Abstract
FGF8 is known to be an important regulator of forebrain development. Here, we investigated the effects of varying the level of Fgf8 expression in the mouse forebrain. We detected two distinct responses, one that was proportionate with Fgf8 expression and another that was not. The latter response, which led to effects on cell survival, displayed a paradoxical relationship to Fgf8 dosage. Either eliminating or increasing Fgf8 expression increased apoptosis, whereas reducing Fgf8 expression had the opposite effect. To explain these counterintuitive observations, we suggest that an FGF8-dependent cell-survival pathway is negatively regulated by intracellular inhibitors produced in proportion to FGF8 concentration. Our data provide insight into the function of FGF8 in forebrain development and underscore the value of using multiple alleles and different experimental approaches to unravel the complexities of gene function in vertebrate development.
Keywords: FGF inhibitor‖Bmp4‖Foxg1‖sprouty‖cell death
It is now well established that the fibroblast growth factor (FGF) family of intercellular signaling molecules plays a central role in vertebrate embryogenesis. At present, 22 different mouse/human genes are classified as FGF family members because the proteins they encode contain a conserved core sequence of ≈120 aa that includes FGF receptor-binding and heparin-binding domains (1). Most FGFs are secreted proteins that bind to high-affinity receptor tyrosine kinases, leading to the activation of multiple signal transduction pathways, including the RAS/MAPK, PLC-γ, PI3 kinase, and STAT1 pathways (2, 3). One response to FGF receptor activation is the production of antagonists of FGF signaling (4–7). Thus, cellular responses to FGF depend on which signaling pathways are activated and the degree to which they are affected by FGF-induced inhibitors.
Fgf8 is an FGF family member that is essential for normal development of the forebrain. At early stages, Fgf8 is expressed along the apex of the anterior neural ridge (ANR), the rostral-most portion of the neural plate, which contains the progenitors of much of the anterior forebrain (telencephalon; ref. 8). After neural tube closure, Fgf8 expression is localized in a domain that encompasses the rostral midline of the telencephalon (9, 10). Surgical removal of the ANR or treatment of forebrain explants with inhibitors of FGF signaling causes a loss of expression of molecular markers of the telencephalon, and beads soaked in FGF8 protein can prevent this effect (11, 12). Moreover, experimentally changing the level of Fgf8 expression at later stages of forebrain development alters telencephalic patterning, suggesting that FGF8 plays a role in specifying positional information in the developing forebrain (13). Analysis of Fgf8 mutants also demonstrates an important role for FGF8 in forebrain development. Although mouse embryos homozygous for an Fgf8-null allele fail to gastrulate and die without forming organs (14, 15), embryos carrying an Fgf8 hypomorphic allele survive to birth and have telencephalic defects (14, 35). Likewise, zebrafish embryos with reduced Fgf8 function have an abnormal telencephalon, with striking defects at the midline (16, 17).
In this study, we set out to determine the consequences of inactivating Fgf8 in the ANR at an early stage of mouse development by using a Cre-based recombination approach. The results showed that FGF8 is required for cell survival in the telencephalon. However, when we compared the effects of genetically eliminating, genetically reducing, or experimentally increasing Fgf8 expression in the forebrain, we found that cell survival was affected in unexpected ways. It decreased when Fgf8 expression was either eliminated or increased, and increased when Fgf8 expression was reduced. We discuss a possible explanation for this dosage sensitivity based on the hypothesis that antagonist(s) of FGF signaling expressed in response to FGF8 specifically inhibit an Fgf8-dependent cell-survival pathway.
Materials and Methods
Production, Genotyping, and Analysis of Embryos.
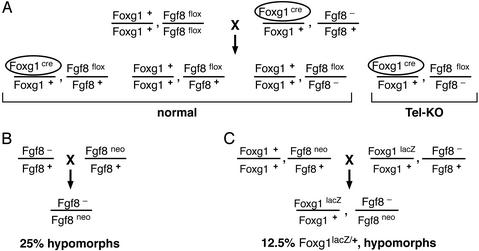
Mutant embryos were generated by the crosses illustrated in Fig. 1. For normal controls, we used littermates that either did not inherit Foxg1cre or inherited a wild-type allele of Fgf8. Noon of the day on which a vaginal plug was observed was considered E0.5. Embryos were genotyped by PCR using DNA from tail or other tissues. Fgf8neo, Fgf8Δ2,3, and cre were detected as described (14, 18). Foxg1lacZ was detected by using primers for lacZ sequences: 5′-GTCTCGTTGCTGCATAAACC-3′ and 5′-TCGTCTGCTCATCCATGACC-3′.
Figure 1.
Crosses used to generate mutant embryos. In these crosses, the Fgf8flox allele has wild-type activity, Fgf8neo is a hypomorphic allele, and Fgf8− is a null allele described as Fgf8Δ2,3 (14). Foxg1cre and Foxg1lacZ are both null alleles.
