Abstract
Processing of the amyloid β protein precursor (AβPP) by the β and γ secretases leads to the production of two small peptides, amyloid β and the AβPP intracellular domain (AID, or called elsewhere AICD). Whereas the role of amyloid β in the pathogenesis of Alzheimer's disease has been studied extensively, only recently has information begun to accumulate as to the role of AID. Functions identified for AID include its ability to trigger apoptosis and a role in regulating gene transcription, particularly in combination with the AβPP binding protein Fe65. Here, we report that AID in combination with Janus kinase interacting protein-1 (JIP-1) can activate gene expression. We demonstrate that the mechanism is different from activation in combination with Fe65 by first showing that although Fe65 enters the nucleus in the absence of full-length AβPP, JIP-1 does not. Additionally, JIP-1-induced activation is Tip60 independent, whereas a complex with AID, Fe65, and Tip60 is formed for Fe65-induced activation. Finally, and probably most interestingly, we show that although the AβPP family members APLP1 and APLP2 (for amyloid β precursor-like protein) can cause activation in combination with Fe65, APLP1 and APLP2 show little or no activation in combination with JIP-1. This activity for the AID fragment may help explain the unique functions of AβPP relative to its other family members, and changes in gene expression found in Alzheimer's disease.
The importance of amyloid β protein precursor (AβPP) in Alzheimer's disease (AD) pathology is well established. After cleavage of AβPP by the β and γ secretases, the Aβ peptide is released and accumulates in amyloid plaques in the brains of patients with AD (1, 2). According to the amyloid hypothesis, it is believed that the Aβ in these plaques has a direct role in AD pathology (3). It is important to note, however, that this has been repeatedly questioned in recent years with evidence that Aβ may even have a protective role (4, 5). There has also been much interest recently in another peptide derived from AβPP, the AβPP intracellular domain (AID) fragment, which extends from the γ-secretase cleavage site to the carboxyl terminal of AβPP. The AID peptide was initially overlooked probably because it is very unstable and difficult to detect (6, 7), and it was overshadowed by the Aβ fragment, which has been at the center of AD theory. The AID peptide was first identified in the brains of patients with AD and was shown to either sensitize or induce cells to undergo apoptosis (6).
Work to understand the function of the AID fragment has greatly increased recently. AβPP is processed in a way similar to Notch, which undergoes a regulated intramembranous proteolysis (8) by the γ secretase to release Notch intracellular domain, which modulates transcription (9, 10). This relationship between AβPP and Notch has led to the question of whether AID also might modulate transcription. Indeed, using a reporter gene system, Cao and Sudhof (11) showed that the AID fragment was able to cause transcriptional activation in combination with the AβPP binding protein Fe65 and the acetyltransferase Tip60. This was followed by a report by Gao and Pimplikar (13) that showed that AID fragments were able to cause down-regulation of the AβPP interacting protein PAT1 and were able to repress retinoic acid-responsive gene expression in a reporter gene system. Work by us has shown that AID binds the Notch inhibitors (13) Numb and Numb-like (Nbl) to cause inhibition of Notch-dependent gene activation (14). This would provide an elegant mechanism in which the γ secretase would provide both positive and negative signaling on the same pathway to moderate Notch-dependent gene activation. Finally, it has most recently been shown that upon overexpression of AβPP, Fe65, and Tip60, a repressor complex assembled on the KAI-1 promoter is replaced by an AID, Fe65, Tip60 complex (15). All of these reports suggest a role for AID in gene regulation.
AβPP is part of a larger gene family that includes amyloid β precursor-like protein-1 (APLP1) and APLP2 (16). Research has been focused on AβPP itself because mutations in it are linked to familial AD, and it is the precursor from which the Aβ peptide is derived (1, 2); however, much less is known about the APLPs. There is evidence that the APLPs shed their extracellular domains (17–19) and can be cleaved by caspases within their cytoplasmic domains at the conserved VEVD motif (20, 21); however, the significance of these cleavages is unknown. We have recently reported that the APLPs, like AβPP, are processed by the γ secretase to release fragments that cause transcriptional activation in combination with Fe65 (22).
Many binding partners have been found for AβPP's carboxyl terminal, with most of them binding to the phosphotyrosine binding (PTB) YENPTY motif. This motif, lying between amino acids 682 and 687 (using AβPP 695 numbering), interacts with X11 (23), Fe65 (24), mDab (25), Shc (26), and Numb and Nbl (14). We have also identified another PTB-containing protein, c-Jun N-terminal kinase (JNK) interacting protein-1 (JIP-1), to bind to this motif (27, 28). JIP-1 was initially identified as an inhibitor of JNK activation (29) but was soon shown to be a scaffold protein that bound various components of the JNK cascade including JNK, MKK7, and mixed-lineage kinase (30). It is tempting to speculate that interaction between JIP-1 and AβPP could provide the molecular basis for stress pathway activation in the brains of patients with AD (31–34); however, there is little evidence to support this notion. Here, we report an alternative functional implication of the AβPP–JIP-1 interaction. We show that AβPP and JIP-1 are able to cause reporter gene activation in a γ-secretase cleavage-dependent manner that depends on Presenilin-1. Furthermore, this activation seems to occur by a mechanism different from what is seen with an Fe65–AβPP complex, as evidenced by microscopy and its independence of Tip60 coexpression. We most strikingly show that strong activation occurs only for the combination of JIP-1 and AβPP but not for JIP-APLP1 or JIP-APLP2 (which do cause strong activation in combination with Fe65) (22). Should endogenous target genes for the JIP–AβPP complex be identified, this may provide an additional basis of AD being linked to AβPP but not the APLPs.
