 gene for use as a selectable marker. Mol Gen Genet. 1994;242:169–176. doi: 10.1007/BF00391010. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- Dynan WS, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SK, Sistrunk ML, Nugent CI, Lundblad V. Telomerase, Ku, and telomeric silencing in Saccharomyces cerevisiae. Chromosoma. 1998;107:352–358. doi: 10.1007/s004120050318. [DOI] [PubMed] [Google Scholar]
- Fan JB, Rochet M, Gaillardin C, Smith CL. Detection and characterization of a ring chromosome in the fission yeast Schizosaccharomyces pombe. Nucleic Acids Res. 1992;20:5943–5945. doi: 10.1093/nar/20.22.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone C, Jackson SP. Ku, a DNA repair protein with multiple cellular functions? Mutat Res. 1999;434:3–15. doi: 10.1016/s0921-8777(99)00006-3. [DOI] [PubMed] [Google Scholar]
- Feldmann H, Winnacker EL. A putative homologue of the human autoantigen Ku from Saccharomyces cerevisiae. J Biol Chem. 1993;268:12895–12900. [PubMed] [Google Scholar]
- Fellerhoff B, Eckardt-Schupp F, Friedl AA. Subtelomeric repeat amplification is associated with growth at elevated temperature in yku70 mutants of Saccharomyces cerevisiae. Genetics. 2000;154:1039–1051. doi: 10.1093/genetics/154.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Larrivee M, Labrecque P, Wellinger RJ. Yeast Ku as a regulator of chromosomal DNA end structure. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- Greider CW. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- Hsu HL, Gilley D, Blackburn EH, Chen DJ. Ku is associated with the telomere in mammals. Proc Natl Acad Sci USA. 1999;96:12454–12458. doi: 10.1073/pnas.96.22.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche T, Martin SG, Gotta M, Gorham HC, Pryde FE, Louis EJ, Gasser SM. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr Biol. 1998;8:653–656. doi: 10.1016/s0960-9822(98)70252-0. [DOI] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Lustig AJ. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 1996;10:1310–1326. doi: 10.1101/gad.10.11.1310. [DOI] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- Milne GT, Jin S, Shannon KB, Weaver DT. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4189–4198. doi: 10.1128/mcb.16.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori T, Hardin JA. Mechanism of interaction between Ku protein and DNA. J Biol Chem. 1986;261:10375–10379. [PubMed] [Google Scholar]
- Naito T, Matsuura A, Ishikawa F. Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat Genet. 1998;20:203–206. doi: 10.1038/2517. [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Cech TR. Reversing time: origin of telomerase. Cell. 1998;92:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Cooper JP, Cech TR. Two modes of survival of fission yeast without telomerase. Science. 1998;282:493–496. doi: 10.1126/science.282.5388.493. [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- Nugent CI, Bosco G, Ross LO, Evans SK, Salinger AP, Moore JK, Haber JE, Lundblad V. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol. 1998;8:657–660. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- Nugent CI, Lundblad V. The telomerase reverse transcriptase: components and regulation. Genes Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- O'Reilly M, Teichmann SA, Rhodes D. Telomerases. Curr Opin Struct Biol. 1999;9:56–65. doi: 10.1016/s0959-440x(99)80008-6. [DOI] [PubMed] [Google Scholar]
- Polotnianka RM, Li J, Lustig AJ. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr Biol. 1998;8:831–834. doi: 10.1016/s0960-9822(98)70325-2. [DOI] [PubMed] [Google Scholar]
- Porter SE, Greenwell PW, Ritchie KB, Petes TD. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:582–585. doi: 10.1093/nar/24.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CM. Synthesis of the telomeric C-strand. Biochemistry. 1997;62:1216–1223. [PubMed] [Google Scholar]
- Reddel RR, Bryan TM, Murnane JP. Immortalized cells with no detectable telomerase activity. Biochemistry. 1997;62:1254–1262. [PubMed] [Google Scholar]
- Singer MS, Gottschling DE. TLC1 template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- Smith GC, Jackson SP. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- Sugawara NF. DNA Sequences at the Telomeres of the Fission Yeast S. pombe. Ph.D. Thesis. Cambridge, MA: Harvard University; 1988. [Google Scholar]
- Teng SC, Zakian VA. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:470–473. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Yu G-L, Bradley JD, Attardi LD, Blackburn EH. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature. 1990;344:126–131. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]