Wax embedding, sectioning (7 μm), and whole-mount RNA in situ hybridization were performed according to standard protocols by using probes from published sources. Nonradioactive section in situ hybridization was performed by using prehybridization steps essentially as described by Neubüser et al. (19) and posthybridization steps essentially as described by Storm et al. (20). In the whole-mount in situ hybridization assays, two normal and two mutant embryos were analyzed by using the Fgf8-ex2,3 probe (Fig. 2 B and C); between three and five embryos of each genotype were analyzed by using each of the other probes (Foxg1, Bmp4, Fgf8-FL, Spry1, Emx2; Figs. 2 and 3), except that only one Foxg1lacz/+ hypomorph was analyzed by using the Fgf8-FL probe (Fig. 3M). In each experimental group, the results were similar. TUNEL reactions were performed on wax sections by using an in situ cell death kit (Roche Applied Science) and detected with nickel-enhanced diaminobenzidine, according to manufacturer's protocols (Vector Laboratories). For each genotype studied, serial sections from between three and five embryos were assayed by TUNEL.
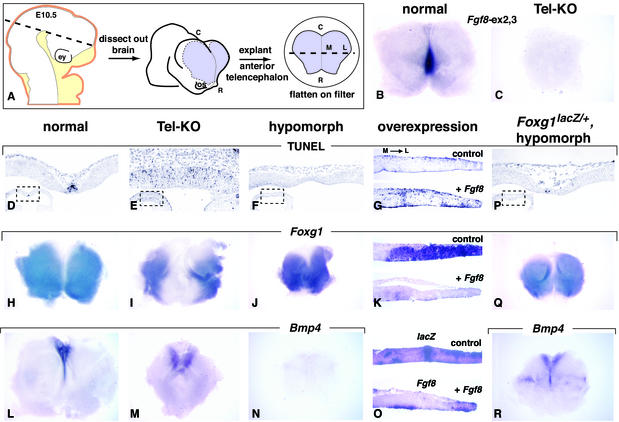
Figure 2.
Response of a Foxg1-dependent cell-survival pathway to different levels of Fgf8 expression. (A) Schematic representation of procedure for isolating E10.5 telencephalic explants. (Left) Surface ectoderm (orange) and craniofacial mesenchyme (yellow), which were dissected away from the neural tube (white). (Center and Right) Forebrain region that was subsequently isolated (purple) and then flattened on a Nucleopore filter. The caudal and rostroventral limits of the explant were the approximate telencephalic/diencephalic border and the optic stalks, respectively. (B and C) RNA in situ hybridization of E10.5 explants using a probe, Fgf8-ex2,3, that hybridizes to sequences deleted by Cre. (D–F and P) TUNEL assays for apoptosis in horizontal sections of E10.5 forebrains of the genotypes indicated. (Lower Left) Low magnification view of the telencephalon, with a dotted box demarcating the area shown at higher magnification. The plane of section is indicated by the dashed line in A (Left). (H–J, L–N, Q, and R) Analysis by whole-mount RNA in situ hybridization of Foxg1 and Bmp4 expression in telencephalic explants from E10.5 embryos of the genotypes indicated. (G, K, and O) Explants from E10.5 wild-type embryos were electroporated with lacZ (control) or Fgf8 expression vectors. Near-adjacent sections of control and of +Fgf8 explants were assayed for TUNEL (G), Foxg1 RNA (K), and lacZ or Fgf8 RNA (O). The representative plane of the section is illustrated by the dashed line in A (Right). C, caudal; ey, eye; L, lateral; M, medial; os, optic stalk; R, rostral.
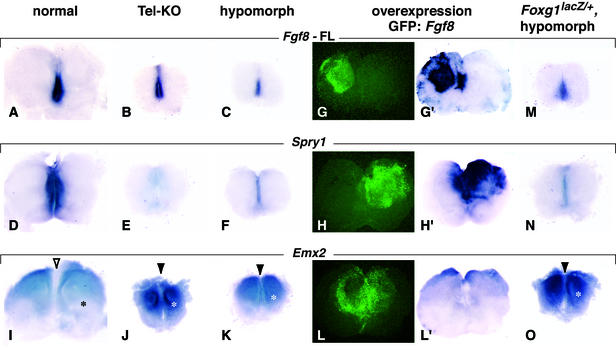
Figure 3.