Methods
DNA Constructs.
Gal4BD-AβPP and Gal4BD-AβPPCT44 constructs containing the Gal4 binding domain (Gal4BD) (Fig. 1a) as well as Tip60 and the Gal4-dependent luciferase reporter construct G5E1b were obtained from Thomas Sudhof (University of Texas, Southwestern Medical Center, Howard Hughes Medical Institute, Dallas). Mutants were made by incorporating mutations into the primers as described (22). Constructing and obtaining the APLP (22), Shc (26), JIP-1 (28), and Nbl (14) constructs have been described. The full-length JIP-1 construct used was the human JIP-1e described (28) and is referred to as JIP-1 throughout this article. All constructs obtained using PCR were confirmed by sequencing. YFP-Fe65 was obtained from Tommaso Russo (Universita di Napoli Federico II, Naples). Untagged Fe65 was constructed by moving the Fe65 insert from YFP-Fe65 to the pcDNA3. Unless otherwise specified, JIP-1 and Fe65 refer to untagged constructs. PS1 D385A was obtained from Dennis J. Selkoe (Harvard Medical School and Brigham and Women's Hospital, Boston). Constructs used for the binding assays in Fig. 5 have been described (27); however, they are labeled differently in this article to maintain consistency as follows: GST-cyt, GST-ΔNPTY, GST-APLP1cyt, and GST-APLP2-cyt have been renamed GST-AID, GST-AIDΔ, GST-ALID1, and GST-ALID2, respectively.
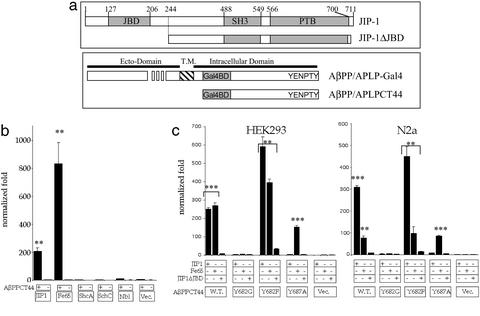
Figure 1.
JIP-1 activates transcription in combination with AID. (a) Schematics of JIP-1 and the AβPP/APLP constructs used. JIP-1 is shown with the location of the JNK binding domain (JBD), SH3 domain, and PTB domain. The construct without the JBD (JIP-1ΔJBD) is also shown. AβPP/APLP constructs with the Gal4 binding domain (Gal4BD) are shown for full-length constructs (AβPP/APLP) and constructs containing the terminal 44 aa (AβPP/APLPCT44). The ecto-domain, transmembrane domain (T.M.), and intracellular domain, along with the location of the conserved YENPTY motif, are also indicated. (b) YFP-tagged proteins (JIP-1, Fe65, etc.) were cotransfected with AβPPCT44 (+) or empty vector pMst (−). Normalized values were compared with YFP (Vec.) cotransfected with pMst yielding a value for normalized fold. (c) In two cell lines (HEK293 and N2a), AβPPCT44 WT mutants or pMst empty vector were cotransfected with untagged JIP-1, Fe65, or JIP-1ΔJBD. Readings normalized for transfection efficiency were compared with JIP-1ΔJBD cotransfected with pMst, yielding a value for normalized fold. Similar experiments with other cell lines including HeLa, Cos-7, and NIH 3T3 yielded similar data but with lower fold activation for both JIP-1 and Fe65. Significance was determined by using a two-tailed Student's t test (**, P < 0.01; ***, P < 0.001).
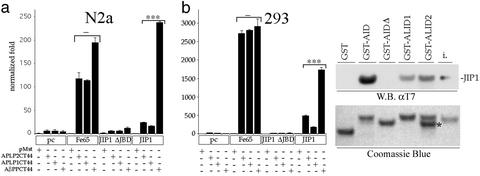
Figure 5.
JIP-1 functions with AβPP but not the APLPs. (a) Gal4BD-tagged ALID2, ALID1, AID (APLP2CT44, APLP1CT44, AβPPCT44, respectively), or empty vector (pMst) were cotransfected into two cell lines (HEK293 or N2a) with empty vector pcDNA3 (pc), Fe65, JIP-1ΔJBD, or JIP-1. Normalization was carried out, and fold activation was calculated relative to the pMst and pcDNA3 sample. Note that although the differences in Fe65 are not significant, the differences in JIP-1 are highly significant. (b) Equal amounts of His/T7-JIP1 were incubated with equal amounts of GST, GST-AID, GST-AIDΔ, GST-ALID1, and GST-ALID2. Samples were washed and subjected to SDS/PAGE. His/T7-JIP1 was detected with T7 antibody. One percent of JIP input is also indicated (i). Coomassie staining was carried out to demonstrate that similar amounts of GST fusion protein were used for each binding assay. A degradation product of ALID2 is indicated (*); however, it did not impact these data because quantification of input protein only included full-length protein, and this degradation product, based on the molecular weight, does not contain the interacting YENPTY motif. Significance was determined by using a two-tailed Student's t test (***, P < 0.001; −, not significant).
Cell Culture and Luciferase Assays.
