Abstract
Previously we identified MIR16 (membrane interacting protein of RGS16) as an integral membrane glycoprotein that interacts with regulator of G protein signaling proteins and shares significant sequence homology with bacterial glycerophosphodiester phosphodiesterases (GDEs), suggesting that it is a putative mammalian GDE. Here we show that MIR16 belongs to a large, evolutionarily conserved family of GDEs with a characteristic putative catalytic domain that shares a common motif (amino acids 92–116) with the catalytic domains of mammalian phosphoinositide phospholipases C. Expression of wild-type MIR16 (renamed GDE1), but not two catalytic domain mutants (E97A/D99A and H112A), leads to a dramatic increase in glycerophosphoinositol phosphodiesterase (GPI-PDE) activity in HEK 293T cells. Analysis of substrate specificity shows that GDE1/MIR16 selectively hydrolyzes GPI over glycerophosphocholine. The GPI-PDE activity of GDE1/MIR16 expressed in HEK 293T cells can be regulated by stimulation of G protein-coupled, α/β-adrenergic, and lysophospholipid receptors. Membrane topology studies suggest a model in which the catalytic GDE domain faces the lumen/extracellular space and the C terminus faces the cytoplasm. Our results suggest that by serving as a PDE for GPI with its activity regulated by G protein signaling, GDE1/MIR16 provides a link between phosphoinositide metabolism and G protein signal transduction.
Keywords: phosphoinositide‖RGS proteins‖phospholipid metabolism‖GDE domain
The glycerophospholipids, phosphatidylinositol, phosphatidylcholine, phosphatidylethanolamine, and phosphatidlylserine, are the major lipids of biological membranes. Of these, phosphatidylinositol (PtdIns) and its derivatives have taken on increasing importance based on their evolving roles as regulatory molecules in signal transduction and membrane trafficking during protein secretion, endocytosis, and cytoskeleton organization (1–4). Therefore, their metabolism and fate are of increasing interest. It is known that PtdIns, as well as other glycerophospholipids, can be deacylated to water-soluble glycerophosphodiesters (GPs) sequentially by phospholipase A and lysophospholipase or by phospholipase B alone (2, 5, and 6) and that GPs can be further hydrolyzed to sn-glycerol 3-phosphate and the corresponding alcohols by GP phosphodiesterases (GDEs). GDE activity toward glycerophosphoinositol (GPI), glycerophosphocholine (GPC), and glycerophosphoethanolamine (GPE) has been found in many mammalian tissues (7–11). Of these activities, glycerophosphoinositol phosphodiesterase (GPI-PDE) is of particular interest because of its participation in phosphoinositide (PI) metabolism.
Recently we identified MIR16 (membrane interacting protein of RGS16) and showed that it binds to RGS16, as well as several other regulator of G protein signaling (RGS) proteins that serve as GTPase activating proteins (GAPs) for trimeric G proteins (12). We further showed that rat MIR16 is an integral membrane glycoprotein and shares significant sequence homology with bacterial GDEs (12), suggesting that it may represent a mammalian GDE.
To further understand the biological functions of MIR16, we investigated its GDE activity and its regulation. Here we report that MIR16 belongs to an evolutionarily conserved family of GDE proteins, that it is a phosphodiesterase (PDE) for GPI, and that its activity can be modulated by stimulation of G protein-coupled receptors (GPCRs). Our characterization of MIR16 as a GPI-PDE should facilitate understanding of the biological activities and turnover of GPI and other water-soluble glycerophospholipids.
Materials and Methods
Cell Culture and Transfection.
HEK 293T, Swiss 3T3, and LβT2 cells (from Pamela Mellon, University of California at San Diego) were cultured in DMEM high glucose containing 10% FCS, 100 units/ml penicillin G, and 100 μg/ml streptomycin sulfate. Culture media were purchased from GIBCO/BRL. HEK 293T cells were transfected as described (13).
Preparation of [3H]Inositol-Labeled GPI.
To monitor PDE activity toward GPI, [3H]GPI was prepared by HPLC purification of deacylated [3H]inositol-labeled cellular lipids. Swiss 3T3 fibroblasts were labeled with myo-[3H]inositol (21 Ci/mmol; 1 Ci = 37 GBq; New England Nuclear) for ≈36 h in M199 medium (GIBCO) supplemented with glutamine, penicillin, streptomycin, and 2% calf serum, after which cold (−20°C) methanol was added and [3H]inositol-labeled lipids were extracted through a two-phase acid extraction procedure [chloroform/methanol/H2O/12.1 M HCl at 1.0/1.0/0.6/0.013 (vol/vol)]. After separation and drying of the lower organic phase, lipids were deacylated using methylamine reagent [40% aqueous methylamine/H2O/n-butyl alcohol/methanol at 36/8/9/47 (vol/vol)] as described (14). The resultant aqueous compounds were extracted with n-butyl alcohol/petroleum ether/ethyl formate [20/4/1 (vol/vol); ref. 14], and the [3H]GPI was separated from other aqueous [3H]inositol-labeled compounds by Partisil 10 SAX HPLC (Jones Chromatography, Mid Glamorgan, U.K.) as described below. To monitor the PDE activity toward GPC, [3H]phosphatidylcholine (1,2-dipalmitoyl-l-3-phosphatidylcholine; 81 Ci/mmol; Amersham Pharmacia) was deacylated and extracted by the procedure described above for [3H]GPI without further purification. The [3H]GPI and [3H]GPC stocks were analyzed by HPLC just before use and were routinely ≈95–98% radiochemically pure.
GPI-PDE Activity Assays.
