Abstract
Omega-6 (ω-6) polyunsaturated fatty acids (PUFA), abundant in the Western diet, are precursors for a number of key mediators of inflammation including the 2-series of prostaglandins (PG). PGE2, a cyclooxygenase (COX) metabolite of arachidonic acid, a ω-6 PUFA, is a potent mediator of inflammation and cell proliferation. Dietary supplements rich in ω-3 PUFA reduce the concentrations of 2-series PG and increase the synthesis of 3-series PG (e.g., PGE3), which are believed to be less inflammatory. However, studies on cellular consequences of increases in 3-series PG in comparison to 2-series PG have not been reported. In this study, we compared the effects of PGE2 and PGE3 on (i) cell proliferation in NIH 3T3 fibroblasts, (ii) expression and transcriptional regulation of the COX-2 gene in NIH 3T3 fibroblasts, and (iii) the production of an inflammatory cytokine, IL-6, in RAW 264.7 macrophages. PGE3, unlike PGE2, is not mitogenic to NIH 3T3 fibroblasts. PGE2 and PGE3 both induce COX-2 mRNA via similar signaling mechanisms; however, compared with PGE2, PGE3 is significantly less efficient in inducing COX-2 gene expression. Furthermore, although both PGE2 and PGE3 induce IL-6 synthesis in RAW 264.7 macrophages, PGE3 is substantially less efficient compared with PGE2. We further show that increasing the ω-3 content of membrane phospholipid results in a decrease in mitogen-induced PGE2 synthesis. Taken together, our data suggest that successful replacement of ω-6 PUFA with ω-3 PUFA in cell membranes can result in a decreased cellular response to mitogenic and inflammatory stimuli.
Prostaglandins (PG) are bioactive lipids derived from the metabolism of membrane polyunsaturated fatty acids (PUFA), and play important roles in a number of biological processes including cell division, immune responses, and wound healing (1). Altered PG production is associated with a variety of illnesses, including acute and chronic inflammation and colon cancer (2). Arachidonic acid (AA), a common omega-6 (ω-6) PUFA, is found esterified at the sn-2 position of membrane phospholipids (3). Cyclooxygenase (COX), also known as PG synthase, is the key enzyme in PG synthesis from AA (2). COX converts AA (released from membrane phospholipids by phospholipases) to PGH2, the common precursor of all prostanoids. Two COX isoforms have been described. COX-1 is constitutively expressed in nearly all cells. The second COX isoform, COX-2, is induced in many cell types by a wide range of proinflammatory or mitogenic agents (2). COX-2-derived PG promote inflammation by increasing vascular permeability and vasodilatation and by directing the synthesis and migration of proinflammatory cytokines into the site of inflammation (4). Aberrant expression of COX-2 has been implicated in the etiology of a number of cancers (5–8). The mitogenic and proinflammatory functions of COX-2 are linked primarily to exaggerated synthesis of PGE2 (9). Interestingly, PGE2 has recently been shown to amplify its own production by inducing COX-2 expression in various cells (10–14). PGE2-dependent amplification of COX-2 is hypothesized to be an important part of sustained proliferative and chronic inflammatory conditions, and may explain the overexpression of COX-2 in tumors.
Because the availability of free AA is a rate-limiting step in the synthesis of PG, modulating AA availability could serve as a means of altering PG synthesis and prevent/inhibit the pathological effects of 2-series PG. Eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) are ω-3 PUFA found primarily in fish oils. EPA and DHA compete with AA both at the level of incorporation into cell membrane phospholipids (15) as well as at the level of substrate for COX pathway (16) generating the 3-series of PG, e.g., PGE3 (Fig. 1A). Dietary supplements of fish oils rich in ω-3 PUFA are used as preventive measures against a number of illnesses, including arthritis and cancer (17). In addition to reducing the concentrations of 2-series PG, fish oil ingestion also results in a 10–50-fold increase in 3-series PG in vivo (18, 19). Although similar in structure (Fig. 1A) and stability (3), the 2-series of PG is considered to be more mitogenic and proinflammatory compared with the 3-series of PG. However, studies directly comparing the effects of 2-series vs. 3-series PG on cellular functions have not been reported. If indeed the 3-series of PG possesses antiinflammatory and antimitogenic properties, it could serve as a therapeutic agent in a number of inflammatory processes.