HEK293T and N2a cells were grown in RPMI 1640 media with 10% heat-inactivated FCS. N2a cells for microscopy were differentiated by growing them in low serum media for 2 days. Compound E (35) was used at a concentration of 3 × 10−8 M, and N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine-t-butyl ester (DAPT) (36) was used at a concentration of 2 × 10−6 M; both were obtained from T. Golde (Mayo Clinic, Jacksonville, FL). For luciferase assays, transfections were done by using Fugene 6 (Roche Biochemicals), with all samples done in 96-well plates in triplicate and being normalized with cotransfected β-galactosidase. For assays in which four DNAs were transfected, the following amounts of DNA were used: 0.03 μg of β-galactosidase; 0.18 μg of G5E1b; 0.20 μg of Gal4BD and AβPP/APLP1/APLP2; 0.20 μg of JIP/Fe65/pcDNA. For assays in which five DNAs were transfected, the following amounts of DNA were used: 0.03 μg of β-galactosidase; 0.15 μg of G5E1b; 0.15 μg of Gal4BD and AβPP/APLP1/APLP2; 0.15 μg of JIP/Fe65/pcDNA; and 0.15 μg of PS1wt/PS1 D385A/Tip60/pcDNA. PS1 knockout cells provided by Jie Shen (Harvard Medical School and Brigham and Women's Hospital, Boston) were used for assays in Fig. 2, with transfections done with Lipofectamine 2000 (Invitrogen) in 24-well plates with all amounts of DNA scaled up by a factor of 5. Cells were lysed after 24–48 h by using reporter lysis 5× buffer (Promega), and luciferase substrate from Promega was used for the assay. Luciferase values were normalized by using a β-galactosidase assay (Tropix, Bedford, MA) following the manufacturer's protocol.
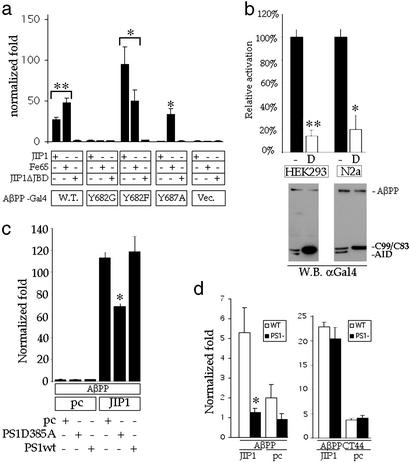
Figure 2.
JIP-1/AβPP transactivation requires a PS1-dependent γ-secretase activity. (a) In HEK293 cells, AβPP or AβPP mutants fused to Gal4 (as indicated in Fig. 1a) or pMst empty vector (Vec.) were cotransfected with untagged JIP-1, Fe65, or JIP-1ΔJBD. Normalization was done as described in Fig. 1c. (b) Experiments similar to those in a were performed, except DAPT (D) or the carrier DMSO (−) was added to the medium 5–7 h after transfection. Data are represented by setting activation in the presence of carrier alone to 100%. Western blotting was done by using an antibody against the Gal4BD. Full-length AβPP-Gal4 (AβPP) and the processed Gal4-tagged C99/C83 and AID are indicated. (c) Gal4-tagged AβPP or pMst empty vector were cotransfected with JIP-1 or pcDNA3 (pc) and pcDNA3, PS1-D385A, or PS1 WT (PS1wt), as described (see Methods). (d) Luciferase experiments were conducted as before with WT and PS1 knockout mouse embryonic fibroblasts being used. (Left) AβPP-Gal4 was used. (Right) AβPPCT44 was used. Significance was determined by using a two-tailed Student's t test (*, P < 0.05; **, P < 0.01). Note that the difference between WT and PS1 KO fibroblasts in the AβPP-Gal4+JIP-1 sample was not significant.
Microscopy.
Cells were transfected by using Fugene 6 such that the ratio of AβPP-GFP/AID-GFP/GFP to FLAG-JIP-1 was 1:1. The FLAG epitope was detected with anti-FLAG mAb (Sigma) and Cy5-conjugated secondary antibody (Jackson ImmunoResearch), whereas AβPP was stained with anti-AβPPCT antibody (Zymed) and Alexa-594 secondary antibody. Nuclei were stained with 4′,6-diamidino-2-phenylindole (Sigma). Images were captured by using a charge-coupled device camera mounted on an Olympus Provis AX70.
Production of Proteins for Binding Assays.
All purification and binding procedures were performed at 4°C, unless otherwise noted. A 2-liter culture of BL21 (DE3) transformed with His/T7-tagged JIP1b 493–707 (His/T7-JIP1) (27) was induced with 0.1 mM isopropyl β-d-thiogalactoside for 5 h at 37°C. The bacteria were collected, washed with PBS, and solubilized by sonication in 20 ml of buffer A (20 mM Tris·HCl, pH 7.4/1 mM EDTA/1 mM DTT/150 mM NaCl) containing 1% (wt/vol) Triton X-100, 2 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM PMSF. The lysates were centrifuged at 20,000 × g for 20 min, and the inclusion bodies in the precipitate were washed four times with buffer A. The inclusion bodies were solubilized with 5 ml of buffer B (20 mM NaPO4, pH 7.4/150 mM NaCl/8 M urea), sonicated, and cleared at 20,000 × g for 20 min. The cleared urea extract was diluted with 5 ml of buffer C (20 mM NaPO4, pH 7.4/150 mM NaCl/10 mM imidazole) and was injected into 400 ml of buffer D (100 mM Tris·HCl, pH 8.0/400 mM L-arginine/10 mM DTT/0.2 mM PMSF) to refold the denatured protein. The refolded protein was dialyzed against 4 liters of water, cleared by centrifugation at 20,000 × g for 20 min, filtrated through a 0.2-μm filter to remove the precipitants, and applied to 0.4 ml of Ni-CAM HC resin equilibrated with 8 ml of buffer C. The bound protein was washed with 4 ml of buffer C and eluted with 2.5 ml of buffer C containing 500 mM imidazole. The eluate was dialyzed three times against 250 ml of buffer A and stored on ice until use. GST, GST-AID, GST-AIDΔ, GST-ALID1, and GST-ALID2 (27) were expressed in BL21, purified on a glutathione Sepharose column, and dialyzed three times against buffer E (20 mM Tris·HCl, pH 8.0/1 mM EDTA/1 mM DTT). Proteins were stored at −80°C until use.