HEK 293T cells were transiently transfected with pCDNA3-MIR16. Forty-eight hours after transfection and after stimulation of the cells where relevant, postnuclear supernatants (PNS) were prepared (15) and used in the enzyme assays. Incubations were routinely carried out in a final volume of 50 μl, which included (unless otherwise indicated) 100 mM Tris/HCl (pH 7.5), 10 mM MgCl2, 2 mg/ml fatty acid free BSA, ≈30,000 dpm [3H]GPI/GPC, 1 mM unlabeled GPI/GPC, and 2 μg of PNS protein, without or with the addition of competing GPs. The incubations (37°C for 30 min, unless otherwise stated) were terminated by addition of cold methanol (−20°C), followed by a two-phase extraction (as above for cell lipid extractions, without acid) and lyophilization of the resultant upper aqueous phase. GPI- and GPC-PDE activities were calculated by a combination of the known cold GPI/GPC in each assay (pmol) and the level of postincubation GPI/GPC hydrolysis seen by HPLC analysis of the 3H-labeled inositol/choline and GPI/GPC (see below; converting % GPI/GPC hydrolyzed to pmol hydrolyzed per min per mg protein). For the GP competition assays, an estimate of the Kapp of each competing compound was calculated knowing the concentration of GPI (1 mM) and the competing GP (10 mM) and the level of inhibition of GPI-PDE activity, and assuming competitive Michaelis–Menten kinetics (without knowing whether this represents an alternative substrate or inhibition) and the dose ratio principle ([GPI]GP/[GPI]0 = [GP] × KappGP), following an extrapolation according to the GPI dose–response curve.
HPLC Separation of [3H]Inositol/Choline-Labeled Compounds.
For the routine anion-exchange HPLC analysis of [3H]GPI and its water-soluble metabolites, a standard Partisil 10 SAX HPLC column elution system and an on-line flow detector (FLO ONE A-525, Packard) were used, with a linear gradient of 0–6.4% buffer B (1.0 M ammonium phosphate/phosphoric acid, pH 3.35), a flow rate of 1.0 ml/min, and a sample size of 1 ml (in water). [3H]inositol-labeled HPLC standards were obtained from New England Nuclear.
For routine anion-exchange HPLC analysis of [3H]GPC and its water-soluble metabolites, an 8.3-cm-long, 3-μm silica column (Perkin–Elmer) elution system and an on-line flow detector (FLO ONE A-525) were used, with a nonlinear, acetonitrile-based gradient of acetonitrile/H2O/100% ethanol/glacial acetic acid/1.0 M ammonium acetate/0.1 M NaH2PO4.H2O [800/127/68/2/3/10 (vol/vol) for buffer A and 400/400/68/44/88/10 (vol/vol) for buffer B; modified from ref. 16]. A flow rate of 0.8 ml/min and a sample size of 300 μl (in buffer A) were used, with the following gradient (% buffer B): 0–6 min, 0%; 6–21 min, 0–20%; 21–36 min, 20–100%. Individual and mixed [3H]choline-labeled HPLC standards were produced from the [3H]phosphatidylcholine stock with (i) bee venom PLA2 (Sigma) in 25 mM Hepes (pH 7.5), 125 mM KCl, and 1 mM Ca2+ for lysophosphatidylcholine (LPC); (ii) deacylation (as described above) for GPC; and (iii) 50% HCl at 95°C for choline phosphate and choline (also from GPC and LPC). The four main standard peaks observed corresponded to those reported previously (16).
Mutagenesis.
The QuikChange site-directed mutagenesis kit (Stratagene) was used to generate GDE1(Glu97Ala/Asp99Ala) and GDE1(His112Ala) mutants with pCDNA3-GDE1/MIR16 as a template, and the sequences of mutants were confirmed by automated sequencing. Sequences of primers used are available on request.
In Vitro Translation.
In vitro translation of MIR16 was carried out using the TNT T7 rabbit reticulocyte Quick Coupled Transcription/Translation System (Promega). The reaction mixture (25 μl), containing pCDNA3-MIR16 plasmid, [35S]methionine (1,000 Ci/mmol, in vivo cell labeling grade, Amersham Pharmacia), and 2 μl of canine pancreatic microsomes (Promega), was incubated at 30°C for 1 h, and the products were analyzed by SDS/PAGE and autoradiography (12).
Proteinase K (PK) Protection Assays.
PK digestion was performed as described (15) on membranes prepared by centrifugation (100,000 × g pellet) from LβT2 cells or on microsomal membranes containing in vitro translated MIR16. Briefly, membranes were suspended in 50 μl of reaction buffer (10 mM Tris⋅HCl, pH 7.8/150 mM KCl/2 mM MgCl2/2 mM CaCl2/200 mM sucrose), and 5 μg of PK (Boehringer Mannheim) was added at room temperature for 30 min. Reactions were stopped with 10 mM PMSF. Proteins were separated by SDS/PAGE and analyzed by immunoblotting (LβT2 membranes) or autoradiography (microsomal membranes).
Immunoblotting.
Proteins were separated by 12% SDS/PAGE and transferred to PVDF membranes (Millipore). Membranes were blocked in TBS/5% calf serum/0.1% Tween 20 and incubated with protein A-purified anti-MIR16 IgG (12) or anti-calnexin serum (from J. J. M. Bergeron, McGill University, Toronto) followed by horseradish peroxidase (HRP)-conjugated goat anti-rabbit (1:3,000) or anti-mouse (1:5,000) IgG (Bio-Rad) and ECL detection (Pierce).
Results
GDE1/MIR16 Belongs to a Family of Mammalian GDEs.