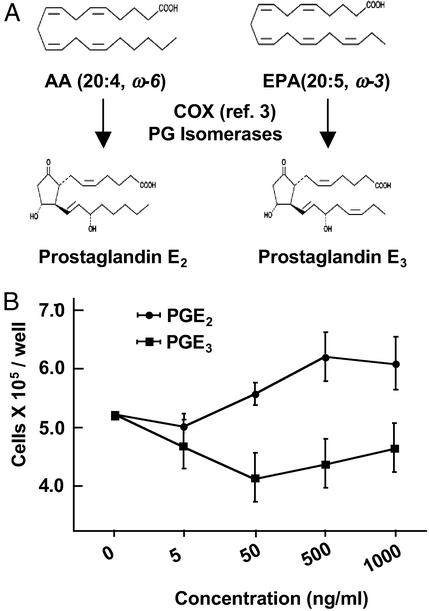
Figure 1.
(A) Structures of AA (20:4, ω-6), EPA (20:5, ω-3), and their oxygenated COX metabolites PGE2 and PGE3, respectively. (B) PGE2, but not PGE3, stimulates NIH 3T3 cell proliferation. Cells were treated with PGE2 or PGE3 at various concentrations. Cell proliferation was measured after 24 h. Data are the means ± SD from triplicate wells.
In this article, we compared the effects of PGE2 and PGE3 on cell proliferation, COX-2 message and protein expression, and transcriptional regulation of COX-2 in NIH 3T3 fibroblasts, and induction of IL-6 in RAW 264.7 macrophages.
Experimental Procedures
Materials.
AA, EPA, PGE2, PGE3, polyclonal rabbit anti-COX-2 antibody, murine COX-2 cDNA probe, and PGE2 EIA kits were purchased from Cayman Chemical (Ann Arbor, MI). DMEM and PBS were supplied by GIBCO. The mouse IL-6 ELISA kit was purchased from BioSource International (Camarillo, CA). A dual-luciferase reporter assay system was purchased from Promega. An RNeasy Mini kit for RNA isolation was purchased from Qiagen (Valencia, CA).
Plasmids.
WT and mutant COX-2 promoter fragments (from −724 to +7) were cloned into HindIII–XhoI sites of pXP2 plasmid, and the expression vector for a kinase-defective JNK1 (pCDNA-DN-JNK1), pCEP4Erk1K71R (encoding dominant-negative Erk1) and pZIPM17 (an expression vector dominant-negative Ha-Ras), were provided by H. Herschman (University of California, Los Angeles) (20).
Cell Culture and Treatment.
NIH 3T3 cells were purchased from the American Type Culture Collection. The cells were maintained in DMEM supplemented with 10% FCS, penicillin (100 units/ml), and streptomycin (100 μg/ml) at 37°C in 10% CO2. RAW 264.7 murine macrophage cells were cultured in endotoxin-free DMEM supplemented with 10% FBS, penicillin (100 units/ml), and streptomycin (100 μg/ml) at 37°C in 5% CO2. In general, NIH 3T3 cells were cultured to 60–70% confluency in 10% FCS-supplemented DMEM. The cells were then washed with PBS and incubated in DMEM supplemented with 0.5% heat-inactivated FCS and penicillin/streptomycin for an additional 18–24 h before experiments.
Northern Analysis.
Cells were treated with PGE2 or PGE3 at various concentrations and for various time durations as described in the figure legends. After the treatment with PG, the cells were washed once with PBS. Total RNA from the cell lysates was extracted by using the RNeasy Mini kit (Qiagen). Ten micrograms of total RNA from each sample was denatured at 65°C for 10 min, electrophoresed on a 1.1% agarose gel, and transferred onto nylon membranes. The membranes were UV cross-linked and hybridized to a random-primed 32P-labeled cDNA probe for murine COX-2 in QuickHyb solution (Amersham Pharmacia) for 1 h at 37°C. After hybridization, the membranes were washed 2 times in 2× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7) and 0.5% SDS for 15 min each at room temperature and for 20 min in 0.1× SSC and 0.5% SDS at 65°C. Washed membranes were then subjected to autoradiography. For loading normalization, Northern membranes were stripped and rehybridized with a cDNA for GAPDH. Quantitation was performed on a PhosphorImager (Molecular Dynamics), and all measurements of COX-2 mRNA were corrected to the internal standard GAPDH and are reported as ratios.
Western Analysis.
NIH 3T3 cells were treated with PGE2 or PGE3 at various concentrations and for various times as described in the figure legends. After the treatment with PG, the cells were washed once with PBS and lysed in passive lysis buffer (Promega). Fifteen micrograms per sample was electrophoresed on an 8% SDS-polyacrylamide gel and then transferred to nitrocellulose membranes. Primary antibodies to COX-2 (Cayman Chemicals) and GAPDH (Chemicon) were used at the manufacturer's recommended dilutions. Secondary horseradish peroxidase-linked anti-rabbit IgG antibody was used at a dilution of 1:12,000. Bands were visualized with an enhanced chemiluminescence detection reagent (Amersham Pharmacia).