Binding Experiments.
Six micrograms of GST proteins was immobilized on 30 μl of glutathione Sepharose beads in 500 μl of buffer E. The beads were washed once with buffer A containing 0.1% (wt/vol) Triton X-100 and mixed with 0.4 μM His/T7-JIP1 in buffer A containing 0.1% (wt/vol) Triton X-100. After 2 h of incubation, the beads were washed three times with the same buffer and then boiled in 30 μl of 2× SDS sample buffer. Ten microliters of each was subjected to Western blotting with T7 antibody (Novagen) to detect the amount of bound JIP-1 and Coomassie brilliant blue staining was used to confirm that equal amounts of GST fusion proteins were used. Quantification was carried out by using nih image.
Results
It has previously been shown that Fe65 and the AβPP intracellular domain combine to form a transcriptionally active complex that can activate gene expression (11). We have previously identified JIP-1 (27, 28), ShcA, ShcC (26), and Numb and Nbl (14) as AβPP binding proteins, and we questioned whether any of these proteins may also function in combination with AβPP to activate gene expression. Using the reporter system used by Cao and Sudhof (11), we cotransfected each AβPP interacting protein along with the carboxyl terminus of AβPP fused to the Gal4BD (AβPPCT44, see Fig. 1a) and the luciferase reporter construct into HEK 293 cells. Fig. 1b shows that although Fe65 and JIP-1 show gene activation, ShcA, ShcC, Nbl, and pcDNA3 do not. Thus, in addition to Fe65, which has previously been identified to bind AID and cause transcriptional activation, we identified JIP-1 to cause gene activation.
We next identified the domains of JIP-1 necessary for gene activation. To do this, we used the JIP-1 PTB domain, JIP-1 SH3 domain, JIP-1 SH3+PTB, and JIP-1 without the JNK binding domain (JIP-1ΔJBD) as well as full-length JIP-1 to carry out the same assay. We found that only full-length JIP-1 is able to cause gene activation, and even the loss of the JNK binding domain alone was able to abolish gene activation (data not shown). We also analyzed AβPP by mutagenesis to show that its binding to JIP-1 was responsible for activation. Point mutations in AβPPCT44 (Fig. 1a) were made where the first tyrosine of the conserved YENPTY motif was mutated to glycine or phenylalanine (Y682G and Y682F, respectively), or the last tyrosine of the motif was mutated to alanine (Y687A). Fig. 1c shows that for WT AβPPCT44 and the Y682F, which bind JIP-1 (and Fe65), there is activation, whereas for Y682G, which does not bind JIP-1 (or Fe65), there is no activation. Additionally, Y687A, which does not bind JIP-1, causes no activation when cotransfected with JIP-1, whereas Fe65, which does bind Y687A (27), is able to cause activation. It is also interesting to note from Fig. 1d that the magnitude of JIP-1 and Fe65's fold activation depends on the cell type, such that, for example, in 293 cells the fold activation is approximately equal, whereas, in N2a cells, JIP-1 is able to cause a much greater fold activation.
We next wanted to ascertain whether the processing of AβPP, with the resulting release of AID, could regulate this transcriptional activation. Full-length constructs of AβPP and AβPP mutants, each with a Gal4BD tag inserted (Fig. 1a), were cotransfected with JIP-1. Fig. 2a shows that the constructs that bind JIP-1 (WT and Y682F) were able to cause gene activation, whereas the ones that do not bind (Y682G and Y687A) do not activate either. We then carried out a similar assay with the AβPP-Gal4BD WT construct, but this time treated the cells with either the γ-secretase inhibitor DAPT (36) or carrier alone (DMSO). Fig. 2b shows that DAPT is able to decrease activation by ≈80%, presumably by inhibiting release of the Gal4BD-tagged fragment at AβPP's carboxyl terminal. Western blotting was used to confirm that DAPT was actually inhibiting AβPP-Gal4BD processing at the concentration used (Fig. 2b). Similar results were obtained with another γ-secretase inhibitor, compound E (35) (data not shown). Finally, to determine whether the γ-secretase processing of AβPP, which causes gene activation in combination with JIP-1, is Presenilin dependent, we performed the assay in the presence of the dominant negative PS1 mutant D385A. PS1 containing this aspartate mutation has been shown to replace endogenous Presenilins and abolish Presenilin-dependent γ-secretase activity (37, 38). Fig. 2c shows that the D385A mutant but not WT PS1 is able to depress gene activation by ≈40%. The partial inhibition can be explained by PS1 D385A not being stably transfected into the cells (which would cause replacement of endogenous Presenilins) and nonuniform transfection into the cells of the PS1 D385A DNA with the other DNAs. To obtain a more interpretable result, we performed the assay in PS1 knockout fibroblast. Fig. 2d shows that there is an ≈80% decrease in activation in the PS1 knockout fibroblast compared with WT control fibroblast (similar to what was seen with DAPT above). To validate this inhibition data, we confirmed that these PS1 knockout cells were capable of undergoing AID-induced reporter gene activation in combination with JIP-1 when the need for γ-secretase processing was removed (i.e., AβPPCT44 cotransfected with JIP-1). In Fig. 2d, AβPPCT44 cotransfected with JIP-1 shows equivalent activation in WT and PS1 knockout cells. These data indicate that AβPP processing by the γ secretase in a Presenilin-dependent manner is required for JIP-1-dependent reporter gene activation by AID.
