Abstract
The Scar/WAVE family of scaffolding proteins organize molecular networks that relay signals from the GTPase Rac to the actin cytoskeleton. The WAVE-1 isoform is a brain-specific protein expressed in variety of areas including the regions of the hippocampus and the Purkinje cells of the cerebellum. Targeted disruption of the WAVE-1 gene generated mice with reduced anxiety, sensorimotor retardation, and deficits in hippocampal-dependent learning and memory. These sensorimotor and cognitive deficits are analogous to the symptoms of patients with 3p-syndrome mental retardation who are haploinsufficient for WRP/MEGAP, a component of the WAVE-1 signaling network. Thus WAVE-1 is required for normal neural functioning.
Dynamic changes in the actin cytoskeleton control a range of cellular events such as wound healing, immune defense, embryonic development, and neuronal outgrowth (1–3). The Rho-family GTPases, Rho, Cdc42, and Rac, mediate these processes by promoting distinct forms of actin remodeling (4). In recent years it has become evident that Cdc42 and Rac relay signals to cytoskeletal sites through the Wiskott–Aldrich-syndrome family of scaffolding proteins (5–7). The Wiskott–Aldrich-syndrome protein (WASp) and its paralog N-WASp respond to Cdc42, whereas three other members of the family, WAVE-1–3 (also called Scar proteins), process signals emanating from Rac (8–11). These multifunctional scaffolding proteins are composed of modular domains. For example, a Cdc42/Rac-interactive binding (CRIB) domain at the amino terminus of WASp and N-WASp binds directly to Cdc42, whereas analogous regions in the WAVE isoforms associate indirectly with Rac, perhaps via an adapter protein called IRSp53 (8, 12). Cross-linking of WASp and WAVE to the actin cytoskeleton occurs thorough a verprolin homology (VPH) domain and a carboxyl-terminal acidic domain that binds to the Arp2/3 complex, a group of seven proteins that facilitate actin remodeling and branching at the leading edges of cells (10, 13–15).
Another function for WAVE proteins may be to integrate information from a variety of signaling pathways. WAVE-1 has been identified as an A kinase-anchoring protein (AKAP) that tethers the cAMP-dependent protein kinase at locations where it has preferential access to a subset of its target substrates (16). The central core of each WAVE protein has multiple proline-rich sequences that provide binding sites for a variety of SH3 proteins including the Abl tyrosine kinase, the Abl-interacting proteins Abi1 and Abi2, and the actin-binding protein profillin (17, 18). Proteomic approaches have identified other binding partners that are positive and negative regulators of WAVE. Rac promotes WAVE-1 activation by causing the release of an inhibitory complex that includes PIR 121, Nap-125 and HSPC300 (17). In contrast, Rac signaling is terminated by a WAVE-1-associated GTPase-activating protein (GAP) called WRP (18). Interestingly WRP (also called MEGAP/srGAP3) was independently identified as a gene that is disrupted in patents suffering from 3p-syndrome mental retardation (19).
Although WASp/WAVE scaffolding proteins share a conserved modular structure and interact with a subset of the same binding partners, they have distinct physiological roles. For instance, the WASp gene is mutated in Wiskott–Aldrich syndrome, a rare X-linked immunodeficiency disease manifested by signaling and cytoskeletal abnormalities in lymphocytes and platelets (7). WASp knockout mice largely recapitulate this immunodeficiency (20). Disruption of the N-WASp gene in mice causes defects in neural tube formation and cardiovascular development that result in embryonic lethality (21). With the exception of WAVE-3, which is inactivated in a patient with ganglioneuroblastoma (22), little is known about the physiological roles of the WAVE isoforms. Therefore, gene targeting was used to inactivate the WAVE-1 gene in mice. In this report we demonstrate that WAVE-1 null mice have a reduced viability, are runted, and exhibit behavioral abnormalities. These behavioral deficits, which include poor balance, reduced coordination, and impaired learning and memory, are comparable to the symptoms of patients with 3p-syndrome mental retardation that is linked to genetic lesions in the WAVE-1-associated protein WRP (19).
Methods
WAVE-1 Genomic Cloning and Sequencing.
Bacterial artificial chromosomes (Incyte Genomics, St. Louis) containing WAVE-1 were digested and ligated into the vector Yplac22 and sequenced by using GPS-1 (New England Biolabs).
Gene Targeting.
A WAVE-1-targeting construct was prepared by homologous recombination in yeast by using Pray-1 and Yplac22 (23). Sequences (300 bp) from within exon 4 and intron 5 were subcloned into Pray-1 flanking the Ura- and Neo-selectable markers. The resulting construct was cotransfected into yeast with Yplac22 vector containing a SacI genomic DNA insert encompassing exons 3–8 of WAVE-1. Homologous recombination in yeast generated a targeting construct replacing exon 4 with the Ura and Neo markers. Chimeric mice were generated at Incyte Genomics according to established protocols. See Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org. Chimeric offspring were bred to C57BL/6 mice (The Jackson Laboratory), establishing germ-line transmission, and bred further to C57BL/6 mice for colony expansion.
PCR Genotyping.
Primers specific for the portion of exon 4 deleted by homologous recombination were used to generate a 421-bp band in wild-type and heterozygous progeny, whereas primers specific to the Ura-selectable marker amplified a 311-bp band from heterozygous and homozygous null mice.
Northern and Southern Blots.
The probe for Southern analysis was a SacI fragment of genomic DNA outside of the targeting construct between exons 2 and 3, whereas for Northern analysis DNA probes were generated from full-length WAVE-1. Genomic DNA from embryonic stem (ES) cells was prepared by incubation in proteinase K buffer (20 mM Tris, pH 7.4/1 mM CaCl2/50% glycerol) at 55°C for 12 h followed by phenol/chloroform extraction and ethanol precipitation. DNA was digested with EarI overnight and separated by electrophoresis in 0.8% agarose gel. Southern blotting was done in 0.4 M NaOH.