Response of Spry1 and Emx2 to different levels of Fgf8 expression. Whole-mount RNA in situ hybridization assays for Fgf8, Spry1, and Emx2 expression in telencephalic explants from E10.5 embryos of the genotypes indicated. The full-length Fgf8 probe (Fgf8-FL) hybridizes to Fgf8 sequences not deleted by Cre. The level of Fgf8 RNA in hypomorphs is lower than in Tel-KO mutants, presumably because the Fgf8-neo fusion RNA, which constitutes ≈60% of transcripts produced from the Fgf8neo allele carried by hypomorphs, is unstable (ref. 14; E. Meyers and G.R.M., unpublished results). Black asterisk and open arrowhead (I) indicate regions in which Emx2 expression is low. White asterisks and filled arrowheads (J, K, and O) indicate regions in which Emx2 expression is increased. To increase Fgf8 expression, telencephalic explants from E10.5 wild-type embryos were electroporated with a 1:1 mixture of Fgf8 and Gfp (22) expression vectors. GFP fluorescence was monitored to detect regions in which Fgf8 was ectopically expressed (G and G′). The effects of ectopic Fgf8 expression (H and L) were assayed by using probes for Spry1 and Emx2 (H′ and L′). Note that in some regions these effects extend beyond the cells that express Fgf8, presumably because FGF8 is secreted. In control experiments (Gfp only, data not shown), the expression patterns of Fgf8, Spry1, and Emx2 were similar to those observed in normal embryos (A, D, and I).
Ectopic Gene Expression.
Telencephalic explants were prepared as described in the legend to Fig. 2A and were either fixed immediately for in situ hybridization analysis or electroporated within 1 h of isolation. Expression vectors were generated by cloning an Fgf8 cDNA (isoform 1 in ref. 9 and isoform B in ref. 21), a Spry1 cDNA (from 5′HamSpry1/Ires-neo, generously provided by Lesley Jarvis and Mark Krasnow, Stanford University), or a lacZ cDNA (from an IRES-lacZ plasmid, pDB1, kindly provided by Doris Brown, University of California, San Francisco) into pCAGGS/ES, an expression plasmid containing a chick β-actin promoter and rabbit β-globin poly(A) (22). We also used the Gfp expression vector described by Stuhmer et al. (22). Plasmid DNA for electroporation was diluted to 1.75–2.5 μg/μl in water before use. Electroporation was performed on explants from CD-1 embryos (Charles River Laboratories) dissected at room temperature in Hibernate E (GIBCO/BRL). They were placed individually, mesenchyme side down, on Whatman nucleopore filters (no. 110414, Fisher), which were then floated on fresh Hibernate E. Electroporation was performed as described with minor modifications (22). One microliter of DNA solution was pipetted onto the explant, and six 5-ms pulses at 30 V were applied. After electroporation, each explant on its filter was floated on fresh Hibernate E until all explants had been processed. They were then transferred to culture medium (75% Optimem I/25% HAM F12, supplemented with 1% FBS/40 mM glucose/Glutamax 1/penicillin-streptomycin; GIBCO/BRL) and incubated at 37°C in 5% CO2. After 24 h of culture, the explants were photographed, fixed overnight in 4% paraformaldehyde, and either processed in whole mount or serially sectioned.
Results
Fgf8 is expressed from early stages of forebrain development, first in a group of cells at the rostral boundary of the neural plate and then, following neural tube closure, in the rostral midline of the telencephalon (9, 10). To obtain embryos in which Fgf8 function was eliminated in the telencephalon at early stages (hereafter referred to as Tel-KO mutants; see Fig. 1A), we used Cre-mediated recombination to delete essential Fgf8 sequences (14), thereby circumventing the early lethality of Fgf8-null homozygotes (15). The Cre protein was produced by Foxg1cre, a null allele generated by inserting a Cre recombinase gene into the Foxg1 coding sequence, which is expressed in the telencephalon, olfactory placodes, and midbrain/hindbrain region (23). To determine the extent of Fgf8 inactivation in the forebrain, we assayed telencephalic explants (Fig. 2A) by whole-mount in situ hybridization by using a probe, Fgf8-ex2,3, that detects Fgf8 sequences deleted by Cre (14). Because Fgf8 expression normally commences before Foxg1 (11), we assume that Fgf8 was transiently expressed in Tel-KO mutants at very early stages of telencephalic development. However, at embryonic day (E) 9.25 (data not shown) and E10.5 (Fig. 2 B and C), Fgf8 RNA was not detected in the telencephalon of Tel-KO mutants.