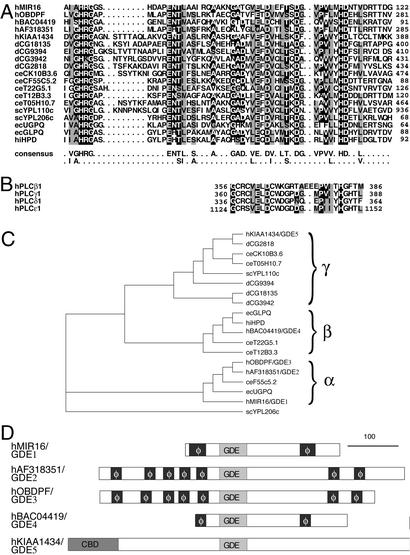
Previously, we showed that MIR16 shares strong homology with the bacterial GDEs GLPQ and HPD (12, 17, 18). To identify additional putative mammalian GDEs, we used the MIR16 protein sequence in a blast search and identified four hypothetical proteins of unknown function that share significant sequence similarity to MIR16 (E values <2 × 10−4) in the human genome, four related sequences in Drosophila melanogaster, and five in the Caenorhabditis elegans genome (ref. 12; Fig. 1A). The most conserved region among all these proteins and the bacterial GDEs resides in a domain of ≈56 residues (corresponding to amino acids 67–122 of MIR16), which we designate the GDE domain (Fig. 1A). The similarity between these domains is 40–85% (pairwise analysis). These results suggest that MIR16 is a member of an evolutionarily conserved family of GDEs, and thus we have renamed MIR16 and its four newly identified human homologs GDE1–GDE5.
Figure 1.
GDE1/MIR16 belongs to a family of GDE phosphodiesterases. (A) Alignment of the GDE domain in GDE1/MIR16 with those in bacterial GDEs (GLPQ and HPD) and selective putative GDEs. Invariant residues are shaded in black, similar residues in gray. Numbers indicate the positions of amino acids. h, Homo sapiens; d, D. melanogaster; ce, C. elegans; ec, Escherichia coli; sc, Saccharomyces cerevisiae; hi, H. influenzae. (B) Alignment of GDE domains with the catalytic X domains of human PI-specific phospholipases C (PI-PLCs). (C) Phylogenetic analysis of the GDE protein family. Sequences of GDE domains were used for the analysis with the clustalw program (www.ebi.ac.uk/clustalw/). Three different groups (α, β, and γ) of proteins were observed. (D) Domain structures of putative members of the GDE family encoded in the human genome. φ, hydrophobic region; GDE, GDE domain; CBD, carbohydrate binding domain.
Based on phylogenetic analysis of the GDE domain, we further classified the family into three distinct groups: α, β, and γ (Fig. 1C). Domain–structure analysis showed that GDE2, GDE3, and GDE4 in groups α and β resemble GDE1/MIR16 in that they appear to be membrane proteins and contain multiple putative transmembrane regions (Fig. 1D). GDE5 (group γ) does not have a transmembrane region but contains an N-terminal carbohydrate binding domain (CBD) (Fig. 1D) that binds to polysaccharides and is often found in glycosylhydrolases (19). The diverse domain structures of these GDEs suggest that they may have diverse subcellular locations, substrates, and methods of regulation.
GDE1 Is a PDE for GPI.
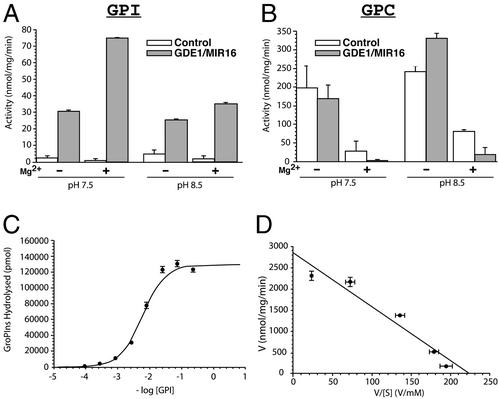
To investigate its PDE activity, GDE1 cDNA was transfected into HEK 293T cells and PNS fractions were assayed for activity on GPI and GPC. HEK 293T cells were chosen because they contain very little endogenous GDE1 as determined by Western blotting with a GDE1/MIR16 antibody (data not shown). Cells transfected with GDE1 showed greatly increased activity (>10-fold) on GPI (Fig. 2A), but not on GPC (Fig. 2B), compared with cells transfected with control vector alone (Fig. 2A). GPI-PDE activity was enhanced by addition of 10 mM Mg2+ (3-fold), but not Ca2+ ([Ca2+]free, 0.1–10 μM; data not shown), to the assay buffer and was higher at pH 7.5 than at pH 8.5 (Fig. 2A).
Figure 2.
Characterization of the GPI-PDE activity of GDE1. HEK 293T cells were transfected with empty vector pCDNA3 (control) or pCDNA3-GDE1. PNS fractions were isolated, and GDE activities were determined by measuring [3H]GPI (A) or [3H]GPC (B) hydrolysis as described in Materials and Methods. (A) A dramatic increase is seen in the PDE activity on GPI of PNS fractions from transfected cells (gray bars), as compared with control cells (white bars). (B) The GPI-PDE activity is higher at pH 7.5 than pH 8.5 and is enhanced by addition of 10 mM Mg2+. Little or no increase is seen in the PDE activity on GPC under the same conditions. The data are from a single experiment carried out in duplicate (mean ± SE) and are representative of two independent experiments. (C) Log dose–response curve of GDE1 activity toward increasing concentrations of [3H]GPI. Samples containing 2 μg of PNS were incubated with the indicated amounts of [3H]GPI for 30 min, and GPI hydrolysis was measured by HPLC. The curve shows a computer-generated free fit of the data to a four-parameter logistic equation by using nonlinear regression analysis. Data are from two combined independent experiments, each carried out in duplicate (n = 4; mean ± SE). (D) The Eadie–Hofstee plot of the same set of data as in C, showing an apparent Km of ≈12 mM and a Vmax of ≈3,000 pmol/mg/min.