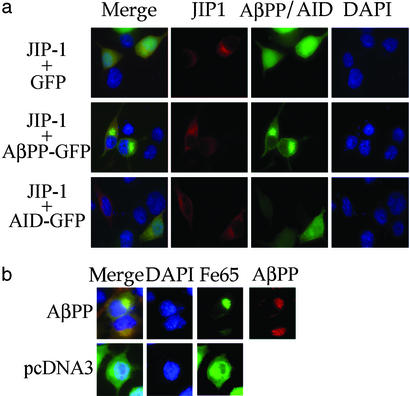
It has been reported by multiple groups that AβPP is able to restrain Fe65 from entering the nucleus and that AID in combination with Fe65 moves into the nucleus presumably to cause gene expression (11, 22, 39). To determine whether a similar phenomenon happens with JIP-1, we cotransfected Flag-tagged JIP-1 along with AβPP-GFP, AID-GFP, or GFP, and JIP-1 was visualized by immunocytochemistry. Fig. 3a shows similar staining for JIP-1 in all samples, with JIP-1 being localized diffusely in the cytoplasm and being mostly excluded from the nucleus. This finding contrasted sharply with Fe65, where AβPP is able to exclude Fe65 from the nucleus, and in AβPP's absence, Fe65 can move freely between the nucleus and cytoplasm with no nuclear boundary being evident from the Fe65 localization (Fig. 3b; see also ref. 22). These data indicate that either the action of JIP-1+AID does not require JIP-1 translocation to the nucleus or there is in fact translocation of JIP-1 also; however, because of its instability or small quantity, it is not significantly higher than the background staining of JIP-1 in the nucleus.
Figure 3.
JIP-1 does not accumulate in the nucleus in the absence of AβPP. (a) Flag-tagged JIP-1 was cotransfected with GFP, AβPP-GFP, or AID-GFP. The Flag epitope was located by immunostaining with anti-Flag primary antibody and Cy5 secondary antibody. Nuclei were stained with 4′,6-diamidino-2-phenylindole. Merged images along with the individual channels are presented. Note that the amount of JIP-1 located in the nucleus is nearly the same regardless of which GFP construct is cotransfected. (b) GFP-Fe65 was cotransfected with either full-length AβPP or pcDNA3. AβPP was immunostained with anti-AβPPCT primary antibody and Alexa-594 secondary antibody. Nuclei were stained with 4′,6-diamidino-2-phenylindole. (Magnifications: ×60.)
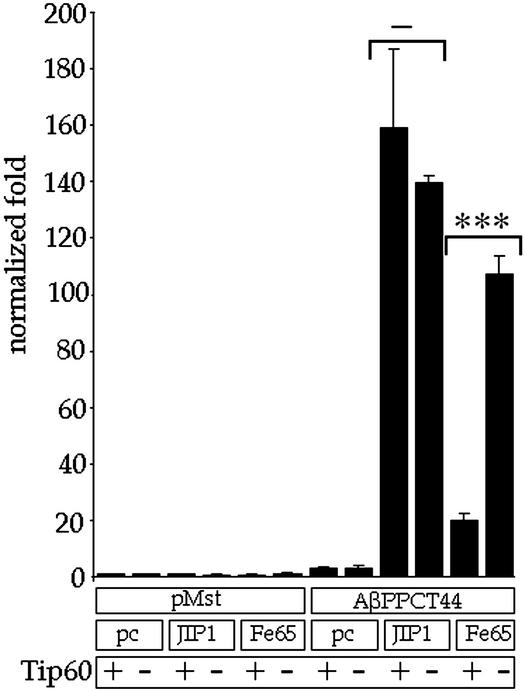
Two reports have indicated that the AβPP + Fe65 complex also contains Tip60 (11, 15). Tip60 is found in a large multimolecular complex (40), which possesses many activities including DNA binding and a histone acetyltransferase. Cao and Sudhof (11) have shown that Tip60 binds to the first PTB domain of Fe65 and Tip60-Gal4BD cotransfected with AβPP, and Fe65 was able to cause Gal4-dependent reporter gene activation. To test the effects of Tip60 on our JIP-1+AID system, we cotransfected JIP-1 or Fe65 with AβPPCT44 and a Gal4-dependent reporter gene with or without Tip60. Luciferase assays revealed (Fig. 4) that although coexpression of Tip60 had little effect on JIP-1-dependent gene activation, it repressed Fe65-dependent activation by >80%. Data were repeated in triplicate three times, and we obtained similar results with four other cell lines. Our data indicate that Tip60 is involved in reporter gene activation by Fe65+AID but not JIP-1+AID.
Figure 4.