To examine the effect of inactivating Fgf8 on cell survival in the forebrain, we analyzed serial sections of E10.5 embryos by using a TUNEL assay. As reported (24), a high level of apoptosis was detected in the telencephalic midline of normal embryos (Fig. 2D and data not shown). In sections of Tel-KO mutants taken at a similar rostrocaudal level, TUNEL-positive cells were detected in a much wider domain than normal (Fig. 2E and data not shown). This effect was not observed in Foxg1cre/+, Fgf8+/− embryos, indicating that the increase in TUNEL-positive cells was not caused by cre expression (data not shown). Given the observed increase in apoptosis in Tel-KO embryos, we expected to detect increased apoptosis in embryos with reduced Fgf8 expression. However, in Fgf8neo/− embryos (hereafter referred to simply as “hypomorphs”; see Fig. 1B), in which transcripts that code for FGF8 protein are produced at a substantially lower than normal level (14), we observed a decrease rather than an increase in TUNEL-positive cells in the telencephalic midline (Fig. 2F and data not shown). Similar results were obtained at E9.5, when the morphology of the rostral telencephalon was similar in normal embryos, Tel-KO mutants, and hypomorphs (data not shown), suggesting that the differences in cell survival were unlikely to be secondary to morphological differences observed at E10.5 (Fig. 2 D–F).
To determine how increasing Fgf8 expression affects apoptosis, we explanted the telencephalon from normal E10.5 embryos and electroporated it with an Fgf8 expression vector (+Fgf8, n = 4) or a lacZ expression vector (control, n = 3). After 24 h of culture, the explants were serially sectioned. Assays for lacZ and Fgf8 expression identified regions of ectopic gene expression (Fig. 2O). In sections of explants electroporated with Fgf8, there were more TUNEL-positive cells than in control explants (Fig. 2G), a phenotype qualitatively similar to what was observed in Tel-KO embryos (compare Fig. 2 G and E). Thus, either eliminating or increasing Fgf8 expression decreased cell survival, whereas reducing Fgf8 expression had the opposite effect.
Previous studies have provided evidence for a molecular pathway that regulates cell survival in the forebrain. At neural plate stages, FGF8 induces and/or maintains Foxg1 expression in the ANR (11, 12). Foxg1 then functions to restrict the expression of Bmp4 to the midline (25). In turn, BMP4 is thought to induce apoptosis (24). In view of our data showing that FGF8 regulates cell survival in the forebrain, we sought to determine what effect changing the level of Fgf8 expression had on this pathway, hereafter referred to as the Foxg1 pathway. Although Foxg1 expression is detected throughout the ANR at early stages, by E10.5 it is excluded from the midline but is detected at high levels in regions lateral to it (Fig. 2H). In Tel-KO mutants, Foxg1 RNA was excluded from a midline domain that was much wider than normal (Fig. 2I). In hypomorphs, however, the opposite effect was observed: Foxg1 RNA was detected at high levels across the midline (Fig. 2J). Increasing Fgf8 expression by electroporation decreased Foxg1 expression lateral to the midline, as observed in both serial sections (Fig. 2K) and whole-mounts (n = 6; data not shown). This phenotype was qualitatively similar to what we observed in Tel-KO mutants (compare Fig. 2 K and I). We next examined whether changes in Bmp4 expression correlated with these changes in Foxg1 expression. At E10.5, Bmp4 RNA, which is normally expressed in the telencephalic midline (Fig. 2L), was detected in a midline domain that appeared wider than normal in Tel-KO mutants (Fig. 2M). In contrast, Bmp4 expression was not detected in hypomorphs (Fig. 2N). Analysis of serial sections of forebrains from normal, Tel-KO mutants and hypomorphs confirmed that the Foxg1 and Bmp4 expression domains in the midline were complementary and that TUNEL was detected in the Bmp4 expression domain (data not shown). Thus, the data on Foxg1 and Bmp4 expression support the hypothesis that FGF8 regulates cell survival in the telencephalon via the Foxg1 pathway and show that either eliminating or increasing Fgf8 expression decreases Foxg1 pathway activity, whereas reducing Fgf8 expression increases it.
One potential explanation for the opposite Foxg1 pathway phenotypes in Tel-KO mutants and hypomorphs is that Tel-KO embryos are heterozygous for a null allele of Foxg1 (i.e., Foxg1cre), whereas hypomorphs are wild type at the Foxg1 locus. To explore this possibility, we generated hypomorphs heterozygous for Foxg1lacZ (see Fig. 1C), a null allele in which Foxg1 is inactivated by insertion of lacZ (26), and assayed Foxg1 pathway activity. In such embryos, the extent of cell death and the domains of Bmp4 and Foxg1 expression (Fig. 2 P–R) differed from what was observed in Tel-KO mutants (Fig. 2 E, I, and M), suggesting that heterozygosity at the Foxg1 locus is not the reason for the opposite Foxg1 pathway phenotypes. Interestingly, the phenotype of Foxg1lacZ/+ hypomorphs more closely resembled that of normal embryos (Fig. 2 D, H, and L) than of hypomorphs wild type at the Foxg1 locus (Fig. 2 F, J, and N). This finding that the hypomorphic phenotype is suppressed by lowering Foxg1 dosage supports the conclusion that increased midline cell survival in hypomorphs is the result of an increase in Foxg1 pathway activity.
