Abstract
Methylation of lysine-79 (K79) within the globular domain of histone H3 by Dot1 methylase is important for transcriptional silencing and for association of the Sir silencing proteins in yeast. Here, we show that the level of H3-K79 methylation is low at all Sir-dependent silenced loci but not at other transcriptionally repressed regions. Hypomethylation of H3-K79 at the telomeric and silent mating-type loci, but not the ribosomal DNA, requires the Sir proteins. Overexpression of Sir3 concomitantly extends the domain of Sir protein association and H3-K79 hypomethylation at telomeres. In mammalian cells, H3-K79 methylation is found at loci that are active for V(D)J recombination, but not at recombinationally inactive loci that are heterochromatic. These results suggest that H3-K79 methylation is an evolutionarily conserved marker of active chromatin regions, and that silencing proteins block the ability of Dot1 to methylate histone H3. Further, they suggest that Sir proteins preferentially bind chromatin with hypomethylated H3-K79 and then block H3-K79 methylation. This positive feedback loop, and the reverse loop in which H3-K79 methylation weakens Sir protein association and leads to further methylation, suggests a model for position-effect variegation.
In eukaryotic cells, DNA is packaged along with histones and other nuclear proteins to form chromatin. Cytologically, chromatin can be broadly classified into condensed heterochromatin and decondensed euchromatin (1–4). In general, heterochromatin is associated with repetitive elements at telomeric and pericentric chromosomal regions, contains few genes, and replicates late in S phase. However, certain chromosomal regions can be either heterochromatic or euchromatic, depending on developmental or environmental conditions.
Heterochromatin in divergent animal species (3, 4) and the fission yeast Schizosaccharomyces pombe (5) has a characteristic pattern of histone modifications, in which the N-terminal histone tails are virtually nonacetylated and lysine-9 (K9) of histone H3 is methylated. Specific nonhistone proteins (e.g., HP1) associate with nucleosomes in which histone H3 is methylated at lysine-9, thereby providing a physical difference between heterochromatin and euchromatin. In the budding yeast Saccharomyces cerevisiae, telomeric, silent mating-type (HM), and ribosomal DNA (rDNA) loci are considered to be heterochromatic (2, 3). These heterochromatic loci have histones that are essentially nonacetylated (6, 7) but are atypical in that histone H3-K9 is not methylated and proteins such as HP1 do not exist.
In general, DNA in heterochromatin is relatively inaccessible to enzymatic probes and is inert for transcription and recombination, whereas euchromatic DNA is accessible and active for these processes. In Drosophila, translocation of euchromatic genes or integration of reporter genes next to heterochromatin results in variegated expression, a phenomenon in which genetically identical cells stochastically express or repress the gene in a heritable manner (1). Like position-effect variegation in Drosophila, genes inserted into S. cerevisiae (8) and S. pombe (9) heterochromatic regions are transcriptionally silenced. These resulting epigenetic on or off states can be transmitted through many cell divisions, although they are metastable and can be switched at low frequency.
Sir (silent information regulator) proteins are required for heterochromatic silencing in S. cerevisiae. Sir2, Sir3, and Sir4 associate with and are required for silencing of telomeric and HM loci (10–12), whereas rDNA silencing is mediated by the RENT complex, which contains Sir2 but not Sir3 or Sir4 (13, 14). At telomeres, the DNA-binding protein Rap1 recruits a Sir2–Sir4 complex, perhaps through an interaction with Sir4 (12, 15, 16), whereupon interactions among Sir3, Sir4, and histones H3 and H4 tails result in spreading of the heterochromatic region away from the telomeres (10, 11). Overexpression of Sir3 increases the size of the heterochromatin region and consequently causes silencing at greater distances from the telomeres (11). Sir2, the key silencing protein at all heterochromatic loci, is a NAD-dependent histone deacetylase (17–20) that is likely to maintain the very low level of histone acetylation that is a hallmark of silenced loci.
In addition to the large number of modifications on histone tails, the globular domain of histone H3 is methylated at lysine-79 (K79) in a wide variety of eukaryotic species (21, 22). In S. cerevisiae, H3-K79 methylation is exclusively mediated by Dot1, a histone methylase that lacks a SET domain (21–23), and a human Dot1 homolog carries out the same enzymatic function (24). Dot1 methylates H3-K79 only in the context of intact nucleosomes (21–23), and Dot1-mediated methylation of H3-K79 in vivo strongly depends on Rad6-dependent ubiquitination of histone H2B at lysine-123 (25, 26). Thus, the ability of Dot1 to methylate histone H3-K79 is strongly influenced by nucleosomal structure.