Cell Proliferation Assay.
The cell growth assay was performed by using the Cell Titer 96 Aqueous One Solution cell proliferation assay (Promega). NIH 3T3 cells were seeded in 96-well plates at low density (10,000 cells per well) in DMEM, allowed to attach overnight, and made quiescent by a 24-h incubation in 0.5% serum medium. Cells were then treated with different concentrations of PGE2 or PGE3. After 24 h, Cell Titer 96 Aqueous One Solution reagent was added to each well, and the plate was incubated for 4 h after which the absorbance was recorded at 490 nm with a 96-well plate reader.
Transient Transfections.
Murine NIH 3T3 cells at 50–60% confluency in 6-well dishes were transfected with 2 μg of total DNA per well by using Superfect (Qiagen) for 2 h. All DNAs were prepared with Endotoxin-Free Plasmid Preparation kits (Qiagen). All transfections also included 0.1 μg per well of pRL-TK (a plasmid encoding Renilla luciferase, used as a transfection efficiency control; from Promega). After this procedure, the cells were washed once with PBS and incubated overnight in DMEM supplemented with 0.5% heat-inactivated FCS. After 24 h, the cells were treated for 6 h with 20% FBS, PGE2, and PGE3. After the treatment, cells were washed twice with PBS and lysed with 1× Passive Lysis buffer (Promega).
Luciferase Assays.
Luciferase activity in cell lysates was measured with the Dual Luciferase kit (Promega). Relative luciferase activity of purified cell extracts was typically represented as (firefly luciferase value/Renilla luciferase value) × 103.
Other Methods.
PGE2 concentrations in the conditioned media were determined with a PGE2 EIA kit (Cayman Chemical). IL-6 concentrations in the supernatants were determined by using an ELISA kit (BioSource International). Protein concentrations of cell extracts were measured by using the Bradford reagent (Bio-Rad).
Results
PGE3 Is Not Mitogenic to NIH 3T3 Fibroblasts.
We first compared the effect of PGE2 and PGE3 on cell proliferation. PGE2 at a concentration of 50 ng/ml or greater stimulated proliferation of NIH 3T3 cells. In contrast, PGE3 up to 1 μg/ml did not affect cell proliferation (Fig. 1B).
PGE2 and PGE3 both Induce COX-2 mRNA in NIH 3T3 Fibroblasts.
PGE2-dependent amplification of COX-2 (10–14) is hypothesized to be an important part of sustained proliferative and chronic inflammatory conditions, and may explain the overexpression of COX-2 in tumors (21). We next examined the effect of PGE3 on COX-2 expression. Interestingly, both PGE2 and PGE3 induce COX-2 message in a time- and dose-dependent fashion (Fig. 2). COX-2 message peaks at 1 h after induction with either PGE2 or PGE3 and is detectable at a concentration as low as 50 ng/ml for both PGE2 and PGE3. However, PGE3 is less potent compared with PGE2. At all concentrations tested, PGE2 induced COX-2 up to 4-fold more than PGE3. Furthermore, although both PGE2 and PGE3 also induce the accumulation of COX-2 protein in a time- and dose-dependent manner (Fig. 3), similar to the mRNA, COX-2 protein levels achieved with PGE2 treatment were substantially higher compared to those achieved with PGE3.
Figure 2.
Time course and dose–response of induction for COX-2 mRNA by PGE2 and PGE3. NIH 3T3 fibroblasts were treated for 1 h with different concentrations of PGE2 or PGE3 (A), or with either PGE2 (1 μg/ml) or PGE3 (1 μg/ml) for different time periods (B). Total RNA was isolated, and Northern analysis for COX-2 and GAPDH was performed as described in Experimental Procedures. (Right) Quantified data normalized to GAPDH.
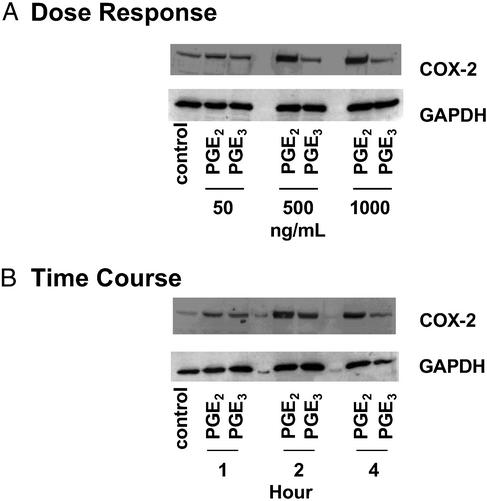
Figure 3.