Another potential explanation for the opposite phenotypes is that in Tel-KO mutants, Fgf8 expression is normal throughout development until Cre-mediated recombination occurs, whereas in hypomorphs, it is lower than normal in all tissues throughout development. To determine whether this early reduction of Fgf8 function causes the hypomorphic phenotype, we generated Foxg1cre/+, hypomorphs by using a cross similar to that illustrated in Fig. 1C, except that we used animals carrying a Foxg1cre rather than a Foxg1lacZ allele. Like hypomorphs, these embryos had reduced Fgf8 expression throughout development. However, because they also carried Foxg1cre, they were converted to Tel-KO mutants when the loxP sites in their Fgf8neo allele were recombined to yield a null allele (14). The Foxg1 pathway phenotype of these embryos was qualitatively similar to that of Tel-KO mutants (e.g., increased apoptosis), albeit more severe (not shown). This indicates that early reduction of Fgf8 expression is not the reason why the midline phenotype of hypomorphs is opposite to that of Tel-KO embryos. Moreover, these data show that the phenotype of hypomorphs is not due to a dominant effect of the neo cassette present in the Fgf8neo allele they carry because Foxg1cre/+ hypomorphs, which also carry Fgf8neo, did not resemble hypomorphs.
Significantly, when we assayed mutant embryos for expression of other genes, we found that they did not respond in the same way to changes in Fgf8 level as did the Foxg1 pathway. We first assayed telencephalic explants with a full-length Fgf8 cDNA probe containing sequences not deleted by Cre. The presence of transcripts in the normal Fgf8 expression domain (Fig. 3 A–C) demonstrated that rostral midline tissue is present in both mutants and that FGF8 activity is not necessary to maintain Fgf8 transcription, confirming what has been observed in limb buds (18). Expression of Spry1 (Fig. 3D), which encodes a member of the Sprouty family of FGF signaling inhibitors (4, 5, 27, 28) and is positively regulated by FGF8 (5), was not detected in Tel-KO mutants (Fig. 3E) and was detected at a lower than normal level in hypomorphs (Fig. 3F). As expected, increasing Fgf8 expression by electroporation (Fig. 3 G and G′) induced Spry1 expression (n = 6; Fig. 3 H and H′). Emx2 expression, which is negatively regulated by FGF8 (10), also displayed a proportionate response to variations in Fgf8 expression. Emx2 expression was detected in a larger domain than normal in Tel-KO mutants and hypomorphs (Fig. 3 I–K) and in a smaller domain when Fgf8 was overexpressed (n = 8; Fig. 3 L and L′). Furthermore, unlike the effects on the Foxg1 pathway, effects on the expression of Spry1 and Emx2 were not suppressed when Foxg1 dosage was lowered (Fig. 3 M–O).
These data demonstrate that some responses to FGF8 are proportionate with the level of Fgf8 expression. Why then does the Foxg1 pathway respond as it does, with both elimination and increase in Fgf8 expression causing increased cell survival? One possibility is that the loss-of-function-like phenotypes in the Foxg1 pathway observed following electroporation of Fgf8 are caused by the resulting induction of FGF signaling antagonists such as Spry1. To explore this possibility, we performed experiments similar to those described above with Fgf8 and lacZ expression vectors (Fig. 2) by using a Spry1 expression vector (Fig. 4A). We found that like ectopic Fgf8, ectopic Spry1 expression caused an increase in the level of apoptosis (Fig. 4B, compare with Fig. 2G), which was correlated with a Spry1-induced decrease in Foxg1 expression (Fig. 4C, compare with Fig. 2K).
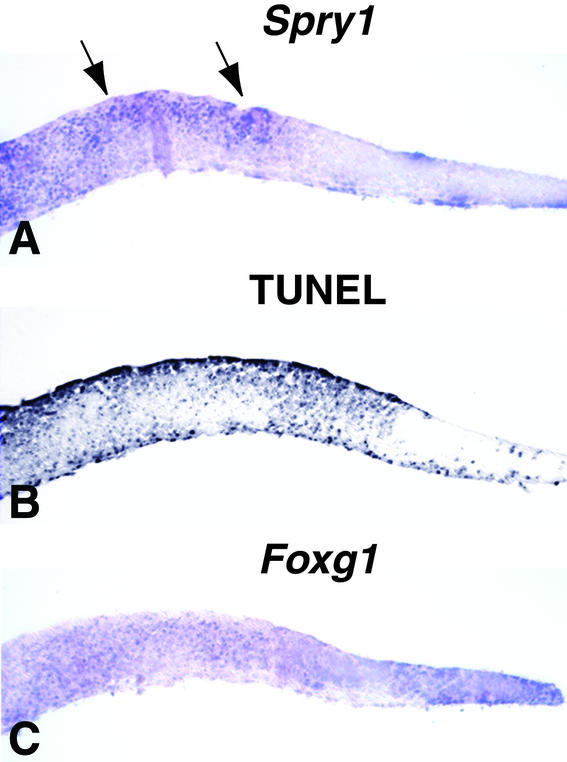
Figure 4.