Dot1-mediated methylation of H3-K79 plays an important role in heterochromatic silencing. Deletion or overexpression of Dot1 affects both telomeric and HM silencing, whereas rDNA silencing is affected only by Dot1 overexpression (27). Mutations of histone H3-K79 or mutations that abolish Dot1 catalytic activity impair telomeric and HM silencing (21, 22). Furthermore, Dot1-dependent methylation and H3-K79 itself are important for Sir protein association at telomeres, with the effects being more dramatic at telomere-distal regions than at telomere-proximal regions. Sir protein association with telomeric regions is affected more strongly in strains with a mutated H3-K79 than in strains lacking Dot1 methylase (21, 22). Opposing models suggest that Sir proteins prefer to associate H3-K79 that is methylated (21) or unmethylated (22), the latter model being supported by the high level of H3-K79 methylation in bulk chromatin (22). However, the precise role of H3-K79 methylation in heterochromatic silencing remains unclear, because silencing is impaired after either loss or overexpression of Dot1, and because H3-K79 methylation patterns in vivo have yet to be described. In addition, it is unknown whether H3-K79 methylation patterns are conserved among eukaryotic species.
Here, we examine the distribution of H3-K79 methylation in both S. cerevisiae and mammalian cells. Our results indicate that H3-K79 methylation is a hallmark of euchromatin. Further, they suggest that Sir proteins maintain an epigenetic, silenced state by preferentially binding chromatin containing undermethylated H3-K79 and then blocking H3-K79 methylation. We propose a model for position-effect variegation involving reciprocal positive-feedback loops involving histone methylation and Sir protein association.
Materials and Methods
Yeast Strains and Plasmids.
The plasmid expressing the Myc-tagged histone H4 (28) was kindly provided by M. Nomura (University of California, Irvine), and it was shuffled into yeast strain UCC1111 (21). The yeast strain containing Flag-H2B as the sole copy of histone H2b has been described (26). Derivatives of UC1111 containing sir2∷KanR, sir3∷KanR, sir4∷KanR, and dot1∷KanR deletion alleles were generated by PCR-based gene replacement of the wild-type loci (29). DMY1864 (sir2∷HIS3, pSIR2-LEU2), DMY1865 (sir2∷HIS3, pRS315), DMY1866 (sir2∷HIS3, pSIR2-H364Y-LEU2), DMY1867 (sir2∷HIS3, pSIR2-G262A-LEU2), DMY1928 (HHT2 HHF2, CEN), and DMY1503 (HHT2 hhf2-K16Q, CEN) were kind gifts from D. Moazed (15). For Sir3 overexpression experiments, plasmids pKAN63 (SIR3, 2Β, LEU2) and pJR104 (SIR3, 2μ, URA3), which were obtained from D. Gross (Louisiana State University, Shreveport), and control 2μ plasmids YCplac181 and YCplac195, were introduced into strain FT4 (30).
Mouse Cell Lines.
Two different mouse cell lines were analyzed, a RAG2−/− pro-B cell line (31) and a RAG1−/− p53−/− pro-T cell line (32). These cell lines are maintained in RPMI medium 1640, supplemented with 20% FCS and 50 μM 2-mercaptoethanol.
Chromatin Immunoprecipitation.
Chromatin immunoprecipitation in yeast cells was performed essentially as described (30). Crosslinked chromatin was immunoprecipitated with 2 μl of antibody that specifically recognized a region of dimethylated H3-K79 (21, 24), 2 μl of antibody that was generated against an H4 peptide acetylated at positions 5, 8, 12, and 16 (Upstate Biotechnology, Lake Placid, NY), or 1 μl of affinity-purified anti-Sir3 antibody (kindly provided by D. Moazed). Chromatin immunoprecipitation in mammalian cells was carried out¶ by using 5 μl of the antibody against dimethylated H3-K79. For all experiments, quantitative PCR analyses were performed in real time as described (34, ¶). The data are presented in arbitrary units that are directly related to the apparent immunoprecipitation efficiency (i.e., the amount of material immunoprecipitated relative to that of the input sample); the error for independent determinations is ±20%.