Time course and dose–response of induction for COX-2 protein by PGE2 and PGE3. NIH 3T3 fibroblasts were treated for 4 h with different concentrations of PGE2 or PGE3 (A), or with either PGE2 (1 μg/ml) or PGE3 (1 μg/ml) for different time periods (B). Total protein was harvested, and 15 μg per condition was subjected to Western blotting with anti-COX-2 antibody as described in Experimental Procedures.
Transcriptional Activation of COX-2 by PGE2 and PGE3.
In transient transfections, using WT COX-2 promoter (−724/+7) linked to a luciferase reporter gene, PGE2 (1 μg/ml) induced COX-2-dependent luciferase activity by >8-fold (Fig. 4A). Although PGE3 also induced COX-2-dependent luciferase activity, it was less effective at inducing COX-2 promoter activity compared to PGE2 (Fig. 4A). Two NF-IL-6 sites and a cyclic-AMP response element (CRE) are present between −724 bp and +7 bp of the COX-2 promoter, and two Ras-mediated signal transduction pathways (Ras/JNK and Ras/ERK) are required for the regulation of COX-2 gene expression in platelet-derived growth factor (PDGF)-treated NIH 3T3 cells (22), mast cells activated by the aggregation of IgE receptors (20), and in MC3T3-E1 cells treated with basic fibroblast growth factor or PDGF (23). We next examined the cis-acting elements and the signal transduction pathways necessary for COX-2 induction by PGE2 and PGE3 in NIH 3T3 fibroblasts. PGE2- and PGE3-dependent luciferase activity was significantly inhibited in COX-2 promoter mutant constructs in which either both NF-IL6 sites were mutated or the CRE site was mutated (Fig. 4B). Mutation of E-BOX or the NFκB site (data not shown) did not affect PGE2- or PGE3-dependent luciferase activity. To determine the signal transduction pathways necessary for COX-2 induction by PGE2 or PGE3, NIH 3T3 cells were cotransfected with a WT COX-2 reporter and with either a control plasmid or plasmids expressing dominant-negative mutants of Ras, JNK, and ERK signal transduction pathways. Similar to the observations made for other ligands in NIH 3T3 cells (19), PGE2- and PGE3-mediated COX-2 gene expression also depends on both Ras/JNK and Ras/ERK signal transduction pathways in NIH 3T3 fibroblasts (Fig. 4C).
Figure 4.
Transcriptional activation of COX-2 by PGE2 and PGE3. NIH 3T3 fibroblasts were transiently transfected, with WT COX-2 reporter construct (A) or with either WT COX-2 promoter or mutant COX-2 promoter constructs (B) or cotransfected with WT COX-2 reporter and a control plasmid or plasmids expressing DN-Ras, DN-JNK, or DN-ERK1 (C). The transfected cells were treated with serum (20%), PGE2, or PGE3 at the concentrations indicated. Cells were harvested after 6 h and assayed for luciferase activity and total protein. Values are means ± SD. *, P < 0.05 vs. PGE2 treatment.
Altering ω-6 Composition of Membrane Phospholipids in NIH 3T3 Fibroblasts Enhances COX-2-Dependent PGE2 Synthesis.
It has been shown that both ω-3 and ω-6 long-chain PUFA added exogenously to cells in culture become part of the membrane phospholipid pool (24, 25). Accumulation of exogenously added PUFA occurs in the phospholipid component of the membranes during the first 8–16 h of exposure, after which there was little or no accumulation. Moreover, ligand-induced PG synthesis in murine fibroblasts and macrophages depends on both the endogenous AA released from membrane phospholipid stores as well as the synthesis and activity of the COX-2 protein (26). To determine whether membrane composition of fatty acids can affect ligand-induced substrate availability and COX-2-dependent PG synthesis, we first incubated NIH 3T3 fibroblasts with AA, EPA, or both. After 24 h, the cells were washed and stimulated with phorbol 12-myristate 13-acetate (PMA) for 6 h to determine COX-2-dependent PG synthesis. Basal PG synthesis was not significantly affected by modifying the membrane phospholipid with AA (10 μM), EPA (10 μM), or both. PGE2 synthesis was significantly increased after PMA stimulation in AA-treated cells (Fig. 5A). However, no such increase was observed in PMA-stimulated cells treated with EPA (Fig. 5A). Interestingly, when cells were treated with AA (10 μM) and EPA (10 μM), the 3-fold increase in PGE2 synthesis observed with AA alone was decreased by >50% (Fig. 5A). To be certain that modification of fatty acid composition of membranes did not affect signals initiated by PMA for induction of COX-2 gene expression, we analyzed COX-2 mRNA levels in NIH 3T3 cells that had been treated with fatty acids. There was no difference in PMA-induced accumulation of COX-2 message after treatment with AA or EPA (Fig. 5B), suggesting that the difference in PGE2 synthesis between the two cell populations was not due to changes in COX-2 expression levels.