Effect of ectopic expression of Spry1 on apoptosis and Foxg1 expression. In parallel with the experiments shown in Fig. 2 G, K, and O, explants from E10.5 wild-type embryos were electroporated with a Spry1 expression vector. Near-adjacent sections of each explant were assayed for Spry1 expression (A), TUNEL (B), or Foxg1 expression (C). Arrows in A indicate regions in which cells expressed the electroporated cDNA. Spry1 expression resulted in an increase in TUNEL-positive cells (B, compare with control explant in Fig. 2G) and a decrease in Foxg1 expression (C, compare with control explant in Fig. 2K).
Discussion
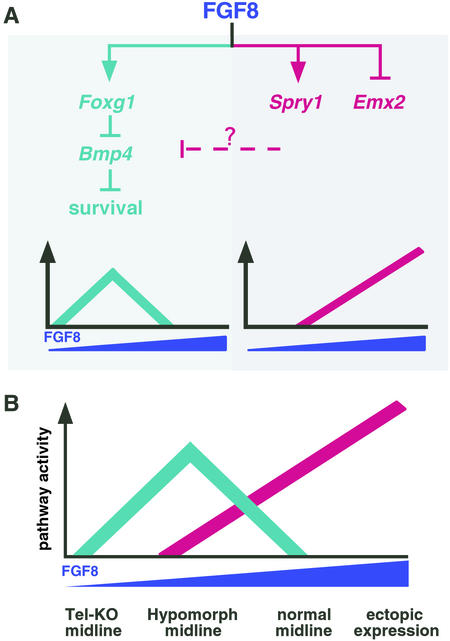
In this study, we compared the phenotypes of a series of forebrains with different levels of FGF8 activity. Fgf8 expression was eliminated in Tel-KO mutants, significantly reduced in hypomorphs that carry both a mild hypomorphic allele and a null allele, and increased in forebrain explants electroporated with an Fgf8 expression vector. The results of our analysis showed that there are two distinct responses to FGF8. One response, which includes effects on Spry1 and Emx2 expression, is proportionate with the level of FGF8 activity (Fig. 5A Right). The second response comprises a Foxg1 pathway that when active leads to cell survival and when inactive leads to apoptosis (Fig. 5A Left). The response of this pathway is not proportionate with the level of FGF8 activity. When FGF8 is either absent or high, pathway activity is low and cell survival decreases. In contrast, when FGF8 is low, pathway activity is high and cell survival increases.
Figure 5.
A model to explain the effects of varying Fgf8 dosage on cell survival. Our data identify two classes of response to variations in Fgf8 expression in the developing mouse forebrain, schematically illustrated in A. The x and y axes represent FGF8 concentration and pathway(s) activity, respectively. One response is proportionate with the level of Fgf8 expression (Right), and the other, a Foxg1 pathway, is not (Left). We suggest that genes, expressed as part of the proportionate response, such as Spry1, encode inhibitors of the Foxg1 cell-survival pathway and that their expression is not subject to negative feedback. (B) Schematic diagram illustrating the combined responses to variation in FGF8 concentration, in which it is assumed that the proportionate response is activated at a higher level of FGF8 than is the Foxg1-dependent response. In the absence of FGF8 (Tel-KO midline), the Foxg1 pathway is inactive and cell survival is reduced. At very low FGF8 concentrations, there is little or no inhibitor activity, so Foxg1 pathway activity is not antagonized and cell survival is high. We suggest that the FGF8 concentration is within this range in the midline of hypomorphs. As FGF8 concentration rises, the inhibitor(s) produced as part of the proportionate response becomes active and begins to inhibit the Foxg1 pathway. Further increases result in more inhibitor activity and decreased cell survival. We suggest that the FGF8 concentration is within this range in the normal midline and in our gain-of-function experiments.
We have considered various explanations for why the Foxg1 pathway responds in this curious fashion and have ruled out heterozygosity at the Foxg1 locus in Tel-KO mutants and reduced levels of functional Fgf8 RNA throughout development in hypomorphs. Thus, we favor a model in which the observed Foxg1 pathway phenotypes result from different levels of FGF8 activity in the telencephalic midline. To explain this dosage sensitivity, we suggest that an FGF8-induced inhibitor(s) produced as part of the proportionate response is induced at a higher threshold of FGF8 than the Foxg1 pathway. Once induced, the inhibitor(s) specifically blocks the Foxg1 cell-survival pathway, but not its own expression. According to this model (Fig. 5B), when FGF8 is low (as in hypomorphs), little or no inhibitor is produced, resulting in high Foxg1 expression, low Bmp4 expression, and high levels of cell survival. When FGF8 is high (following electroporation), inhibitor levels are high enough to block the pathway, resulting in low Foxg1 expression, high Bmp4 expression, and decreased cell survival.