Antibodies.
Characterization of antibodies used in this study have been described: WAVE-1 [VO101 (18)], pan-WAVE [VO59 (16)], tubulin (Sigma), and AKAP150 (24).
Body Weight Measurements.
Littermates of all genotypes (+/+, n = 13; +/−, n = 20; −/−, n = 10) were weighed from 2 to 15 weeks of age.
In Situ Hybridization.
The hybridization protocol was as described (25), using an 824-bp cDNA fragment (nucleotides 1,365–2,186 of mouse WAVE-1 cDNA).
Immunohistochemistry.
Coronal cryosections from brains of mice were incubated with WAVE-1 antiserum (1:2,000) and monoclonal anti-calbindin D-28K (clone CB-955, 4 μg/ml, Sigma) or preimmune rabbit and normal mouse IgG followed by FITC donkey anti-rabbit and Texas red donkey anti-mouse (both 1:100, Jackson ImmunoResearch) and deep-red fluorescent Nissl stain (NeuroTrace 640/660, Molecular Probes). The same fields from corresponding sections were selected for all genotypes.
Animals.
Animals used for behavioral analysis were the offspring from heterozygotes of the third-generation backcrossing (+/+, n = 8; +/−, n = 16; −/−, n = 8), and those for biochemical and histochemical analyses were the offspring of third- and fourth-generation backcrossing. Mice were housed individually during the behavioral-testing, and tests were administered in order of increasing stress levels.
Behavioral Tests.
The rotarod, open-field, passive-avoidance, Morris water maze, and novel-object recognition tests were performed as described (26–28).
Inclined Screen.
In the inclined-screen test the mice were placed on a 182-cm-long inclined screen (36° incline) and allowed to explore for 3 min. The total distance moved and mean velocity of movement were recorded with a Noldus Instruments (Leesburg, VA) EthoVision video tracking system. Missteps were defined as placing any paw between the grids of the screen.
Balance Beam.
Mice were placed in the middle of a horizontal beam (88.5 cm long, 1.8 cm in diameter) 30.5 cm high. Total distance moved and mean velocity of movement in two trials of 2 min each were recorded using Noldus Instruments' EthoVision video tracking system.
Elevated Zero Maze.
The elevated zero maze (Hamilton–Kinder) consists of two enclosed areas and two open areas and has a diameter of 53.34 cm. Mice were placed in the closed part of the maze and allowed free access for 10 min. A video tracking system (Noldus Instruments) was used to calculate the time spent in the open and closed areas.
Wire Hanging.
To evaluate muscle strength, front- and hind-paw strength was evaluated with the wire hanging test. Mice were placed on a horizontal cotton wire (diameter, 1 mm) 60 cm above a surface. The length of time that the mice held onto the wire (latency) was recorded.
Results
Generation of the WAVE-1 Mice.
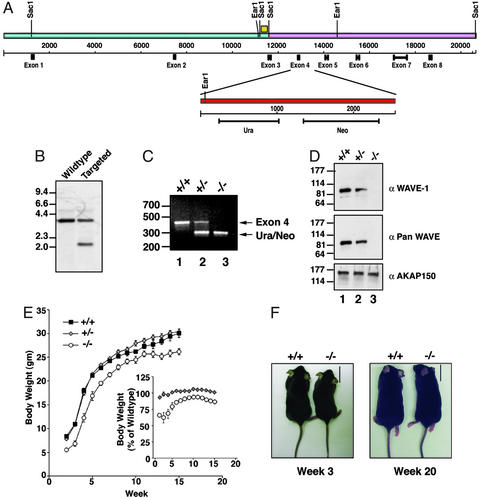
The WAVE-1 gene (20.6 kb) was isolated from bacterial artificial chromosomes by SacI digestion, sequenced, and found to encode the protein within eight exons (Fig. 1A). This information was used to construct a targeting vector that inserted uracil and neomycin resistance markers at 3′ end of exon 4 (Fig. 1A Upper). This introduced an in-frame stop codon at amino acid 175 in WAVE-1 and deleted 95 bp including the splice donor site of exon 4 (Fig. 1A Lower). The targeting vector was electroporated into mouse ES cells, and 190 neomycin-resistant clones were screened for the altered WAVE-1 gene. First, DNA from isogenic ES cells was digested with NotI, and samples containing a single copy of the neomycin-resistance gene were detected by Southern blot by using a specific probe (data not shown). Second, DNA from positive ES cell clones was digested with EarI and screened by Southern blot with a probe from outside of the targeting vector to confirm homologous recombination (Fig. 1B). ES cells from one double-positive colony were used to generate chimeric mice. Genotyping of the F2 offspring was performed by PCR. Primers to the 3′ portion of exon 4 detected a 421-bp band in wild-type and heterozygous progeny (Fig. 1C, lanes 1 and 2). Primers to the uracil marker detected a 311-bp band in the targeted DNA that was present only in heterozygous and homozygous null progeny (Fig. 1C, lanes 2 and 3).
Figure 1.