The GPI-PDE activity was substrate-concentration dependent (Fig. 2C), and GPI hydrolysis was essentially linear over the standard 30-min incubation period at 37°C (data not shown). The Eadie–Hofstee plot shows that this GDE1 has an apparent Km of ≈12 mM and a Vmax of ≈3,000 pmol/mg/min for GPI (Fig. 2D) under these conditions. The only GDE1 products seen from [3H]GPI and [3H]GPC by HPLC analysis were [3H]inositol and [3H]choline, respectively. No accumulation of either 3H-labeled inositol monophosphates or choline phosphate was seen. Based on these results, we conclude that GDE1 is a PDE for GPI.
Substrate Competition Assay of GDE1.
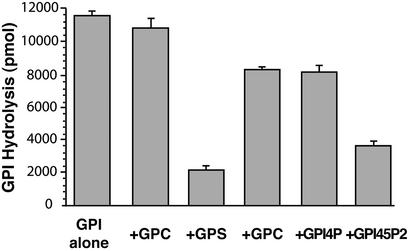
To further establish the substrate specificity of GDE1, competition studies were performed by adding a 10-fold excess of unlabeled GPs to the PDE assay for GPI. Under these conditions GPC had no significant effect, GPE and GPI 4-phosphate (GPI4P) blocked GPI-PDE activity by ≈30%, and GPI 4,5-bisphosphate (GPI45P2) and glycerophosphoserine (GPS) by ≈70% and ≈80%, respectively (Fig. 3). With the assumption of competitive Michaelis–Menten kinetics, extrapolation of these data indicate Kapp values for GPE and GPI4P in the ≈30 mM range and those for GPI45P2 and GPS in the ≈3–5 mM range. Hence, the competitive preference of GDE1 based on calculation of Kapp seen is GPI45P2 ≈ GPS > GPI (see above) > GPI4P ≈ GPE ≫ GPC, indicating a general preference for GPIs. Whether other GPs are alternative substrates or GPI-PDE inhibitors remains to be determined.
Figure 3.
Effects of various GPs on GPI hydrolysis by GDE1. PNS fractions (prepared as in Fig. 1) were incubated with 1 mM [3H]GPI for 30 min in the presence or absence of 10 mM competing GP, as indicated, and the level of GPI hydrolysis was measured by HPLC. Data are from a single experiment carried out in triplicate (mean ± SE) and are representative of two independent experiments. Values from the pCDNA3 empty vector control were subtracted from each data value. GPI45P2 and GPS, as well as GPI4P and GPE (to a lesser extent), but not GPC, significantly (P < 0.005; Student's t test) inhibit the hydrolysis of GPI by GDE1.
Essential Residues for the GPI-PDE Activity of GDE1.
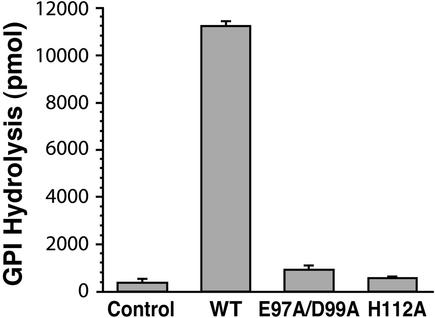
In the course of the blast search, we discovered that a common sequence motif is shared between the C-terminal half (amino acids 92–116) of the GDE domains and the N terminus of catalytic X domains of mammalian PI-PLCs (Fig. 1B), which hydrolyze phosphatidylinositol-4,5-bisphosphate (PtdIns45P2), producing two important second messengers, inositol 1,4,5-trisphosphate and diacylglycerol (20). Several residues in this motif (Glu-341, Asp-343, and His-356) have been identified as important catalytic sites of PLCδ1, based on crystal structure and mutagenesis studies. The presence of these residues in the GDE domain of GDE1 suggests that they may be critical for PDE activity. To determine whether this is the case, we generated two mutants, GDE1(Glu97Ala/Asp99Ala) and GDE1(His112Ala). When these mutants were expressed in HEK 293 cells PDE activity was negligible (Fig. 4). By immunoblotting there was no detectable difference in the expression levels of mutant and wild-type GDE1 in HEK 293T cells (data not shown). These results demonstrate that (i) the GDE domain of GDE1 represents its catalytic domain and that (ii) Glu-97/Asp-99 and His-112 are essential for its GDE activity.
Figure 4.
GPI-PDE activities of wild type and GDE1 mutants. HEK 293T cells were transfected with pCDNA3 empty vector (Control), pCDNA3-GDE1 (WT), or pCDNA3-GDE1 E97A/D99A or H112A, and PNS fractions were assayed for GPI-PDE activity as described in Materials and Methods. Both mutants (E97A/D99A and H112A) had lost their activity. Data are from a single experiment carried out in triplicate (mean ± SE) and are representative of three independent experiments.
Orientation of the Catalytic Domain of GDE1.