Figure 5.
Effect of membrane phospholipid composition on PMA-induced COX-2 mRNA levels and COX-2-dependent PGE2 synthesis in NIH 3T3 cells. Membrane fatty acid composition of NIH 3T3 cells was modified by treating NIH 3T3 cells with either AA (10 μM) or EPA (10 μM), or with both AA (10 μM) and EPA (10 μM) for 24 h. Cells were then stimulated with or without PMA (50 ng/ml) for an additional 6 h. Cells were lysed, supernatants were collected for PGE2 determination (A), and total RNA was isolated for Northern analysis (B). Data are means ± SD from triplicate plates. *, P < 0.05 vs. control with PMA.
Effect of PGE2 and PGE3 on IL-6 Secretion in RAW 264.7 Macrophages.
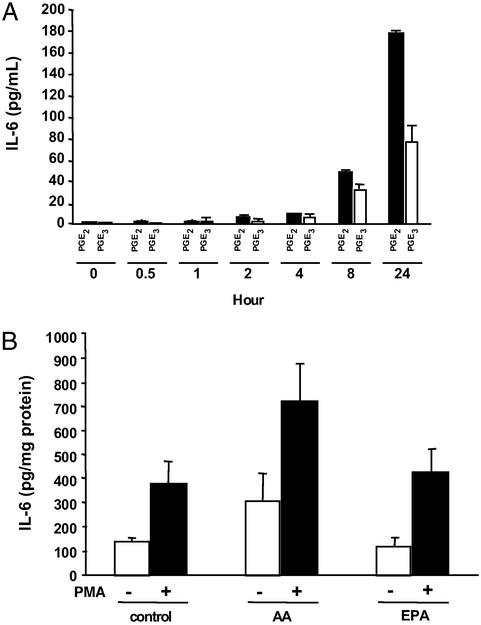
PGE2 modulates production of proinflammatory cytokines by macrophages (27). However, the effect of PGE3 on inflammatory cytokines has not been studied. We first studied the time course of IL-6 secretion after treatment with exogenous PGE2 or PGE3 in RAW 264.7 macrophages. Both PGE2 and PGE3 stimulated the secretion of IL-6 in RAW 264.7 macrophages. However, the effect of PGE3 was, once again, significantly lower than PGE2 in inducing IL-6 secretion in macrophages (Fig. 6A). Moreover, conditioned media from PMA-treated NIH 3T3 cells whose membranes had been modified with AA (as described in Fig. 5A) induced higher levels of IL-6 secretion from target RAW 264.7 macrophages, compared with conditioned media from PMA-treated NIH 3T3 cells whose membranes had been modified with EPA (Fig. 6B).
Figure 6.
(A) Effect of PGE2 and PGE3 on IL-6 secretion in RAW 264.7 macrophages. Macrophages were treated with either PGE2 (50 ng/ml) or PGE3 (50 ng/ml). IL-6 in the supernatants was determined by ELISA at the indicated time points. (B) Conditioned medium from NIH 3T3 fibroblasts treated with AA and stimulated with PMA induces IL-6 secretion in RAW 264.7 macrophages. Membrane fatty acid composition of NIH 3T3 cells was modified by incubating the cells with AA (10 μM) or EPA (10 μM) for 24 h. Exogenous fatty acid was removed by washing the cells with PBS. Cells were stimulated with or without PMA (50 ng/ml) for 6 h, and the conditioned medium was used to treat the RAW 264.7 macrophages for 24 h. IL-6 secretion by the macrophages was measured by ELISA. Each value represents the mean ± SD.
Discussion
PGE2 plays a critical role in both acute and chronic inflammation. The most compelling evidence for the direct role of PGE2 in inflammation came from studies in animal models of inflammation. Selective neutralization of PGE2, in an animal model of Carrageenan-induced paw inflammation, prevented tissue edema and hyperalgesia in affected paws (28). Furthermore, in an adjuvant-induced arthritis model, administration of neutralizing PGE2 antibody reversed edema in the affected paw (28). These observations clearly demonstrate that PGE2 is necessary for the progression of acute and chronic inflammatory conditions in vivo. Several reports have shown that ω-3 fatty acid supplementation can lead to an increased synthesis of 3-series PG in vivo; however, the cellular effects of increasing the synthesis of 3-series PG have not been investigated. Moreover, very few studies have determined the effect of PGE3 on inflammatory processes. In one study, PGE3 had 67% lower oedemogenic effect compared with PGE2 in mice (29); however, the molecular mechanism by which these structurally similar molecules cause differential effects has not been determined.