Results
H3-K79 Is Hypomethylated at All Silenced Loci in S. cerevisiae.
We determined the levels of H3-K79 methylation at a variety of genomic loci by chromatin immunoprecipitation. We arbitrarily defined the level of H3-K79 methylation to be 1.0 at two intergenic regions (I and VIII) lacking ORFs. A variety of intergenic and protein-coding regions located 5–20 kb away from the right end of chromosome VI (regions E–L) show comparable levels of H3-K79 methylation (Fig. 1A; values range from 0.8 to 1.5 except for region L, which is 2.5). In contrast, the levels of H3-K79 methylation at four regions (A–D) less than 2.5 kb from the telomeric end are ≈10-fold lower (Fig. 1A). The domain of low H3-K79 methylation corresponds well to the domain of heterochromatin, as defined by telomeric repression (8), inaccessibility to enzymatic probes (35), and Sir protein association (11). Methylation of H3-K79 at the telomere, although 10-fold lower than other genomic regions, is 8- to 10-fold above that observed in dot1 or H3-K79A mutant strains, which represents the experimental background (Fig. 1B). In addition to the telomeric regions, six positions within the silent HMRa locus and three positions within the rDNA locus are hypomethylated at H3-K79, with values ranging from 0.1 to 0.2 (Fig. 1C). Thus, all silenced loci are hypomethylated at H3-K79.
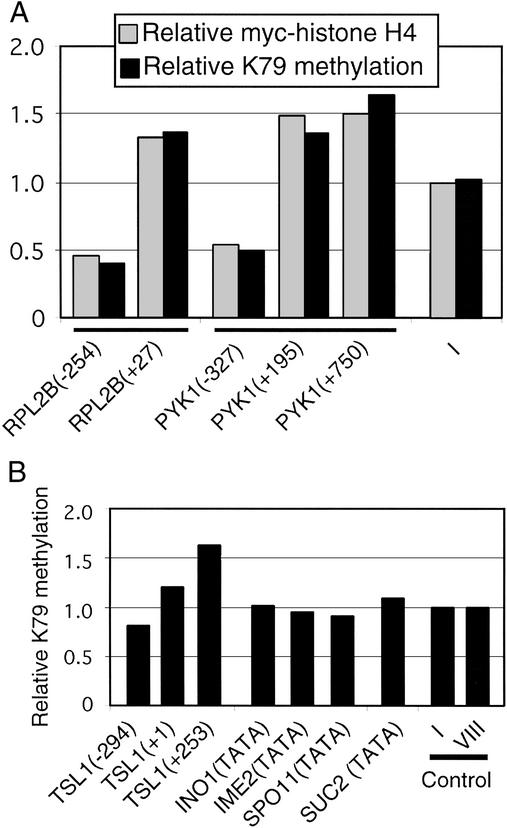
Figure 1.
H3-K79 methylation is low at heterochromatic loci. (A) Relative levels of H3-K79 methylation at regions (A–L) at the indicated positions (drawn to scale) with respect to the telomeres and linked genes on chromosome VI. ORF-free regions on chromosomes I and VIII serve as controls and were arbitrarily defined as having H3-K79 levels of 1.0. (B) H3-K79 methylation in wild-type, dot1, and H3-K79 mutant strains at the indicated regions from the telomere of chromosome VI; note difference in scale. (C) H3-K79 methylation at the indicated regions of HMRa, HMRE, and rDNA loci, along with the ORF-free region controls.
Hypomethylation of H3-K79 Is Associated with Heterochromatic Regions, Not Transcriptional Repression or Histone Deacetylation.
To address directly whether hypomethylation of H3-K79 is specifically associated with Sir-mediated heterochromatin, we analyzed well characterized regions of the yeast genome that are transcriptionally active, inactive, or repressed. At the highly expressed RPL2B and PYK1 genes, protein-coding regions show typical methylated H3-K79 levels, whereas promoter regions show slightly reduced levels (0.4–0.5; Fig. 2A). However, chromatin immunoprecipitation experiments in strains containing Myc-tagged H4 (Fig. 2A) or Flag-tagged histone H2B (data not shown) show that the RPL2B and PYK1 promoter regions have 2- to 3-fold lower levels of histone occupancy. After normalizing to levels of histone occupancy, promoter and protein-coding regions of these active genes have very similar levels of H3-K79 methylation.