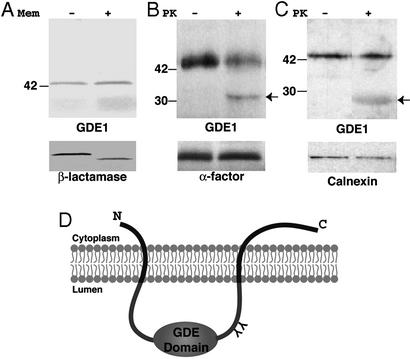
Previously we showed that GDE1/MIR16 is an ≈43-kDa integral membrane glycoprotein, because it remains associated with membranes after alkaline (pH 11) extraction and is sensitive to endoglycosidase H in LβT2 pituitary cells (12). Hydropathy analysis of the GDE1 amino acid sequence indicates that it contains two hydrophobic regions, a putative signal sequence at the N terminus (amino acids 8–45) and a putative membrane-spanning region close to the C terminus (amino acids 243–276) (12). No difference in the mobility of GDE1 was found when it was in vitro translated in the presence or absence of canine microsomal membranes, suggesting that the putative N-terminal signal sequence of GDE1/MIR16 is not cleaved during membrane translocation (Fig. 5A) and probably serves as a membrane anchor. Assuming that both the N- and C-terminal hydrophobic regions of GDE1 are transmembrane domains, the location of two putative N-glycosylation sites at amino acids 168 and 198 in close proximity to the GDE domain (ref. 12; Fig. 1C) suggests that this region faces the lumen/extracellular space and the C terminus faces the cytoplasm (Fig. 5D).
Figure 5.
Membrane topology of GDE1. (A) GDE1 and β-lactamase were in vitro translated in the presence (+) or absence (−) of microsomal membranes, and the 35S-labeled products were analyzed by SDS/PAGE and autoradiography. No difference in mobility is seen when GDE1 is translated in the presence of microsomal membranes, whereas the mobility of β-lactamase, used as a positive control, shifts. (B and C) PK protection analysis of in vitro translated and endogenous GDE. GDE1 and yeast α factor were in vitro translated in the presence of microsomal membranes. Microsomal membranes (B) and LβT2 cell membranes (C) were treated with PK as described in Materials and Methods. An ≈32-kDa fragment is seen after digestion of both in vitro translated (B) and endogenous (C) GDE1, confirming that the bulk of the enzyme faces the lumen. As controls, yeast α factor (a secretory peptide; B) and calnexin (an integral membrane protein facing the lumen of the endoplasmic reticulum; C) were protected from digestion by PK. (D) Proposed membrane topology of GDE1 with the catalytic domain of GDE1 facing the lumen and both the N and C termini facing the cytoplasm. The N-terminal hydrophobic region of GDE1 is not cleaved during membrane translocation. Y, N-glycosylation site.
To investigate the membrane topology of GDE1 we used PK protection assays. If our proposed model (Fig. 5D) is correct, the GDE domain would be protected from PK digestion, whereas the C terminus would be accessible to PK and be removed and an ≈31-kDa product (corresponding to the protected fragment, amino acids 46–242) would be expected after PK digestion. However, if the GDE domain faces the cytoplasm an ≈8-kDa product corresponding to the protected C terminus (amino acids 277–331) would be expected. When GDE1 was in vitro translated in the presence of microsomal membranes, an ≈32-kDa fragment was seen after digestion with PK (Fig. 5B). Similar results were obtained for endogenous GDE1 when membrane fractions from LβT2 cells were used in the same assay (Fig. 5C). Taken together, these results support a model for the topology of GDE1 (Fig. 5D) whereby its C terminus faces the cytoplasm and the GDE domain faces either the lumen (intracellular membranes) or extracellular space (plasma membrane).
The GPI-PDE Activity of GDE1 Can Be Regulated by Stimulation of GPCRs.
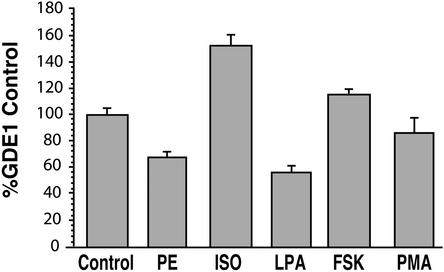
Several enzymes involved in phospholipid metabolism (e.g., phospholipases A2, C, and D) are known to be regulated by trimeric G proteins (20–22). The fact that GDE1/MIR16 was originally identified by its interaction with RGS proteins (12), which are GTPase activating proteins (GAPs) for trimeric G proteins (23), suggested it might also be regulated by G proteins. To gain information on this point, HEK 293T cells were transfected with GDE1 and treated with various agonists for endogenous GPCRs, and the PNS was tested for PDE activity on GPI. Isoproterenol, an agonist for Gαs-coupled β-adrenergic receptors, increased the GPI-PDE activity of GDE1 by ≈50% compared with untreated cells (Fig. 6). Two other agonists, phenylephrine for Gαi/q-coupled α-adrenergic receptors and lysophosphatidic acid for Gαi/q/12/13-coupled lysophospholipid receptors, reduced the activity by 30% and 40%, respectively (Fig. 6). As controls, forskolin, an adenylyl cyclase activator, and PMA (phorbol 12-myristate 13-acetate), a protein kinase C activator, did not significantly affect GDE1 activity (Fig. 6). These results indicate that the PDE activity of GDE1 on GPI can be modulated both positively and negatively by G protein signaling pathways and that this regulation does not depend on protein kinase A or C.
Figure 6.
Effects of various stimulants on GDE1 GPI-PDE activity. HEK 293T cells transfected with GDE1 were serum starved overnight and treated or not (Control) with the indicated stimulants for 10 min. PNS fractions were prepared and used in the GPI-PDE activity assay. PE, phenylephrine (10 μM); ISO, isoproterenol (10 μM); LPA, lysophosphatidic acid (10 μM); FSK, forskolin (10 μM); PMA, phorbol 12-myristate 13-actate (200 nM). Data (mean ± SE) from two to four independent experiments, each carried out in duplicate, are expressed as a percentage of the untreated control after subtraction of the pCDNA3 empty vector control.