We have shown that fish oil supplementation can significantly increase the concentration of EPA and DHA in plasma and tissues, thereby affecting the ω-3/ω-6 ratios (30, 31). EPA and DHA can compete and partially replace and/or redistribute AA in a specific time- and dose-dependent manner (24). Once incorporated into cellular membranes, EPA and DHA can compete with AA and reduce substrate availability for 2-series PG synthesis (25). Our data demonstrating that PGE2 synthesis is affected by altering the fatty acid composition of the membranes (Fig. 5A) are consistent with the idea that PG production can be modulated by dietary manipulation of PUFA.
The COX-2 gene is quiescent in most cells, including fibroblasts and macrophages. However, transcription of the COX-2 gene is dramatically activated by a wide variety of ligands, in many cell types (reviewed in ref. 32). In a number of cell and animal models, induction of COX-2 has been shown to promote cell growth, inhibit apoptosis, and enhance cell motility and adhesion. Interestingly, PGE2 has been shown to amplify its own production by inducing COX-2 expression in various cells (10, 14). Four PGE2 receptor subtypes, EP1, EP2, EP3, and EP4 (EP, endorphin), have been identified (33). By using EP-deficient mutant mice, it has been shown that PGE2 signaling is most likely transduced through the G protein-coupled EP2 receptor, which stimulates more production of PGE2 by boosting COX-2 expression through a positive-feedback mechanism (34). Thus, the PGE2 signal through the EP2 receptor may contribute to COX-2 effects observed in chronic inflammatory conditions and in colon tumors from both human and animal models (34).
The effect of PGE3 on COX-2 expression has not been studied. In fact, we originally hypothesized that COX metabolites derived from ω-3 PUFA, such as PGE3, may lack inflammatory and mitogenic properties due to their inability to induce COX-2 expression. Interestingly, although PGE3 is not mitogenic to NIH 3T3 cells, to our surprise, we found that both PGE2 and PGE3 induced COX-2 mRNA via similar signaling mechanisms (Fig. 4). However, at similar concentrations, PGE3 was a weak inducer of COX-2 compared with PGE2. Thus, it seems that although PGE2-mediated COX-2 expression can cause a positive-feedback mechanism of COX-2 induction and PGE2 synthesis (34), PGE3-mediated COX-2 expression can result in a negative-feedback mechanism and may decrease COX-2 induction and PGE2 synthesis. Thus, manipulating PG synthesis could attenuate the induction of COX-2 and the inflammatory response by dietary fatty acids. We have recently shown that a short-term dietary intervention with fish oil supplements in men with prostate cancer leads to a significant increase in the ω-3/ω-6 fatty acid ratios in plasma and adipose tissue (31). Furthermore, although this study included only a small number of subjects, COX-2 expression and PG production in prostatic tissue after the dietary intervention were significantly reduced in some patients (31). The potential for this diet to prevent the development and progression of prostate cancer by way of altered COX-2 expression and PG production in prostatic tissue is under further study.
A number of studies have implicated a positive association between endogenous PGE2 production and release of inflammatory cytokine IL-6 (27, 35–37). Because macrophages are a major source of PGE2 during inflammation and because they have receptors for and respond to PGE2, we conducted experiments to see whether exogenous PGE3 could also play a role in IL-6 synthesis in macrophages. Our data show that macrophages respond to exogenous PGE3 by synthesizing IL-6 albeit to a far lesser extent than PGE2, suggesting that PGE3 is less effective than PGE2 in eliciting inflammatory cytokine production by the macrophages. Our studies suggest that COX-dependent conversion of ω-3 PUFA in inflamed or diseased tissues may result in a subdued inflammatory cytokine production compared with COX-dependent conversion of ω-6 PUFA.
What is the mechanism for the observed differences between the two structurally similar PG? One explanation for the observed results may be that PGE3 is promiscuous and may act through other EP receptors. We have recently completed a collaborative study with Scientists from The Merck Frosst Centre for Therapeutic Research in Montreal, Canada, in which we tested the affinities of PGE2 and PGE3 toward all four human EP receptors (EP1, EP2, EP3-III, and EP4). Interestingly in these studies, Ki values for PGE3 were slightly higher than those for PGE2 for all four EP receptors (data not shown), suggesting that the differential effects between the two structurally similar PG are not due to promiscuous receptor usage but rather may be, in part, due to the differences in their affinities for the EP receptors. Alternatively, another explanation for the observed results may be that PGE2 and PGE3 have short but significantly different enough half-lives, which could lead to apparent differences in their effects.