Inactivation of WAVE-1 gene and body-weight phenotype. (A) Schematic diagram of the mouse WAVE-1 gene (Upper). The location of the relevant restriction sites and exons for the WAVE-1-coding regions are indicated. The SacI genomic fragment used to generate the targeting vector (pink) is indicated. The site of homologous recombination (Lower, red) that inserted the Ura/Neo markers and deleted the exon 4–intron 5 boundary is highlighted. The yellow box marks the location of a nucleotide sequence that was used as a probe to detect homologous recombination. (B) Screening of DNA from ES cell colonies to detect homologous recombination by Southern blot analysis. Samples are indicated above each lane, and DNA sizes are indicated. (C) Genotyping of F2 progeny by PCR using primers specific for exon 4 (upper band) and the Ura/Neo markers (lower band). Samples from wild-type (+/+), heterozygous (+/−), and knockout (−/−) mice are marked above each lane, and DNA sizes are indicated. (D) WAVE protein expression in brain extracts from wild-type (+/+), heterozygous (+/−), and knockout (−/−) mice was assessed by immunoblot. Analysis of samples with WAVE-1-specific antibody (Top), pan-WAVE antibody (Middle), or unrelated AKAP150 antibody (Bottom) is presented. Molecular weight markers are indicated. (E) Body weight of wild-type (black squares), heterozygous (gray diamonds), and knockout (white circles) mice from weeks 2–15. (E Inset) The body weight of heterozygous and knockout mice expressed as a percentage of the wild-type body weight is shown. (F) Representative photographs of wild-type (+/+) and knockout (−/−) mice at weeks 3 (Left) and 20 (Right). (Scale bar, 1.7 cm.)
WAVE-1 protein levels were reduced in brain extracts isolated from heterozygous mice and were absent in samples from homozygous null mice as assessed by immunoblot with an isoform-specific antibody (Fig. 1D Top, lanes 2 and 3). Similar results were obtained with the antibody that detected all WAVE isoforms, indicating that there were no compensatory increases in expression levels of WAVE-2 or WAVE-3 in the brains of WAVE-1 knockout mice (Fig. 1D Middle, lanes 2 and 3). Additional immunoblots with a monoclonal antibody against the first 100 amino acids of WAVE-1 did not detect low molecular weight fragments in extracts from homozygous null mice, confirming that there was no expression of a truncated WAVE-1 form (data not shown). Control immunoblots demonstrated that equivalent levels of a neuronal marker protein, AKAP150, were present in each lane (Fig. 1D Bottom). Collectively, the results show that the WAVE-1 gene has been disrupted by homologous recombination, and homozygous null mice lack the protein.
The mating of heterozygous mice seemed to produce lower-than-expected numbers of homozygous null progeny (Table 1). This apparently reflected a reduced viability of the WAVE-1 knockout mice, because progeny were obtained in the correct Mendellian ratios when wild-type and heterozygous mice were crossed (Table 1). An obvious feature of the surviving WAVE-1 knockout mice was their reduced body size (Fig. 1 E and F Left). Weekly body-weight measurements indicated that the size differences were most evident between 2 and 6 weeks of age (Fig. 1 E Inset and F Left), yet there was no difference in the daily food intake of the WAVE-1 knockout mice (data not shown). By week 7 and onward the WAVE-1 null mice weighed 90% of their wild-type and heterozygous counterparts (Fig. 1 E Inset and F Right).
Table 1.
A record of offspring from heterozygous crosses
| Cross | +/+ | +/− | −/− | Total |
|---|---|---|---|---|
| +/− × +/− | 25 | 57 | 18 | 100 |
| +/+ × +/− | 92 | 90 | 182 |
The numbers represent individual mice.
The Regional Distribution of WAVE-1 in Mouse Brain.
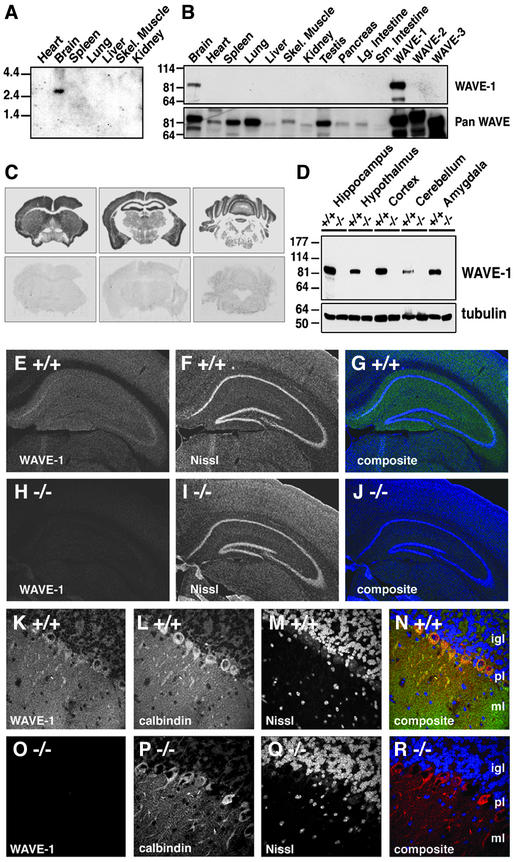
As a prelude to more detailed phenotypic characterization, we established the expression pattern of mouse WAVE-1. Northern blot analysis with probes to the WAVE-1-coding region identified a single mRNA species of 3.2 kb that was expressed exclusively in brain (Fig. 2A). Immunoblot experiments with antibodies specific for WAVE-1 confirmed this observation (Fig. 2B Upper), whereas complementary experiments detected WAVE-2 and WAVE-3 in most tissues (Fig. 2B Lower). The regional distribution of WAVE-1 in the brains of wild-type mice was determined by in situ hybridization, immunoblot, and immunohistochemistry techniques. WAVE-1 mRNA was detected in most brain regions including the cortex, hippocampus, and cerebellum by in situ hybridization with an antisense probe (Fig. 2C Upper). WAVE-1 protein was detected by immunoblot in extracts from the hippocampus, cortex, hypothalamus, and amygdala; less of the protein was present in cerebellar extracts (Fig. 2D Upper). Equal loading of each sample was confirmed by staining for tubulin (Fig. 2D Lower), and extracts obtained from knockout mice were negative for WAVE-1 (Fig. 2D Upper). Immunofluorescence techniques detected WAVE-1 throughout the hippocampus, but it was enriched in the subiculum and the CA3 region (Fig. 2 E and G). WAVE-1 was not detected in hippocampal sections from knockout mice (Figs. 2 H and J). Nissl staining was used as a marker for the cell bodies of hippocampal neurons (Fig. 2 F and I). Immunocytochemical analysis of cerebellar sections detected the majority of the WAVE-1 signal in Purkinje cells and the molecular layers (Fig. 2 K and N). WAVE-1 was not detected in cerebellar sections from knockout mice (Figs. 2 O and R). Purkinje cells were counterstained with antibodies against the marker protein calbindin (Figs. 2 L and P), and the granular layer was identified by Nissl staining (Figs. 2 M and Q). Together, these studies demonstrate that WAVE-1 is expressed in regions of the brain that process higher-order neural skills such as locomotion, coordination, and learning and memory. These later studies did not detect any obvious abnormalities in the cellular organization of hippocampus and cerebellum in WAVE-1 knockout mice. Preliminary examination of additional brain sections, however, suggested an enlargement of the third and lateral ventricles in the WAVE-1 knockout mice (data not shown).