Discussion
In this study we found that GDE1/MIR16, originally identified as a membrane glycoprotein that binds RGS16 (12), belongs to a new family of proteins identified by the presence of an evolutionarily conserved GDE domain. Accordingly, we have renamed the protein GDE1. We have shown that GDE1 prefers GPI and some of its phosphorylated derivatives (e.g., GIP4P and GPI45P2) as potential substrates. Expression of wild-type GDE1, but not two catalytic GDE domain mutants, dramatically increased the GPI-PDE activity in HEK 293T cells. Furthermore, the activity of GDE1 was found to be modulated by stimulation of several GPCRs. Collectively, these findings support the conclusions that GDE1/MIR16 is a bona fide GPI-PDE and that it is regulated by GPCR signaling.
The interaction between GDE1 and selective RGS proteins, including RGS16, prompted us to investigate the potential regulation of the GPI-PDE activity of GDE1 by G protein-mediated signaling pathways. Our results demonstrate that the enzyme activity of GDE1 can be regulated both positively and negatively (depending on the receptor agonist used) by stimulation of GPCRs. Similar dual regulation of enzyme activity by G proteins has been observed for adenylyl cyclase and PLCβ (20, 24). Our results suggest that GDE1 might serve as a downstream effector in G protein signaling, in which the enzymatic activity of GDE1/MIR16 in PI metabolism could be regulated through its interaction with RGS proteins. Alternatively, G proteins may also bind directly to GDE1 and regulate its activity. The interaction between GDE1 and RGS proteins provides yet another example of the role of RGS proteins in linking G protein signaling to other signaling pathways (PI metabolism in this case). RGS proteins have previously been shown to integrate G protein signaling with other signaling pathways, such as Rho signaling, ion channel regulation, and growth factor receptor signaling and trafficking (23, 25–28).
Although no mammalian PDE for GPI has been previously identified, GPI-PDE enzyme activity (3.1.4.44) has been detected in several rat tissues (11). The enzymatic properties we define here for GDE1 expressed in HEK 293T cells are similar to those reported previously. Both have maximal enzymatic activity around pH 7.5 and are activated by 10 mM Mg2+. We previously showed that GDE1/MIR16 is expressed in multiple rat tissues and is localized to intracellular membrane compartments (pre/mid-Golgi) in rat pituitary and to the plasma membrane in liver and kidney (12). This finding is consistent with the fact that the enzyme activity reported previously (11) was found in multiple rat tissues (e.g., liver, pancreas, brain, spleen, kidney, and intestinal mucosa) and was enriched in microsomal fractions of rat pancreas.
Our sequence analysis revealed that the C-terminal half of the GDE domain shows a striking similarity to the catalytic domains of mammalian PI-PLCs and shares a common motif with PI-PLCs, implying that they may have similar catalytic mechanisms and/or substrates. Based on data from crystal structure, enzyme kinetics, and mutagenesis studies, a sequential catalytic mechanism was proposed for PI-PLCδ1: (i) a phosphotransfer step involving His-356 and Glu-341 as general acid/base catalysts that generates a stable cyclic phosphodiester intermediate, followed by (ii) a phosphohydrolysis step that generates the inositol phosphate product (29–31). Both His-356 and Glu-341 of PLCδ1 are strictly conserved in the corresponding positions of all of the GDEs, and mutation of the corresponding residues abolished the PDE activity of GDE1. It is of interest that PLCδ1 can also hydrolyze with lower activity the water-soluble GPI45P2 and GPI (29), the preferred substrates of GDE1, in addition to its main physiological substrate, phosphatidylinositol-4,5-bisphosphate (PtdIns45P2). Whether GDE1 or any of the other GDEs can use PtdIns45P2 and/or the other PIs as substrates remains to be determined.
Substrate competition experiments showed that, in addition to GPI45P2, GPS significantly inhibited the hydrolysis of GPI by GDE1. Whether this relative preference for GPS might indicate an inhibitory activity of GPS, rather than an alternative substrate, remains to be determined.
What are the potential cellular functions of GDE1? The main substrates of GDE1 appear to be GPI and its phosphorylated forms, GPI4P and GPI45P2, which are deacylation products of the membrane PIs formed by the sequential action of PLA1/PLA2 and lysophospholipases (32, 33). As a GPI-PDE that further hydrolyzes GPI to its final catabolic products, inositol and glycerol 3-phosphate, GDE1 is most likely involved in the remodeling of membrane PIs and regulation of membrane phospholipid composition, and hence in intracellular membrane trafficking (3), as well as in the modulation of G protein-mediated signaling. GPIs are signaling molecules implicated in the modulation of multiple cellular functions (33). They can modulate the activities of trimeric G proteins: GPI4P inhibits Gαs-mediated adenylyl cyclase activation (34), and both GPI and GPI4P inhibit the G protein-mediated activation of PLA2 (33, 35). In addition, GPI4P has recently been shown to modulate the activity of the small GTPases Rac and Rho and thus to be involved in the reorganization of the actin cytoskeleton during formation of ruffles (Rac) and stress fibers (Rho) (36). The findings that GDE1 can bind to RGS proteins and that the GPI-PDE activity of GDE1 can be regulated through stimulation of GPCR suggest that there is a bidirectional interplay between GPI-PDE activity and G protein signaling.