We conclude that (i) PGE3, unlike PGE2, is not mitogenic to NIH 3T3 fibroblasts; (ii) PGE2 and PGE3 regulate transcription of the COX-2 gene in NIH 3T3 fibroblasts via similar signaling mechanisms; (iii) PGE2 and PGE3 both induce the secretion of IL-6 protein in RAW 264.7 macrophages; and (iv) the differential effects of PGE2 and PGE3 on cell proliferation and inflammation do not depend on their ability to induce COX-2 and IL-6 but rather may depend on the degree of COX-2 and IL-6 induction. Finally, our data suggest that successful replacement of ω-6 PUFA with ω-3 PUFA in cell membranes can result in a decreased cellular response to mitogenic and inflammatory stimuli.
Acknowledgments
We thank Dr. Kathleen Metters at the Merck Frosst Centre for Therapeutic Research (Montreal) for providing the affinities of PGE ligands at the recombinant prostanoid receptors. This work was supported by the Julia and Delfino Fund for Excellence in Breast Cancer Research at the University of California, Los Angeles (UCLA), Jonsson Comprehensive Cancer Center and the Revlon–UCLA Women's Cancer Research Program.
Abbreviations
- AA
arachidonic acid
- EPA
eicosapentaenoic acid
- COX
cyclooxygenase
- PUFA
polyunsaturated fatty acids
- PG
prostaglandins
- DHA
docosahexaenoic acid
- EP
PGE2 receptor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Herschman H R, Xie W, Reddy S. BioEssays. 1995;17:1031–1037. doi: 10.1002/bies.950171207. [DOI] [PubMed] [Google Scholar]
- 2.Herschman H R. Biochim Biophys Acta. 1996;1299:125–140. doi: 10.1016/0005-2760(95)00194-8. [DOI] [PubMed] [Google Scholar]
- 3.Hansen H S. World Rev Nutr Diet. 1983;42:102–134. doi: 10.1159/000408352. [DOI] [PubMed] [Google Scholar]
- 4.Tilley S L, Coffman T M, Koller B H. J Clin Invest. 2001;108:15–23. doi: 10.1172/JCI13416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta S, Srivastava M, Ahmed N, Bostwick D G, Mukhtar H. Prostate. 2000;42:73–78. doi: 10.1002/(sici)1097-0045(20000101)42:1<73::aid-pros9>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 6.Tucker O N, Dannenberg A J, Yang E K, Zhang F, Teng L, Daly J M, Soslow R A, Masferrer J L, Woerner B M, Koki A T, Fahey T J. Cancer Res. 1999;59:987–990. [PubMed] [Google Scholar]
- 7.Hwang D, Scollard D, Byrne J, Levine E. J Natl Cancer Inst. 1998;90:455–460. doi: 10.1093/jnci/90.6.455. [DOI] [PubMed] [Google Scholar]
- 8.Tsujii M, Kawano S, DuBois N. Proc Natl Acad Sci USA. 1997;94:3336–3340. doi: 10.1073/pnas.94.7.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao Y, Prescott S M. J Cell Physiol. 2002;190:279–286. doi: 10.1002/jcp.10068. [DOI] [PubMed] [Google Scholar]
- 10.Minghetti L, Polazzi E, Nicolini A, Creminon C, Levi G. Eur J Neurosci. 1997;9:934–940. doi: 10.1111/j.1460-9568.1997.tb01444.x. [DOI] [PubMed] [Google Scholar]
- 11.Murakami M, Kuwata H, Amakasu Y, Shimabara S, Nakatani Y, Atsumi G, Kudo I. J Biol Chem. 1997;272:19891–19897. doi: 10.1074/jbc.272.32.19891. [DOI] [PubMed] [Google Scholar]
- 12.Wong E, DeLuca C, Boily C, Charleston S, Cromlish W, Denis D, Kargman S, Kennedy B P, Oullet M, Skorey K, et al. Inflamm Res. 1997;46:51–59. doi: 10.1007/s000110050063. [DOI] [PubMed] [Google Scholar]
- 13.Pang L, Hoult J R S. Biochem Pharmacol. 1997;53:493–500. doi: 10.1016/s0006-2952(96)00737-x. [DOI] [PubMed] [Google Scholar]