Figure 2.
H3-K79 methylation occurs at comparable levels at many nonheterochromatic loci, including those that are transcriptionally repressed and contain deacetylated histones. (A) Relative levels of H3-K79 methylation (black bars) and Myc-tagged histone H4 (gray bars) at the indicated positions of active and control loci. (B) H3-K79 methylation at the indicated genomic regions of inactive or repressed genes.
We also examined H3-K79 methylation at TSL1, an ethanol-induced gene that is inactive in glucose medium, at the SUC2 promoter, which is repressed by the Cyc8-Tup1 corepressor complex, and at three promoters repressed by targeted recruitment of the Rpd3 histone deacetylase complex (INO1, IME2, and SPO11). Owing to the targeted recruitment of the Rpd3 complex, histone acetylation levels at the INO1, IME2, and SPO11 promoters are extremely low and roughly comparable to those observed at silenced loci (7, 36, 37). The SUC2 promoter shows reduced levels of H3, but not H4, acetylation (37, 38). In all these cases, levels of methylated H3-K79 are comparable to those of transcriptionally active or randomly chosen loci (Fig. 2B). Thus, transcriptional repression and histone deacetylation are not sufficient to cause hypomethylation of H3-K79, and such hypomethylation is restricted to heterochromatic loci.
Sir Proteins Are Important for Hypomethylation of H3-K79 at Telomeres and Silent Mating-Type Loci but Not at the rDNA Locus.
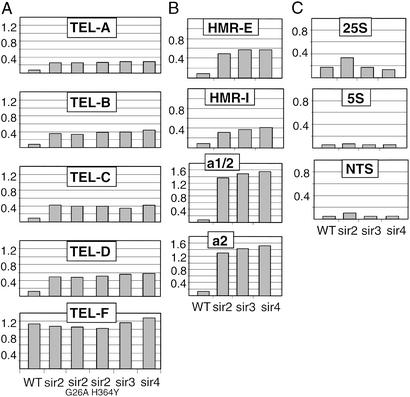
The restriction of H3-K79 hypomethylation to heterochromatic loci strongly implicates Sir silencing proteins as playing an important role. Loss of Sir2, Sir3, or Sir4 results in a 3- to 4-fold increase in H3-K79 methylation at telomeric regions (Fig. 3A) and a 2.5- to 10-fold increase at different regions within the HMRa locus (Fig. 3B). The same effect is observed in strains containing the G262A or H364Y derivatives of Sir2 (Fig. 3A), which abolish the NAD-dependent histone deacetylase activity and strongly reduce Sir protein association at telomeres (15, 39). However, at the four telomeric and at two of the four HMRa regions examined, the level of H3-K79 methylation in sir mutant strains is 2- to 3-fold below that observed at typical genomic regions. This partial Sir dependence of H3-K79 hypomethylation seems to be related to sequences (and hence associated proteins) that recruit the Sir complex to the silenced loci. The HMRa regions (a1/2 and a2) showing complete Sir dependence of H3-K79 methylation are not involved in Sir protein recruitment, whereas the HMRa regions (E and I) showing partial Sir dependence are critical for Sir protein recruitment. Similarly, telomeric region D has a higher level of H3-K79 methylation than regions A–C in a sir2 mutant strain, presumably because it located further from the Rap1 sites at the telomeres. We imagine that proteins involved in Sir recruitment might be responsible for the modest Sir-independent reduction of H3-K79 methylation. In this regard, Dot1 and H3-K79 are more important for Sir protein association at telomere-distal sequences than at telomere-proximal sequences (21, 22). Taken together, these results indicate that the Sir proteins are important but not completely sufficient for hypomethylation of H3-K79 at telomeric and silent mating-type loci.
Figure 3.
Hypomethylation of H3-K79 at the telomeric and HM, but not rDNA, loci depends on Sir proteins and Sir2 histone deacetylase activity. H3-K79 methylation at the indicated regions (see Fig. 1) in wild-type and sir mutant strains for telomeric (A), HM (regions 1, 3, 4, and 6) (B), and rDNA (C) loci.