Our membrane topology studies indicate that the catalytic GDE domain of GDE1 faces the lumen and the C terminus faces the cytoplasm. This topology is supported by our unpublished findings that RGS16 coimmunoprecipitates with the GFP-tagged C terminus, but not the N terminus of GDE1. Although unusual, it is not without precedent that enzymes involved in PI metabolism have their catalytic domains oriented toward the lumen and/or extracellular space. For example, most reactions in glycosylphosphoinositol biosynthesis after the transfer of the first mannose proceed on the luminal side of the endoplasmic reticulum (37), and the synthesis of inositol phosphorylceramide from PI and ceramide precursors takes place on the luminal side of Golgi membranes (38).
Our membrane topology model suggests that GDE1 mainly hydrolyzes GPIs from the luminal side. These luminal GPIs could come from PIs in either the luminal or the cytoplasmic leaflets of the membranes, because PIs and their intermediate breakdown product, LPI, can flip from the cytoplasmic to the luminal leaflet of membranes (39). Furthermore, the hydrolysis of luminal GPI by GDE1 is likely to be highly related to the regulation of the concentration of cytosolic GPIs, because GPIs are water-soluble molecules and can move across the lipid-bilayer membrane through transporters. A transporter for GPI has been identified in yeast (40) and has been shown to transfer GPI across the plasma membrane (41, 42). Similarly, extracellular GPI4P has been shown to cross the plasma membrane and to rapidly reach equilibrium with the cytosol (43).
In summary, we have demonstrated that GDE1/MIR16, previously identified as an RGS-interacting protein, belongs to a large GDE protein family and is capable of hydrolyzing GPIs, producing inositol and glycerol 3-phosphate. Furthermore, this GPI-PDE activity can be regulated by GPCR signaling. It now becomes important to define the cellular functions of each member of the mammalian GDE protein family and the specific pathways and mechanisms underlying the regulation of GDE1 by G protein signaling, i.e., which G proteins are involved and whether the regulation is direct or indirect.
Acknowledgments
We thank Cristiano Iurisci and Elena Fontana (Consorzio Mario Negri Sud) for help with the biochemical analyses and several of the figures, respectively. This work was supported by National Institutes of Health Grants DK17780 and CA58689 (to M.G.F.) and grants to D.C. from the Italian Association for Cancer Research (Milan), the Italian Foundation for Cancer Research (Milan), and Telethon Italy (E.841).
Abbreviations
- GPCR
G protein-coupled receptor
- GP
glycerophosphodiester
- GDE
GP phosphodiesterase
- GPC
glycerophosphocholine
- GPE
glycerophosphoethanolamine
- GPI
glycerophosphoinositol
- GPI4P
GPI 4-phosphate
- GPI45P2
GPI 4,5-bisphosphate
- GPS
glycerophosphoserine
- MIR16
membrane interacting protein of RGS16
- PDE
phosphodiesterase
- GPI-PDE
GPI phosphodiesterase
- PI
phosphoinositide
- PI-PLC
PI-specific phospholipase C
- PK
proteinase K
- PNS
postnuclear supernatant
- RGS
regulator of G protein signaling
References
- 1.Liscovitch M, Cantley L C. Cell. 1994;77:329–334. doi: 10.1016/0092-8674(94)90148-1. [DOI] [PubMed] [Google Scholar]
- 2.Divecha N, Irvine R F. Cell. 1995;80:269–278. doi: 10.1016/0092-8674(95)90409-3. [DOI] [PubMed] [Google Scholar]
- 3.Huijbregts R P, Topalof L, Bankaitis V A. Traffic. 2000;1:195–202. doi: 10.1034/j.1600-0854.2000.010301.x. [DOI] [PubMed] [Google Scholar]
- 4.Simonsen A, Wurmser A E, Emr S D, Stenmark H. Curr Opin Cell Biol. 2001;13:485–492. doi: 10.1016/s0955-0674(00)00240-4. [DOI] [PubMed] [Google Scholar]
- 5.Wang A, Dennis E A. Biochim Biophys Acta. 1999;1439:1–16. doi: 10.1016/s1388-1981(99)00063-3. [DOI] [PubMed] [Google Scholar]
- 6.Takemori H, Zolotaryov F N, Ting L, Urbain T, Komatsubara T, Hatano O, Okamoto M, Tojo H. J Biol Chem. 1998;273:2222–2231. doi: 10.1074/jbc.273.4.2222. [DOI] [PubMed] [Google Scholar]
- 7.Baldwin J J, Cornatzer W E. Biochim Biophys Acta. 1968;164:195–204. doi: 10.1016/0005-2760(68)90146-x. [DOI] [PubMed] [Google Scholar]