Figure 2.
Tissue-specific expression of mouse WAVE-1. (A) Northern blot of wild-type mouse tissues (indicated above each lane) using a probe specific for WAVE-1. Size markers are indicated. (B) Immunoblot analysis of WAVE-1 protein expression using an isoform-specific antibody (Upper) and an antibody that recognizes all WAVE isoforms (Lower) in wild-type mouse tissue samples. Tissue sources are indicated above each lane. Control extracts from human embryonic kidney 293 cells expressing recombinant WAVE-1, WAVE-2, or WAVE-3 (last three lanes on the far right) demonstrate antibody selectivity. (C) In situ hybridization of coronal sections from a wild-type mouse. Hybridization with an anti-sense probe for WAVE-1 (Upper) or a sense probe (Lower) are indicated. (D Upper) Immunoblot analysis of WAVE-1 expression in specific brain regions (indicated above each lane) of wild-type (+/+) or knockout (−/−) mice. (D Lower) Tubulin expression used as a loading control. Molecular weight markers are indicated. (E–J) Immunohistochemical localization of WAVE-1 in coronal hippocampal sections from wild-type (E) or knockout (H) mice. Costaining with Nissl for wild-type (F) and knockout (I) sections is shown. Composite images of WAVE-1 (green) and Nissl (blue) staining are shown for wild-type (G) and knockout (J) sections. (K–R) Immunohistochemical localization of WAVE-1 in coronal cerebellar sections from wild-type (K) and knockout (O) mice. Costaining of Purkinje cells with calbindin (L and P) and the granule cell layer with Nissl (M and Q) are presented. Composite images of WAVE-1 (green), calbindin (red), and Nissl (blue) staining are shown for wild-type (N) and knockout (N) sections. The internal granule cell (igl), Purkinje cell (pl), and molecular (ml) layers are indicated on the composite images.
Characterization of Sensorimotor Deficits in WAVE-1 Knockout Mice.
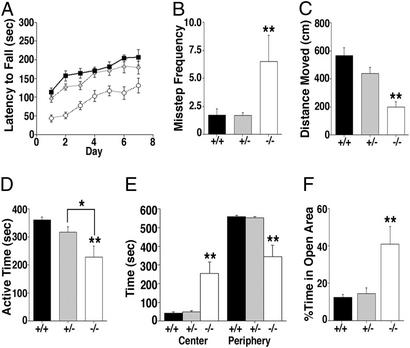
WAVE-1 null mice exhibited clear differences in their behavior (Figs. 3 and 4). Rotarod performance evaluated sensorimotor function in age-matched mice (8–12 weeks old). The latency of each mouse to fall during an increasing rotational speed protocol was measured three times daily for 1 week. WAVE-1 null mice performed poorly and fell with an average latency of 43 ± 9 sec (n = 8) on day 1, whereas wild-type and heterozygous mice remained on the rotorod for 114 ± 11 (n = 8) and 96 ± 10 sec (n = 16), respectively (Fig. 3A). The knockout mice were unable to compensate fully for this deficiency after training. In fact, by day 7 their latency to fall (131 ± 20 sec) was approximately the same as the wild-type mice on day 1 (Fig. 3A). These observations inferred that WAVE-1 null mice had deficits in motor coordination and/or muscular strength. Both attributes were assessed separately. The inclined-screen and balance-beam tests evaluated motor coordination. WAVE-1 null mice were 3.8-fold more prone to misstep while climbing the wire lattice of the inclined screen than heterozygous or wild-type mice (Fig. 3B). The balance beam tests motility on an elevated narrow bar. Mice with impaired balance travel less distance on the beam. Accordingly, the WAVE-1 null mice only traveled 198.2 ± 38.3 cm, whereas wild-type and heterozygous mice were more motile, traveling 568.4 ± 54.1 and 439.3 ± 44.3 cm, respectively (Fig. 3C). These experiments further demonstrate that WAVE-1 knockout mice exhibit deficits in sensorimotor function that include impaired motor coordination and balance. The wire hanging test evaluates muscular strength and endurance. Mice were suspended by either their front or hind paws from a cotton string (1 mm in diameter), and their fall latency was recorded. Each genotype performed identically in this test (i.e., front-paw wire hanging: +/+ = 4.7 ± 2.2 sec, +/− = 5.4 ± 2.1 sec, and −/− = 5.4 ± 1.6 sec; P = 0.95, −/− vs. +/− and +/+), suggesting that muscular strength was unaffected in the WAVE-1 null mice.
Figure 3.