We do not know the identity of the proposed Foxg1 pathway-specific inhibitor(s). However, members of the Sprouty family are good candidates because Sprouty genes are expressed in the midline of the telenecephalon (5) (Fig. 3D and data not shown), they are induced by FGF signaling (Fig. 3H′; refs. 4, 5, and 27), and they function intracellularly as pathway-specific FGF-signaling antagonists (28–31). Furthermore, sprouty in Drosophila has been implicated as an inhibitor of cell-survival pathways (32). Because our data show that Spry1 expression in the telencephalon is regulated by FGF8 (Fig. 3 E, F, and H′), SPROUTY1 may play a role in determining the level of cell survival in response to FGF8. In support of this hypothesis, electroporation of a Spry1 expression vector inhibits Foxg1 expression and induces cell death in forebrain explants (Fig. 4).
Although FGF8 has been found to be essential for cell survival in other developmental settings, including the first branchial arch (33), limb bud (18, 34), and midbrain/hindbrain boundary region (C. L. Chi, S. Martinez, W. Wurst, and G.R.M., unpublished results), there has been no indication from those studies of the complex relationship between cell survival and Fgf8 dosage reported here. This might be because a mechanism that is less sensitive to inhibition by intracellular inhibitors is used to regulate cell survival in those regions. Our observations illustrate how variation in gene dosage can lead to very different phenotypes and point to the importance of understanding how pathways downstream of signaling molecules such as FGF8 are integrated during development.
Acknowledgments
We thank Jean Hébert and Susan McConnell for providing the Foxg1cre/+ mouse line before publication, and Esang Lai for Foxg1lacZ/+ mice. We also thank Salvador Martinez for employing his artistic talents to create the diagrams in Fig. 2A. We thank E. Lai, B. Hogan, and A. Simeone for providing plasmids from which probes for in situ hybridization were prepared, and Christina Petersen, Zenaida Serrano, Jon Watanabe, and Riitta Rantala for excellent technical assistance. We also thank Jean Hébert, Holly Ingraham, Mark Lackner, Barbara Panning, and our colleagues in the Martin laboratory for their insightful comments on the manuscript. E.E.S. was the recipient of a postdoctoral fellowship from the Leukemia Society of America. This work was supported by National Institutes of Health Grants R01 CA78711 (to G.R.M.) and RO1 NS34661 and K02 MH01046 (to J.L.R.R), as well as by funds from Nina Ireland.
Abbreviations
- FGF
fibroblast growth factor
- ANR
anterior neural ridge
References
- 1. Ornitz, D. M. & Itoh, N. (2001) Genome Biol.2, http://genomebiology.com/1465-6906/2/REVIEWS3005. [DOI] [PMC free article] [PubMed]
- 2.Sahni M, Raz R, Coffin J D, Levy D, Basilico C. Development (Cambridge, UK) 2001;128:2119–2129. doi: 10.1242/dev.128.11.2119. [DOI] [PubMed] [Google Scholar]
- 3.Szebenyi G, Fallon J F. Int Rev Cytol. 1999;185:45–106. doi: 10.1016/s0074-7696(08)60149-7. [DOI] [PubMed] [Google Scholar]
- 4.Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow M A. Cell. 1998;92:253–263. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- 5.Minowada G, Jarvis L A, Chi C L, Neubuser A, Sun X, Hacohen N, Krasnow M A, Martin G R. Development (Cambridge, UK) 1999;126:4465–4475. doi: 10.1242/dev.126.20.4465. [DOI] [PubMed] [Google Scholar]
- 6.Furthauer M, Lin W, Ang S L, Thisse B, Thisse C. Nat Cell Biol. 2002;4:170–174. doi: 10.1038/ncb750. [DOI] [PubMed] [Google Scholar]
- 7.Tsang M, Friesel R, Kudoh T, Dawid I B. Nat Cell Biol. 2002;4:165–169. doi: 10.1038/ncb749. [DOI] [PubMed] [Google Scholar]
- 8.Cobos I, Shimamura K, Rubenstein J L, Martinez S, Puelles L. Dev Biol. 2001;239:46–67. doi: 10.1006/dbio.2001.0423. [DOI] [PubMed] [Google Scholar]