- 8.Dawson R M C. Biochem J. 1956;62:689–693. doi: 10.1042/bj0620689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lloyd-Davies K A, Michell R H, Coleman R. Biochem J. 1972;127:357–368. doi: 10.1042/bj1270357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webster G R, Marples E A, Thompson R H S. Biochem J. 1957;65:374–377. doi: 10.1042/bj0650374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson R M, Hemington N, Richards D E, Irvine R F. Biochem J. 1979;182:39–49. doi: 10.1042/bj1820039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng B, Chen D, Farquhar M G. Proc Natl Acad Sci USA. 2000;97:3999–4004. doi: 10.1073/pnas.97.8.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook M, Frisch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 14.Clarke N G, Dawson R M. Biochem J. 1981;195:301–306. doi: 10.1042/bj1950301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin P, Le-Niculescu H, Hofmeister R, McCaffery J M, Jin M, Hennemann H, McQuistan T, De Vries L, Farquhar M G. J Cell Biol. 1998;141:1515–1527. doi: 10.1083/jcb.141.7.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pomfret E A, daCosta K A, Schurman L L, Zeisel S H. Anal Biochem. 1989;180:85–90. doi: 10.1016/0003-2697(89)90091-2. [DOI] [PubMed] [Google Scholar]
- 17.Tommassen J, Eiglmeier K, Cole S T, Overduin P, Larson T J, Boos W. Mol Gen Genet. 1991;226:321–327. doi: 10.1007/BF00273621. [DOI] [PubMed] [Google Scholar]
- 18.Munson R S, Jr, Sasaki K. J Bacteriol. 1993;175:4569–4571. doi: 10.1128/jb.175.14.4569-4571.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sauer J, Sigurskjold B W, Christensen U, Frandsen T P, Mirgorodskaya E, Harrison M, Roepstorff P, Svensson B. Biochim Biophys Acta. 2000;1543:275–293. doi: 10.1016/s0167-4838(00)00232-6. [DOI] [PubMed] [Google Scholar]
- 20.Rebecchi M J, Pentyala S N. Physiol Rev. 2000;80:1291–1335. doi: 10.1152/physrev.2000.80.4.1291. [DOI] [PubMed] [Google Scholar]
- 21.Xie Z, Ho W-T, Spellman R, Cai S, Exton J H. J Biol Chem. 2002;277:11979–11986. doi: 10.1074/jbc.M109751200. [DOI] [PubMed] [Google Scholar]
- 22.Balsinde J, Balboa M A, Insel P A, Dennis E A. Annu Rev Pharmacol Toxicol. 1999;39:175–189. doi: 10.1146/annurev.pharmtox.39.1.175. [DOI] [PubMed] [Google Scholar]
- 23.De Vries L, Zheng B, Fischer T, Elenko E, Farquhar M G. Annu Rev Pharmacol Toxicol. 2000;40:235–271. doi: 10.1146/annurev.pharmtox.40.1.235. [DOI] [PubMed] [Google Scholar]
- 24.Hanoune J, Defer N. Annu Rev Pharmacol Toxicol. 2001;41:145–174. doi: 10.1146/annurev.pharmtox.41.1.145. [DOI] [PubMed] [Google Scholar]
- 25.De Vries L, Farquhar M G. Trends Cell Biol. 1999;9:138–144. doi: 10.1016/s0962-8924(99)01515-9. [DOI] [PubMed] [Google Scholar]
- 26.Hollinger S, Hepler J R. Pharmacol Rev. 2002;54:527–559. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- 27.Lou X, Yano H, Lee F, Chao M V, Farquhar M G. Mol Biol Cell. 2001;12:615–627. doi: 10.1091/mbc.12.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng B, Ma Y C, Ostrom R S, Lavoie C, Gill G N, Insel P A, Huang X Y, Farquhar M G. Science. 2001;294:1939–1942. doi: 10.1126/science.1064757. [DOI] [PubMed] [Google Scholar]
- 29.Wu Y, Perisic O, Williams R L, Katan M, Roberts M F. Biochemistry. 1997;36:11223–11233. doi: 10.1021/bi971039s. [DOI] [PubMed] [Google Scholar]
- 30.Ellis M V, James S R, Perisic O, Downes C P, Williams R L, Katan M. J Biol Chem. 1998;273:11650–11659. doi: 10.1074/jbc.273.19.11650. [DOI] [PubMed] [Google Scholar]
- 31.Essen L O, Perisic O, Cheung R, Katan M, Williams R L. Nature. 1996;380:595–602. doi: 10.1038/380595a0. [DOI] [PubMed] [Google Scholar]
- 32.Irvine R F, Hemington N, Dawson R M. Biochem J. 1978;176:475–484. doi: 10.1042/bj1760475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corda D, Iurisci C, Berrie C P. Biochim Biophys Acta. 2002;1582:52–69. doi: 10.1016/s1388-1981(02)00137-3. [DOI] [PubMed] [Google Scholar]
- 34.Iacovelli L, Falasca M, Valitutti S, D'Arcangelo D, Corda D. J Biol Chem. 1993;268:20402–20407. [PubMed] [Google Scholar]
- 35.Corda D, Falasca M. Anticancer Res. 1996;16:1341–1350. [PubMed] [Google Scholar]
- 36. Mancini, R., Piccolo, E., Mariggiò, S., Filippi, B., Iurisci, C., Pertile, P., Berrie, C. P. & Corda, D. (2003) Mol. Biol. Cell, in press (10.1091/mbc.E02-04-0179). [DOI] [PMC free article] [PubMed]
- 37.Maeda Y, Watanabe R, Harris C L, Hong Y, Ohishi K, Kinoshita K, Kinoshita T. EMBO J. 2001;20:250–261. doi: 10.1093/emboj/20.1.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levine T P, Wiggins C A, Munro S. Mol Biol Cell. 2000;11:2267–2281. doi: 10.1091/mbc.11.7.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kol M A, de Kruijff B, de Kroon A I. Semin Cell Dev Biol. 2002;13:163–170. doi: 10.1016/s1084-9521(02)00044-7. [DOI] [PubMed] [Google Scholar]
- 40.Patton-Vogt J L, Henry S A. Genetics. 1998;149:1707–1715. doi: 10.1093/genetics/149.4.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patton J L, Pessoa-Brandao L, Henry S A. J Bacteriol. 1995;177:3379–3385. doi: 10.1128/jb.177.12.3379-3385.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hawkins P T, Stephens L R, Piggott J R. J Biol Chem. 1993;268:3374–3383. [PubMed] [Google Scholar]
- 43.Berrie C P, Iurisci C, Corda D. Eur J Biochem. 1999;266:413–419. doi: 10.1046/j.1432-1327.1999.00870.x. [DOI] [PubMed] [Google Scholar]