Altered sensorimotor and anxiety levels in WAVE-1 null mice. (A) Rotorod test. The latency to fall during rotarod testing (sec) of wild-type (black squares), heterozygous (gray diamonds), and knockout (white circles) mice is indicated. Mice were tested three times per day over a 7-day period. Daily group averages are represented. Although all groups improved their ability to stay on the rotating rod with training (P < 0.01), WAVE-1 knockout mice fell earlier than heterozygous and wild-type mice (P < 0.01, −/− vs. +/− and +/+, Tukey–Kramer). (B) Inclined-screen test. The frequency of missteping during the inclined-screen test is shown. WAVE-1 knockout mice (−/−) misstepped more often than heterozygous (+/−) and wild-type (+/+) mice (**, P < 0.01 −/− vs. +/− and +/+, Tukey–Kramer). (C) Balance-beam test. Distance moved on a balance beam is shown. WAVE-1 knockout mice (−/−) showed reduced sensorimotor activity as compared with heterozygous (+/−) and wild-type (+/+) mice (**, P < 0.01 −/− vs. +/− and +/+, Tukey–Kramer). (D) Open-field test. The total active time during a 10-min open-field test is presented. WAVE-1 knockout mice (−/−) were significantly less active than heterozygous (+/−) and wild-type (+/+) mice (**, P < 0.01 −/− vs. +/+, and *, P < 0.05 −/− vs. +/−, Tukey–Kramer). (E) Total time in the center and total time spent at the periphery of the open field are indicated for each genotype (marked below each group). WAVE-1 knockout (−/−) mice spent significantly more time in the center and less time in the periphery than heterozygous (+/−) and wild-type (+/+) mice (**, P < 0.01 −/− vs. +/− and +/+, Tukey–Kramer). There was a significant effect of zone in wild-type and heterozygous (P < 0.01) but not in WAVE-1 knockout (P = 0.155) mice. (F) Elevated zero maze. The percentage of time spent in the open areas of the elevated zero maze is shown. WAVE-1 knockout mice (−/−) spent significantly more time in the open areas of the zero maze than heterozygous (+/−) and wild-type (+/+) mice (**, P < 0.01 −/− vs. +/− and +/+, Tukey–Kramer). Error bars represent SEM values.
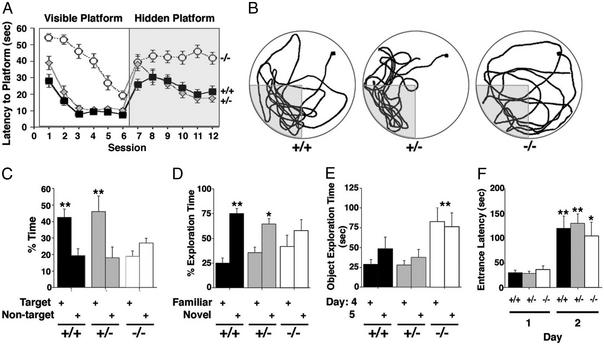
Figure 4.
WAVE-1 null mice are impaired in spatial and nonspatial but not in emotional learning and memory. (A) Morris water maze. Time to swim to the platform (latency) was measured for wild-type (black squares), heterozygous (gray diamonds), and WAVE-1 null (white circles) mice. The mice were tested in two daily sessions of three trials each. Sessions 1–6 (Left) represent learning trials for the visible platform, and sessions 7–12 (Right) represent learning trials for the submerged hidden platform. Session averages ± SEM are represented. Although all groups learned to locate the visible platform location (P < 0.01), WAVE-1 knockout mice required significantly more time to reach the platform (P < 0.01 −/− vs. +/+ and +/−, Tukey–Kramer). In contrast, wild-type (P < 0.05) and heterozygous (P < 0.01) mice learned to locate the hidden platform, but the WAVE-1 knockout did not (P = 0.833). (B) Representative swim-path traces for wild-type (+/+), heterozygous (+/−), and knockout (−/−) mice during the probe trial (platform removed). The start position is marked by a black square, and the target area is indicated by the shaded area. (C) The percentage of time spent in target vs. nontarget areas during the probe trial for wild-type (+/+), heterozygous (+/−), and knockout (−/−) mice is indicated. The wild-type and heterozygous mice spent significantly more time in the target area than any of the nontarget areas (P < 0.01, Tukey–Kramer) than the knockout (−/−) mice. (D and E) Novel-object recognition. (D) The percentage of object-exploration time spent with the familiar and novel objects is presented. Wild-type (+/+, P < 0.01) and heterozygous (+/−, P < 0.05) mice spent a significantly greater percentage of time exploring the novel object, indicating intact object recognition. WAVE-1 knockout mice (−/−) did not spend a greater percentage of time exploring the novel object (P = 0.375). (E) The total times spent exploring both objects. Wave-1 knockout (−/−) mice spent significantly more time exploring both objects on days 4 and 5 than wild-type (+/+) and heterozygous (+/−) mice (**, P < 0.01 −/− vs. +/− and +/+, Tukey–Kramer). (F) Passive-avoidance test. The latency to enter the dark chamber on days 1 and 2 is shown. All groups took significantly longer to enter the dark chamber on day 2 than day 1, and there were no genotype differences (*, P < 0.05, and **, P < 0.01, day 2 vs. day 1).
WAVE-1 Knockout Mice Exhibit Reduced Anxiety.
Evaluation of exploratory activity in a novel open arena revealed additional deficits. Mice were placed into a Plexiglas enclosure, and their movement was recorded accurately. WAVE-1 knockout mice were less active, moving only for a total of 229 ± 39 sec over the 10-min duration of the test (Fig. 3D). Wild-type and heterozygous mice moved for 361 ± 10 and 318 ± 18 sec over the same time period, respectively (Fig. 3D). Significantly, the knockout mice spent more time in the center of the open field (+/+ = 41 ± 7 sec, +/− = 48 ± 8 sec, and −/− = 255 ± 60 sec), indicating that they have reduced anxiety (Fig. 3E). This was confirmed further by tests performed in the elevated zero maze. WAVE-1 null mice spent 41 ± 9% of their time in the open areas and showed no preference for the enclosed (safe) area (Fig. 3F). In contrast, wild-type and heterozygous mice spent little time in the anxiety-provoking open areas (+/+ = 12 ± 2% and +/− = 14 ± 3%). Control measurements confirmed that the average velocity in the zero maze was similar for each genotype (data not shown). These two tests show that WAVE-1 knockout mice exhibit reduced anxiety levels, which often is associated with perturbation of neural networks in the amygdala (29).