Tip60 is not involved in AβPP/JIP-1 transcription. AβPPCT44 or empty vector (pMst) was cotransfected with pcDNA3 (pc), JIP-1, or Fe65. Each of these combinations was also cotransfected with either Tip60 (+) or empty vector pcDNA3 (−). Normalization was done with fold activation being calculated relative to the cotransfection of pMst and pcDNA3. Significance was determined by using a two-tailed Student's t test (***, P < 0.001; −, not significant).
We have previously shown that not only Gal4BD-AID but also the carboxyl-terminal tails of APLP1 and APLP2, termed ALID1 and ALID2 (for AβPP-like intracellular domain), fused to the Gal4BD are able to cause robust reporter gene activation in combination with Fe65 (22). We therefore questioned whether, using the same system, JIP-1 also was able to cause activation in combination with APLP1 or APLP2. Fig. 5a shows that although with Fe65 there is strong activation for all three AβPP family members, for JIP-1 there is only strong activation for AβPP, particularly in neuroblastoma-derived N2a cells. There are two possibilities as to why there is very low activation with the APLPs. First, it could be that the APLPs do not bind JIP-1 as well as AβPP. This would lead to decreased or no activation as was seen with the AβPP/AID YENPTY motif mutants, where the nonbinders did not cause gene activation. Alternatively, although the APLPs bind JIP-1 well, they have less activating ability than AβPP because of properties of their primary or secondary structure. Binding assays were performed where equal amounts of JIP-1 was allowed to bind equal amounts of purified AID, ALID1, or ALID2. Fig. 5b shows that ALID1 binds only 28% and ALID2 binds only 46% the amount of JIP-1 that AID binds. These data indicate that only AβPP, and not the APLPs, participates in transcriptional activation mediated by JIP-1 because of JIP-1's ability to bind more strongly to AβPP/AID.
Discussion
In this article, we show that coexpression of JIP-1 and AβPP/AID is able to cause reporter gene activation, which requires binding between AβPP/AID and JIP-1. Although the same system has been used to show that Fe65 and AβPP/AID form a complex that causes transcriptional activation (11), we use three lines of evidence to show that the mechanism for JIP-1 is different. First, there is no relocation of JIP-1 to the nucleus. Second, Tip60 does not seem to modulate JIP-1+AID gene activation as it does for Fe65+AID. Last, the APLPs show little or no ability to activate transcription in combination with JIP-1.
Regarding the subcellular localization of JIP-1 and Fe65, we have found that although AβPP overexpression is needed to restrict Fe65 to the cytoplasm (22), JIP-1 remains mostly outside the nucleus regardless of the presence or absence of AβPP (Fig. 3). This finding was surprising because as seen by microscopy AID enters the nucleus (Fig. 3) and AID's site of action is in the nucleus because the Gal4BD binds the Gal4 binding element of the reporter gene. We have also previously shown that AβPP binds JIP-1 in vitro by GST pull-down, in cell lysates by immunoprecipitation, and in living cells by fluorescence resonance energy transfer (27, 28). These observations suggest that in vivo JIP-1 and AID interact predominantly in the cytosol and not in the nucleus. This finding would suggest that the mechanism of activation might involve modification of AID or an AID binding partner, whereas AID is bound to JIP-1 in the cytoplasm and then has AID move into the nucleus without JIP-1. An attractive candidate for causing this modification is JNK. In fact, in our mapping of the JIP-1 domains necessary for gene activation, we found that deletion of the JNK binding domain (JIP-1ΔJBD, Fig. 1a) abolished gene activation (Figs. 1c, 2a, and 5). It is important to note an alternative possibility as to why we did not find JIP-1 translocation to the nucleus. Small amounts of the AID+JIP-1 complex may be sufficient to cause activation, and this small amount may not have been detectable over background staining. Indeed, for Notch also, although it was known that a Notch fragment functioned within the nucleus (41), it was only later shown by microscopy that Notch intracellular domain translocated to the nucleus (42).
We have also found that although AID+Fe65 gene activation involves Tip60 (11, 15), AID+JIP-1 activation seems to be Tip60 independent. An additional surprise in our data was that although in the report by Cao and Sudhof (11) they used Tip60 in combination with Fe65 to enhance gene activation, we detected depression. Considering that Tip60 is part of a multimolecular complex, the difference can perhaps be explained as follows. In the experiment of Cao and Sudhof (11), where Tip60 was fused to the Gal4BD, the overexpressed Tip60 is directly recruited together with AID and Fe65 to the Gal4 binding element on the reporter gene and enhances transcription. In our experiment, however, where we used untagged Tip60, most of the overexpressed Tip60 is not recruited directly to the reporter gene; rather, other components of the Tip60 multimolecular complex are distributed between the large number of free Tip60 molecules, and transcription at the reporter gene is inhibited.