- 14.Tjandrawinata R R, Dahiya R, Hughes-Fulford M. Br J Cancer. 1997;75:1111–1118. doi: 10.1038/bjc.1997.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lands W E, Libelt B, Morris A, Kramer N C, Prewitt T E, Bowen P, Schmeisser D, Davidson M H, Burns J H. Biochim Biophys Acta. 1992;1180:147–162. doi: 10.1016/0925-4439(92)90063-s. [DOI] [PubMed] [Google Scholar]
- 16.Culp B R, Bradley T G, Lands W E. Prostaglandins Med. 1979;3:269–278. doi: 10.1016/0161-4630(79)90068-5. [DOI] [PubMed] [Google Scholar]
- 17.Connor W E. Am J Clin Nutr. 2000;71,Suppl.:171S–175S. doi: 10.1093/ajcn/71.1.171S. [DOI] [PubMed] [Google Scholar]
- 18.Fischer S, von Schacky C, Schweer H. Biochim Biophys Acta. 1988;963:501–508. doi: 10.1016/0005-2760(88)90318-9. [DOI] [PubMed] [Google Scholar]
- 19.Knapp H R. Prostaglandins. 1990;39:407–423. doi: 10.1016/0090-6980(90)90122-c. [DOI] [PubMed] [Google Scholar]
- 20.Reddy S T, Wadleigh D J, Herschman H R. J Biol Chem. 2000;275:3107–3113. doi: 10.1074/jbc.275.5.3107. [DOI] [PubMed] [Google Scholar]
- 21.Fosslien E. Ann Clin Lab Sci. 2000;30:3–21. [PubMed] [Google Scholar]
- 22.Xie W, Herschman H R. J Biol Chem. 1996;271:31742–31748. doi: 10.1074/jbc.271.49.31742. [DOI] [PubMed] [Google Scholar]
- 23.Wadleigh D J, Herschman H R. Biochem Biophys Res Commun. 1999;264:865–870. doi: 10.1006/bbrc.1999.1606. [DOI] [PubMed] [Google Scholar]
- 24.Hatala M A, Rayburn J, Rose D P. Lipids. 1994;29:831–837. doi: 10.1007/BF02536250. [DOI] [PubMed] [Google Scholar]
- 25.Belury M A, Patrick K E, Locniskar M, Fisher S M. Lipids. 1989;24:423–429. doi: 10.1007/BF02535150. [DOI] [PubMed] [Google Scholar]
- 26.Reddy S T, Herschman H R. J Biol Chem. 1994;269:15473–15480. [PubMed] [Google Scholar]
- 27.Williams J A, Shacter E. J Biol Chem. 1997;272:25693–25699. doi: 10.1074/jbc.272.41.25693. [DOI] [PubMed] [Google Scholar]
- 28.Portanova J P, Zhang Y, Anderson G D, Hauser S D, Masferrer J L, Seibert K, Gregory S A, Isakson P C. J Exp Med. 1996;184:883–891. doi: 10.1084/jem.184.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawkes J S, James M J, Cleland L G. Agents Actions. 1992;35:85–87. doi: 10.1007/BF01990956. [DOI] [PubMed] [Google Scholar]
- 30.Bagga D, Capone S, Wang H, Chap L, Lill M, Heber D, Glaspy J. J Natl Cancer Inst. 1997;89:1123–1131. doi: 10.1093/jnci/89.15.1123. [DOI] [PubMed] [Google Scholar]
- 31.Aronson W J, Glaspy J A, Reddy S T, Reese D, Heber D, Bagga D. Urology. 2001;58:283–288. doi: 10.1016/s0090-4295(01)01116-5. [DOI] [PubMed] [Google Scholar]
- 32.Herschman H R. Cancer Metastasis Rev. 1994;13:241–256. doi: 10.1007/BF00666095. [DOI] [PubMed] [Google Scholar]
- 33.Coleman R A, Smith W L, Narumiya S. Pharmacol Rev. 1994;46:205–229. [PubMed] [Google Scholar]
- 34.Sonoshita M, Takaku K, Sasaki N, Sugimoto Y, Ushikubi F, Narumiya S, Oshima M, Taketo M M. Nat Med. 2001;7:1048–1051. doi: 10.1038/nm0901-1048. [DOI] [PubMed] [Google Scholar]
- 35.Kozawa O, Suzuki A, Tokuda H, Kaida T, Uematsu T. Bone. 1998;22:355–360. doi: 10.1016/s8756-3282(97)00293-7. [DOI] [PubMed] [Google Scholar]
- 36.Millet I, McCarthy T L, Vignery A. J Bone Miner Res. 1998;13:1092–1100. doi: 10.1359/jbmr.1998.13.7.1092. [DOI] [PubMed] [Google Scholar]
- 37.Hinson R M, Williams J A, Shacter E. Proc Natl Acad Sci USA. 1996;93:4885–4890. doi: 10.1073/pnas.93.10.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]