Analysis of Learning and Memory Deficits.
The Morris water maze evaluates hippocampal-dependent spatial learning and memory (30). Initially mice were trained for six sessions to swim to a visible platform (cued training). During sesions 7–12 the mice were tested to determine whether they could swim to a submerged hidden platform (acquisition). Finally, the hidden platform was removed (probe trial) to measure spatial-memory retention. Each genotype learned to locate the visible platform, yet the WAVE-1 knockout mice took longer to reach it (Fig. 4A, sessions 1–6). This may be a consequence of the reduced swim speed of the WAVE-1 knockout mice (+/+ = 17 ± 0.4 cm/sec, +/− = 16 ± 0.3 cm/sec, and −/− = 11.4 ± 0.7 cm/sec) and is consistent with their poor performance in the tests of sensorimotor function (Figs. 3 A–D). Importantly, the WAVE-1 null mice failed to learn where the platform was hidden during the acquisition phase of the trial (Fig. 4A, sessions 7–12). This was also evident during the probe-trial phase of the test in which wild-type and heterozygous mice preferentially searched the target quadrant for the platform (Fig. 4 B Left and Center traces and C), whereas WAVE-1 null mice swam around randomly (Fig. 4 B Right trace and C). The performance of the WAVE-1 knockout mice during the acquisition and the probe trial suggests that they harbor hippocampal-dependent learning and memory deficits.
The cognitive impairments in WAVE-1 null mice were not restricted to spatial-learning memory. Novel-object recognition tests evaluate hippocampal-dependent nonspatial learning and memory (31). Mice were habituated to an enclosure for 5 min on 3 consecutive days. On day 4 the enclosure contained two objects, and the mice were allowed to explore them. There was no significant difference in the time spent with either object for all three genotypes (data not shown). On day 5, one object was replaced with a novel object, and the other object was replaced with a replica. Recognition of the familiar object was scored by preferential exploration of the novel object. Although wild-type and heterozygous mice preferentially explored the novel object (+/+ = 75 ± 5% and +/− = 65 ± 6% of the exploration time), the WAVE-1 knockout had no preference for either object (Fig. 4D). Importantly, the WAVE-1 knockout mice spent three times longer than their littermates exploring both objects yet were unable to distinguish between them (Fig. 4E). This emphasizes that the WAVE-1 knockout mice exhibit cognitive deficits and provides further evidence that they have reduced anxiety levels.
Finally, emotional learning and memory was assessed using the passive-avoidance test. On day 1 the mice were placed into a step-through box consisting of a brightly lit area connected by a sliding door to a dark compartment. Mice prefer the dark and quickly entered the dark compartment (Fig. 4F). After entering, the sliding door closed, and the mice received a slight foot shock (0.3 mA, 1 sec). The latency to reenter the dark compartment was measured the next day, and no genotype differences were recorded (Fig. 4F). This suggests that emotional learning and memory are not altered in WAVE-1 knockout mice.
Discussion
Our initial characterization of WAVE-1 homozygous null mice has identified three phenotypic abnormalities. First of all, lower numbers of homozygous null animals were recovered, but it remains to be determined conclusively whether this observation was significant. One contributing factor was that WAVE-1 null pups exhibited a higher-than-normal incidence of postnatal death. In fact, almost one third of the homozygous null progeny died within 24–48 h of birth, although autopsies failed to identify a cause of death. Additional pups may have died in utero, although this theory remains to be investigated. A second phenotypic abnormality was the reduced size of the WAVE-1 mice. Their runted stature was most dramatic around 2–6 weeks after birth. During this period the body weight of WAVE-1 knockout mice approached 60% of their littermates. Runted phenotypes have also been observed in knockout mice lacking WAVE-1-binding partners such as the Abl tyrosine kinase or the RIIβ regulatory subunit of the cAMP-dependent protein kinase (32–34). Thus, interruption of kinase anchoring in brain regions that coordinate weight homeostasis such as the hypothalamus (35) might contribute to the reduced stature of the homozygous null mice. This hypothesis may be tested by interbreeding WAVE-1 knockouts with mice lacking Abl or RIIβ.
Perhaps our most intriguing observations are the range of behavioral abnormalities detected in the WAVE-1 knockout mice. Poor performance in the rotarod, inclined-screen, and balance-beam tests reflect deficits in sensorimotor function that often are indicative of a perturbed cerebellar physiology. In fact, similar behavioral abnormalities have been recorded in the weaver, stager, and lurcher mice, which have documented defects in Purkinje cells (36–39). These cells represent the sole synaptic output from the cerebellum (40). Although our immunohistochemical analyses suggest that Purkinje cell morphology is intact in the knockout mice, it is reasonable to assume that the loss of WAVE-1 abrogates the acquisition of these sensorimotor skills. Likewise, a loss of WAVE-1 from regions of the hippocampus and cortex may underlie the cognitive deficits that were exposed in the Morris water maze. WAVE-1 knockout mice were slower in learning to find the platform during the visible phase of the water-maze trial, although it is likely that their reduced swim speed may have been a contributing factor. However, they consistently failed to locate the submerged platform during the hidden platform sessions, strongly suggesting deficits in spatial learning and memory. A more definitive example of hippocampal-dependent cognitive impairment was provided by the novel-object recognition test in which WAVE-1 null mice spent twice as much time exploring the objects on day 4 but still failed to remember them the next day. Collectively, these findings define a physiological role for WAVE-1 in the facilitation of behavioral traits that are regulated by a variety of brain regions.