Finally, we found that only JIP-1 along with AID can cause gene activation, whereas little or no gene activation was found in combination with ALID1 or ALID2. This is potentially relevant to both the physiologic role of AβPP and the pathologic role of AβPP in AD. First, regarding the physiologic role of AβPP, work using knockout mice has revealed that disruption of AβPP, APLP1, or APLP2 individually causes minor abnormalities that are distinct for the different AβPP family members (43, 44). Additionally, although mice with both AβPP and APLP1 disrupted are normal, AβPP plus APLP2 or APLP1 plus APLP2 double knockout mice die soon after birth (44). These differences between the different AβPP family members indicate that there are overlapping and nonoverlapping functions (at least during development) for the different family members. The ability of AβPP/AID but not the APLPs/ALIDs to cause activation with JIP may be one of the modulators of the nonredundant functions carried out by AβPP and not the APLPs. Second, AD is characterized as being a disease of processing and being linked to AβPP but not the APLPs. The Aβ fragment fits these criteria. There is an Aβ fragment generated from AβPP with no equivalent Aβ-like fragments from the APLPs, and Aβ is produced as a result of processing. Considering that the AID fragment is produced by processing and its ability to induce gene activation with JIP-1 is not shared with the ALIDs, it is possible that the functional consequences of the AID/JIP-1 interaction, including its transcription-modulating properties, may also be important in the pathology of AD. Understanding more fully the mechanism of AID+JIP-1 signal transduction and discovery of the target genes involved will help provide directions in AD research and clues to the complex pathways involved in AD.
Acknowledgments
We thank Dr. T. Sudhof, Dr. T. Russo, Dr. D. J. Selkoe, Dr. J. Shen, and Dr. T. Golde for materials provided. For technical assistance we thank Y. M. Zhao and for helpful advice and discussions we thank the members of the D'Adamio Laboratory. We also express our sincere appreciation to Dr. T. Dragic for extensive usage of equipment. This work was supported by an Irene Diamond Foundation grant (to L.D.) and Medical Scientist Training Program Grant T32GM07288 (to M.H.S.).
Abbreviations
- AβPP
amyloid β protein precursor
- AID
AβPP intracellular domain
- AD
Alzheimer's disease
- PTB
phosphotyrosine binding
- APLP
amyloid β precursor-like protein
- JNK
c-Jun N-terminal kinase
- JIP-1
JNK interacting protein-1
- DAPT
N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine-t-butyl ester
References
- 1.Sisodia S S, St. George-Hyslop P H. Nat Rev Neurosci. 2002;3:281–290. doi: 10.1038/nrn785. [DOI] [PubMed] [Google Scholar]
- 2.Koo E H. Traffic. 2002;3:763–770. doi: 10.1034/j.1600-0854.2002.31101.x. [DOI] [PubMed] [Google Scholar]
- 3.Hardy J, Selkoe D J. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 4.Obrenovich M E, Joseph J A, Atwood C S, Perry G, Smith M A. Neurobiol Aging. 2002;23:1097–1099. doi: 10.1016/s0197-4580(02)00038-6. [DOI] [PubMed] [Google Scholar]
- 5.Smith M A, Casadesus G, Joseph J A, Perry G. Free Radical Biol Med. 2002;33:1194–1199. doi: 10.1016/s0891-5849(02)01021-3. [DOI] [PubMed] [Google Scholar]
- 6.Passer B, Pellegrini L, Russo C, Siegel R M, Lenardo M J, Schettini G, Bachmann M, Tabaton M, D'Adamio L. J Alzheimer's Dis. 2000;2:289–301. doi: 10.3233/jad-2000-23-408. [DOI] [PubMed] [Google Scholar]
- 7.Cupers P, Orlans I, Craessaerts K, Annaert W, De Strooper B. J Neurochem. 2001;78:1168–1178. doi: 10.1046/j.1471-4159.2001.00516.x. [DOI] [PubMed] [Google Scholar]
- 8.Brown M S, Ye J, Rawson R B, Goldstein J L. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 9.Artavanis-Tsakonas S, Rand M, Lake R J. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 10.Kopan R, Goate A. Neuron. 2002;33:321–324. doi: 10.1016/s0896-6273(02)00585-8. [DOI] [PubMed] [Google Scholar]
- 11.Cao X, Sudhof T C. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 12.Gao Y, Pimplikar S W. Proc Natl Acad Sci USA. 2001;98:14979–14984. doi: 10.1073/pnas.261463298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spana E P, Doe C Q. Neuron. 1996;17:21–26. doi: 10.1016/s0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 14.Roncarati R, Sestan N, Scheinfeld M H, Berechid B E, Lopez P A, McGlade J C, Meucci O, Rakic P, D'Adamio L. Proc Natl Acad Sci USA. 2002;99:7102–7707. doi: 10.1073/pnas.102192599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baek S H, Ohgi K A, Rose D W, Koo E H, Glass C K, Rosenfeld M G. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- 16.Bayer T A, Cappai R, Masters C L, Beyreuther K, Multhaup G. Mol Psychiatry. 1999;4:524–528. doi: 10.1038/sj.mp.4000552. [DOI] [PubMed] [Google Scholar]