- 9.Crossley P H, Martin G R. Development (Cambridge, UK) 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- 10.Crossley P H, Martinez S, Ohkubo Y, Rubenstein J L. Neuroscience. 2001;108:183–206. doi: 10.1016/s0306-4522(01)00411-0. [DOI] [PubMed] [Google Scholar]
- 11.Shimamura K, Rubenstein J L. Development (Cambridge, UK) 1997;124:2709–2718. doi: 10.1242/dev.124.14.2709. [DOI] [PubMed] [Google Scholar]
- 12.Ye W, Shimamura K, Rubenstein J L, Hynes M A, Rosenthal A. Cell. 1998;93:755–766. doi: 10.1016/s0092-8674(00)81437-3. [DOI] [PubMed] [Google Scholar]
- 13.Fukuchi-Shimogori T, Grove E A. Science. 2001;294:1071–1074. doi: 10.1126/science.1064252. [DOI] [PubMed] [Google Scholar]
- 14.Meyers E N, Lewandoski M, Martin G R. Nat Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 15.Sun X, Meyers E N, Lewandoski M, Martin G R. Genes Dev. 1999;13:1834–1846. doi: 10.1101/gad.13.14.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shanmugalingam S, Houart C, Picker A, Reifers F, Macdonald R, Barth A, Griffin K, Brand M, Wilson S W. Development (Cambridge, UK) 2000;127:2549–2561. doi: 10.1242/dev.127.12.2549. [DOI] [PubMed] [Google Scholar]
- 17.Shinya M, Koshida S, Sawada A, Kuroiwa A, Takeda H. Development (Cambridge, UK) 2001;128:4153–4164. doi: 10.1242/dev.128.21.4153. [DOI] [PubMed] [Google Scholar]
- 18.Sun X, Mariani F, Martin G R. Nature. 2002;418:501–508. doi: 10.1038/nature00902. [DOI] [PubMed] [Google Scholar]
- 19.Neubüser A, Koseki H, Balling R. Dev Biol. 1995;170:701–716. doi: 10.1006/dbio.1995.1248. [DOI] [PubMed] [Google Scholar]
- 20.Storm E E, Kingsley D M. Development (Cambridge, UK) 1996;122:3969–3979. doi: 10.1242/dev.122.12.3969. [DOI] [PubMed] [Google Scholar]
- 21.MacArthur C A, Lawshe A, Xu J, Santos-Ocampo S, Heikinheimo M, Chellaiah A T, Ornitz D M. Development (Cambridge, UK) 1995;121:3603–3613. doi: 10.1242/dev.121.11.3603. [DOI] [PubMed] [Google Scholar]
- 22.Stühmer T, Anderson S A, Ekker M, Rubenstein J L. Development (Cambridge, UK) 2002;129:245–252. doi: 10.1242/dev.129.1.245. [DOI] [PubMed] [Google Scholar]
- 23.Hébert J M, McConnell S K. Dev Biol. 2000;222:296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- 24.Furuta Y, Piston D W, Hogan B L. Development (Cambridge, UK) 1997;124:2203–2212. doi: 10.1242/dev.124.11.2203. [DOI] [PubMed] [Google Scholar]
- 25.Dou C L, Li S, Lai E. Cereb Cortex. 1999;9:543–550. doi: 10.1093/cercor/9.6.543. [DOI] [PubMed] [Google Scholar]
- 26.Xuan S, Baptista C A, Balas G, Tao W, Soares V C, Lai E. Neuron. 1995;14:1141–1152. doi: 10.1016/0896-6273(95)90262-7. [DOI] [PubMed] [Google Scholar]
- 27.Furthauer M, Reifers F, Brand M, Thisse B, Thisse C. Development (Cambridge, UK) 2001;128:2175–2186. doi: 10.1242/dev.128.12.2175. [DOI] [PubMed] [Google Scholar]
- 28.Nutt S L, Dingwell K S, Holt C E, Amaya E. Genes Dev. 2001;15:1152–1166. doi: 10.1101/gad.191301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casci T, Vinos J, Freeman M. Cell. 1999;96:655–665. doi: 10.1016/s0092-8674(00)80576-0. [DOI] [PubMed] [Google Scholar]
- 30.Gross I, Bassit B, Benezra M, Licht J D. J Biol Chem. 2001;276:46460–46468. doi: 10.1074/jbc.M108234200. [DOI] [PubMed] [Google Scholar]
- 31.Impagnatiello M A, Weitzer S, Gannon G, Compagni A, Cotten M, Christofori G. J Cell Biol. 2001;152:1087–1098. doi: 10.1083/jcb.152.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bergmann A, Agapite J, McCall K, Steller H. Cell. 1998;95:331–341. doi: 10.1016/s0092-8674(00)81765-1. [DOI] [PubMed] [Google Scholar]
- 33.Trumpp A, Depew M J, Rubenstein J L, Bishop J M, Martin G R. Genes Dev. 1999;13:3136–3148. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moon A M, Capecchi M R. Nat Genet. 2000;26:455–459. doi: 10.1038/82601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garel S, Huffman K J, Rubenstein J L R. Development. U.K.: Cambridge; 2003. , in press. [DOI] [PubMed] [Google Scholar]