The molecular role of WAVE proteins are to provide molecular platforms that assemble protein networks of actin-binding proteins, adapter proteins, signaling enzymes, and the Arp 2/3 complex (10, 12, 16–18). Thus the removal of a core organizational component such as WAVE-1 is likely to impede the assembly of these molecular machines. Interestingly, missence mutations in WASp that cause Wiskott–Aldrich syndrome have been mapped to a protein-docking module that performs an analogous scaffolding function (41). Currently it is unclear whether spatial perturbation of some or all of the WAVE-1-binding partners contribute to the aberrant behavioral phenotypes that we observed. One newly discovered WAVE-1-binding partner is WRP (also called MEGAP or SrGAP-3), a GAP that selectively terminates Rac signaling in neurons (18, 19, 42). Haploinsufficiency of WRP has been linked to 3p syndrome, a severe form of mental retardation in humans with symptoms that include reduced growth, low IQ, atactic gait, and jerky arm movements (19). These symptoms are remarkably similar to the impaired cognitive and sensorimotor functions that we report for the WAVE-1 knockout mice. Thus WAVE-1 may facilitate normal neuronal network connectivity by localizing WRP for its role in the regulation of Rac signaling. Our preliminary analyses suggest that WRP protein levels are unchanged in WAVE-1 knockout mice (S.H.S. and J.D.S., unpublished observation); however, the correct localization of the GAP may be a more important factor. Interestingly, lesions in genes encoding other effectors of Rho-family GTPases have been linked to mental retardation. These include the GAP oligophrenein 1, a guanine nucleotide-exchange factor for Rac or Cdc42 called αPIX, and the Rac/Cdc42 effector kinase Pak3 (43–45). This body of work highlights the importance of Rho-family GTPases in the control of cytoskeletal remodeling events necessary for normal cognitive processes (4). Thus disruption of actin-based signaling scaffolds that contribute to the formation of synaptic connections may interrupt neuronal responses and be a causative factor in certain disease states. Accordingly, WAVE-1 knockout mice may provide an animal model to study the molecular details underpinning cognitive and sensorimotor impairments in humans.
Supplementary Material
Acknowledgments
We thank Chris Bond and Charles Meshul for expert advice and Anthony LeFevour, Justine Curley, and Chris Mayer for excellent technical assistance. This work was supported by National Institutes of Health Grants AG20079 (to J.R.) and DK44239 (to J.D.S.).
Abbreviations
- WASp
Wiskott–Aldrich-syndrome protein
- AKAP
A kinase-anchoring protein
- GAP
GTPase-activating protein
- ES
embryonic stem
References
- 1.Machesky L M, Insall R H. J Cell Biol. 1999;146:267–272. doi: 10.1083/jcb.146.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop A L, Hall A. Biochem J. 2000;348:241–255. [PMC free article] [PubMed] [Google Scholar]
- 3.Bear J E, Krause M, Gertler F B. Curr Opin Cell Biol. 2001;13:158–166. doi: 10.1016/s0955-0674(00)00193-9. [DOI] [PubMed] [Google Scholar]
- 4.Hall A. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 5.Higgs H N, Pollard T D. J Biol Chem. 1999;274:32531–32534. doi: 10.1074/jbc.274.46.32531. [DOI] [PubMed] [Google Scholar]
- 6.Millard T H, Machesky L M. Trends Biochem Sci. 2001;26:198–199. doi: 10.1016/s0968-0004(01)01788-1. [DOI] [PubMed] [Google Scholar]
- 7.Snapper S B, Rosen F S. Annu Rev Immunol. 1999;17:905–929. doi: 10.1146/annurev.immunol.17.1.905. [DOI] [PubMed] [Google Scholar]
- 8.Kim A S, Kakalis L T, Abdul-Manan N, Liu G A, Rosen M K. Nature. 2000;404:151–158. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- 9.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner M W. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 10.Machesky L M, Insall R H. Curr Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 11.Miki H, Suetsugu S, Takenawa T. EMBO J. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miki H, Yamaguchi H, Suetsugu S, Takenawa T. Nature. 2000;408:732–735. doi: 10.1038/35047107. [DOI] [PubMed] [Google Scholar]
- 13.Machesky L M, Mullins R D, Higgs H N, Kaiser D A, Blanchoin L, May R C, Hall M E, Pollard T D. Proc Natl Acad Sci USA. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marchand J B, Kaiser D A, Pollard T D, Higgs H N. Nat Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- 15.Robinson R C, Turbedsky K, Kaiser D A, Marchand J B, Higgs H N, Choe S, Pollard T D. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 16.Westphal R S, Soderling S H, Alto N M, Langeberg L K, Scott J D. EMBO J. 2000;19:4589–4600. doi: 10.1093/emboj/19.17.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eden S, Rohatgi R, Podtelejnikov A V, Mann M, Kirschner M W. Nature. 2002;418:790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- 18.Soderling S H, Binns K L, Wayman G A, Davee S M, Ong S H, Pawson T, Scott J D. Nat Cell Biol. 2002;4:970–975. doi: 10.1038/ncb886. [DOI] [PubMed] [Google Scholar]