- 17.Paliga K, Peraus G, Kreger S, Durrwang U, Hesse L, Multhaup G, Masters C L, Beyreuther K, Weidemann A. Eur J Biochem. 1997;250:354–363. doi: 10.1111/j.1432-1033.1997.0354a.x. [DOI] [PubMed] [Google Scholar]
- 18.Xu K P, Zoukhri D, Zieske J D, Dartt D A, Sergheraert C, Loing E, Yu F S. Am J Physiol. 2001;281:C603–C614. doi: 10.1152/ajpcell.2001.281.2.C603. [DOI] [PubMed] [Google Scholar]
- 19.Lo A C, Thinakaran G, Slunt H H, Sisodia S S. J Biol Chem. 1995;270:12641–12645. doi: 10.1074/jbc.270.21.12641. [DOI] [PubMed] [Google Scholar]
- 20.Galvan V, Chen S, Lu D, Logvinova A, Goldsmith P, Koo E H, Bredesen D E. J Neurochem. 2002;82:283–294. doi: 10.1046/j.1471-4159.2002.00970.x. [DOI] [PubMed] [Google Scholar]
- 21.Cowan C M, Thai J, Krajewski S, Reed J C, Nicholson D W, Kaufmann S H, Roskams A J. J Neurosci. 2001;21:7099–7109. doi: 10.1523/JNEUROSCI.21-18-07099.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheinfeld M H, Ghersi E, Laky K, Fowlkes B J, D'Adamio L. J Biol Chem. 2002;277:44195–44201. doi: 10.1074/jbc.M208110200. [DOI] [PubMed] [Google Scholar]
- 23.Borg J P, Ooi J, Levy E, Margolis B. Mol Cell Biol. 1996;16:6229–6241. doi: 10.1128/mcb.16.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiore F, Zambrano N, Minopoli G, Donini V, Duilio A, Russo T. J Biol Chem. 1995;270:30853–30856. doi: 10.1074/jbc.270.52.30853. [DOI] [PubMed] [Google Scholar]
- 25.Trommsdorff M, Borg J P, Margolis B, Herz J. J Biol Chem. 1998;273:33556–33560. doi: 10.1074/jbc.273.50.33556. [DOI] [PubMed] [Google Scholar]
- 26.Tarr P E, Roncarati R, Pelicci G, Pelicci P G, D'Adamio L. J Biol Chem. 2002;277:16798–16804. doi: 10.1074/jbc.M110286200. [DOI] [PubMed] [Google Scholar]
- 27.Matsuda S, Yasukawa T, Humma Y, Ito Y, Niikura T, Hiraki T, Hirai S, Ohno S, Kita Y, Kawasumi M, et al. J Neurosci. 2001;21:6597–6607. doi: 10.1523/JNEUROSCI.21-17-06597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheinfeld M H, Roncarati R, Vito P, Lopez P A, Abdallah M, D'Adamio L. J Biol Chem. 2002;277:3767–3775. doi: 10.1074/jbc.M108357200. [DOI] [PubMed] [Google Scholar]
- 29.Dickens M, Rogers J S, Cavanagh J, Raitano A, Xia Z, Halpern J R, Greenberg M E, Sawyers C L, Davis R J. Science. 1997;277:693–696. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- 30.Whitmarsh A J, Cavanagh J, Tournier C, Yasuda J, Davis R J. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 31.Zhu X, Raina A K, Rottkamp C A, Aliev G, Perry G, Boux H, Smith M A. J Neurochem. 2001;76:435–441. doi: 10.1046/j.1471-4159.2001.00046.x. [DOI] [PubMed] [Google Scholar]
- 32.Shoji M, Iwakami N, Takeuchi S, Waragai M, Suzuki M, Kanazawa I, Lippa C F, Ono S, Okazawa H. Mol Brain Res. 2000;85:221–233. doi: 10.1016/s0169-328x(00)00245-x. [DOI] [PubMed] [Google Scholar]
- 33.Reynolds C H, Utton M A, Gibb G M, Yates A, Anderton B H. J Neurochem. 1997;68:1736–1744. doi: 10.1046/j.1471-4159.1997.68041736.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhu X, Castellani R J, Takeda A, Nunomura A, Atwood C S, Perry G, Smith M A. Mech Aging Dev. 2001;123:39–46. doi: 10.1016/s0047-6374(01)00342-6. [DOI] [PubMed] [Google Scholar]
- 35.Seiffert D, Bradley J D, Rominger C M, Rominger D H, Yang F, Meredith J E, Jr, Wang Q, Roach A H, Thompson L A, Spitz S M, et al. J Biol Chem. 2000;275:34086–34091. doi: 10.1074/jbc.M005430200. [DOI] [PubMed] [Google Scholar]
- 36.Dovey H F, John V, Anderson J P, Chen L Z, de Saint Andrieu P, Fang L Y, Freedman S B, Folmer B, Goldbach E, Holsztynska E J, et al. J Neurochem. 2001;76:173–181. doi: 10.1046/j.1471-4159.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- 37.Wolfe M S, Xia W, Ostaszewski B L, Diehl T S, Kimberly W T, Selkoe D J. Nature. 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 38.Kim S H, Leem J Y, Lah J J, Slunt H H, Levey A I, Thinakaran G, Sisodia S S. J Biol Chem. 2001;276:43343–43350. doi: 10.1074/jbc.M108245200. [DOI] [PubMed] [Google Scholar]
- 39.Minopoli G, de Candia P, Bonetti A, Faraonio R, Zambrano N, Russo T. J Biol Chem. 2001;276:6545–6550. doi: 10.1074/jbc.M007340200. [DOI] [PubMed] [Google Scholar]
- 40.Ikura T, Ogryzko V V, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 41.Schroeter E H, Kisslinger J A, Kopan R. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 42.Sestan N, Artavanis-Tsakonas S, Rakic P. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- 43.von Koch C S, Zheng H, Chen H, Trumbauer M, Thinakaran G, van der Ploeg L H, Price D L, Sisodia S S. Neurobiol Aging. 1997;18:661–669. doi: 10.1016/s0197-4580(97)00151-6. [DOI] [PubMed] [Google Scholar]
- 44.Heber S, Herms S, Gajic V, Hainfellner J, Aguzzi A, Rulicke T, von Kretzschmar H, von Koch C, Sisodia S, Tremml P, et al. J Neurosci. 2000;20:7951–7963. doi: 10.1523/JNEUROSCI.20-21-07951.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]