- 19.Endris V, Wogatzky B, Leimer U, Bartsch D, Zatyka M, Latif F, Maher E R, Tariverdian G, Kirsch S, Karch D, Rappold G A. Proc Natl Acad Sci USA. 2002;99:11754–11759. doi: 10.1073/pnas.162241099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snapper S B, Rosen F S, Mizoguchi E, Cohen P, Khan W, Liu C H, Hagemann T L, Kwan S P, Ferrini R, Davidson L, Bhan A K, Alt F W. Immunity. 1998;9:81–91. doi: 10.1016/s1074-7613(00)80590-7. [DOI] [PubMed] [Google Scholar]
- 21.Snapper S B, Takeshima F, Anton I, Liu C H, Thomas S M, Nguyen D, Dudley D, Fraser H, Purich D, Lopez-Ilasaca M, et al. Nat Cell Biol. 2001;3:897–904. doi: 10.1038/ncb1001-897. [DOI] [PubMed] [Google Scholar]
- 22.Sossey-Alaoui K, Su G, Malaj E, Roe B, Cowell J K. Oncogene. 2002;21:5967–5974. doi: 10.1038/sj.onc.1205734. [DOI] [PubMed] [Google Scholar]
- 23.Storck T, Kruth U, Kolhekar R, Sprengel R, Seeburg P H. Nucleic Acids Res. 1996;24:4594–4596. doi: 10.1093/nar/24.22.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colledge M, Dean R A, Scott G K, Langeberg L K, Huganir R L, Scott J D. Neuron. 2000;27:107–119. doi: 10.1016/s0896-6273(00)00013-1. [DOI] [PubMed] [Google Scholar]
- 25.Gu G, Rojo A A, Zee M C, Yu J, Simerly R B. J Neurosci. 1996;16:3035–3044. doi: 10.1523/JNEUROSCI.16-09-03035.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raber J, Wong D, Buttini M, Orth M, Bellosta S, Pitas R E, Mahley R W, Mucke L. Proc Natl Acad Sci USA. 1998;95:10914–10919. doi: 10.1073/pnas.95.18.10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raber J, Wong D, Yu G Q, Buttini M, Mahley R W, Pitas R E, Mucke L. Nature. 2000;404:352–354. doi: 10.1038/35006165. [DOI] [PubMed] [Google Scholar]
- 28.Raber J, Bongers G, LeFevour A, Buttini M, Mucke L. J Neurosci. 2002;22:5204–5209. doi: 10.1523/JNEUROSCI.22-12-05204.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davidson R J. Biol Psychiatry. 2002;51:68–80. doi: 10.1016/s0006-3223(01)01328-2. [DOI] [PubMed] [Google Scholar]
- 30.Morris R G, Garrud P, Rawlins J N, O'Keefe J. Nature. 1982;297:681–683. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- 31.Rampon C, Tang Y P, Goodhouse J, Shimizu E, Kyin M, Tsien J Z. Nat Neurosci. 2000;3:238–244. doi: 10.1038/72945. [DOI] [PubMed] [Google Scholar]
- 32.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 33.Schwartzberg P L, Stall A M, Hardin J D, Bowdish K S, Humaran T, Boast S, Harbison M L, Robertson E J, Goff S P. Cell. 1991;65:1165–1175. doi: 10.1016/0092-8674(91)90012-n. [DOI] [PubMed] [Google Scholar]
- 34.Cummings D E, Brandon E P, Planas J V, Motamed K, Idzerda R L, McKnight G S. Nature. 1996;382:622–626. doi: 10.1038/382622a0. [DOI] [PubMed] [Google Scholar]
- 35.Dallman M F, Akana S F, Levin N, Walker C D, Bradbury M J, Suemaru S, Scribner K S. Ann NY Acad Sci. 1994;746:22–31. doi: 10.1111/j.1749-6632.1994.tb39206.x. [DOI] [PubMed] [Google Scholar]
- 36.Lalonde R. Exp Brain Res. 1987;68:417–420. doi: 10.1007/BF00248807. [DOI] [PubMed] [Google Scholar]
- 37.Lalonde R. Exp Brain Res. 1987;65:479–481. doi: 10.1007/BF00236322. [DOI] [PubMed] [Google Scholar]
- 38.Lalonde R. Brain Res. 1994;639:351–353. doi: 10.1016/0006-8993(94)91753-1. [DOI] [PubMed] [Google Scholar]
- 39.Caston J, Vasseur F, Stelz T, Chianale C, Delhaye-Bouchaud N, Mariani J. Brain Res Dev Brain Res. 1995;86:311–316. doi: 10.1016/0165-3806(95)00037-e. [DOI] [PubMed] [Google Scholar]
- 40.De Schutter E, Bower J M. J Neurophysiol. 1994;71:375–400. doi: 10.1152/jn.1994.71.1.375. [DOI] [PubMed] [Google Scholar]
- 41.Volkman B F, Prehoda K E, Scott J A, Peterson F C, Lim W A. Cell. 2002;111:565–576. doi: 10.1016/s0092-8674(02)01076-0. [DOI] [PubMed] [Google Scholar]
- 42.Wong K, Ren X R, Huang Y Z, Xie Y, Liu G, Saito H, Tang H, Wen L, Brady-Kalnay S M, Mei L, et al. Cell. 2001;107:209–221. doi: 10.1016/s0092-8674(01)00530-x. [DOI] [PubMed] [Google Scholar]
- 43.Billuart P, Bienvenu T, Ronce N, des Portes V, Vinet M C, Zemni R, Carrie A, Beldjord C, Kahn A, Moraine C, Chelly J. Pathol Biol. 1998;46:678. [PubMed] [Google Scholar]
- 44.Allen K M, Gleeson J G, Bagrodia S, Partington M W, MacMillan J C, Cerione R A, Mulley J C, Walsh C A. Nat Genet. 1998;20:25–30. doi: 10.1038/1675. [DOI] [PubMed] [Google Scholar]
- 45.Kutsche K, Yntema H, Brandt A, Jantke I, Nothwang H G, Orth U, Boavida M G, David D, Chelly J, Fryns J P, et al. Nat Genet. 2000;26:247–250. doi: 10.1038/80002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.