At the rDNA loci (Fig. 3C), H3-K79 methylation is unaffected by loss of Sir3 or Sir4, a result expected from the fact that Sir3 and Sir4 associate weakly with rDNA and are not required for rDNA silencing. Loss of Sir2 increases H3-K79 methylation ≈2-fold at the 25S and spacer (NTS) regions but does not affect H3-K79 methylation at the 5S region. However, even in the affected regions of rDNA, H3-K79 methylation is significantly below the level at most genomic regions, even though Sir2 is required for rDNA silencing. Although the possibility is untested, we suspect that non-Sir2 components of the RENT complex (Net1 and/or Cdc14) that is required for rDNA silencing are largely responsible for H3-K79 hypomethylation at the rDNA locus. In this regard, Sir2 does not affect H3-K4 methylation at the rDNA locus (40).
Interplay Between H4-K16 Acetylation and H3-K79 Methylation by Means of Their Effects on Sir Protein Association.
Sir2 preferentially deacetylates the histone H4 tail at lysine-16 in vitro (17, 20), and strains containing the K16Q derivative of H4 are defective for heterochromatic silencing and Sir protein association (11, 15, 16). Similarly, loss of Dot1 and mutations of H3-K79 impair telomeric silencing and Sir protein association (21, 22). We therefore investigated the relationship between H4 acetylation and H3-K79 methylation at the telomere.
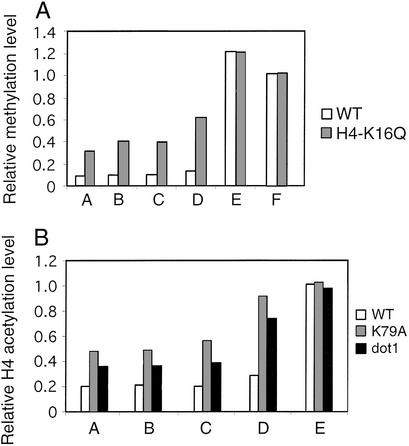
In strains containing the H4-K16Q derivative as the sole source of histone H4, telomeric loci show increased H3-K79 methylation to a roughly comparable extent as observed after loss of Sir2 (Fig. 4A). Conversely, dot1 and H3-K79 mutant strains have increased histone H4 acetylation at telomeres (Fig. 4B; the antibody was made against tetraacetylated H4 and hence measures overall H4 acetylation, not H4-K16 acetylation specifically). As expected from previous results on Sir protein occupancy (21, 22), the effect of the dot1 deletion is less pronounced than that of the H3-K79A mutation. Thus, H3-K79 methylation is important for deacetylation of H4, and H4-K16 acetylation is important for methylation of H3-K79. Presumably, these antagonistic effects are a consequence of both histone modifications on Sir protein association.
Figure 4.
Interplay between H4-K16 acetylation and H3-K79 methylation. (A) H3-K79 methylation at the indicated telomeric regions (see Fig. 1) in wild-type (white bars) and H4-K16Q mutant (gray bars) strains. (B) H4 acetylation (lysines 5, 8, 12, and 16) at the indicated telomeric regions in wild-type (white bars), dot1 (gray bars), and H3-K79 (black bars) mutant strains.
Sir3 Overexpression Extends the Telomeric Domain of Sir Protein Association and H3-K79 Hypomethylation.
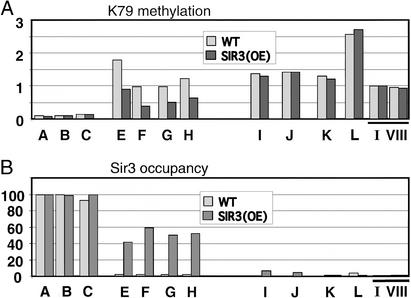
The heterochromatic domain at the telomeric region can be extended by overexpression of Sir3 (11, 12). We addressed whether such an extended heterochromatic domain would also affect the domain of H3-K79 hypomethylation (Fig. 5). As expected, Sir3 association with sequences 5–7 kb from the chromosomal end (fragments E and F) occurs in Sir3-overexpressed strains but not in wild-type strains. Interestingly, these same genomic regions show reduced H3-K79 methylation after Sir3 overexpression. Reduced H3-K79 methylation is not observed at telomere-proximal locations (which are heterochromatic even in wild-type strains) or at regions beyond the extended heterochromatin (as determined by Sir3 occupancy). Thus, Sir3 (and presumably heterochromatic) spreading from the telomere concomitantly results in the spreading of the domain of H3-K79 hypomethylation.
Figure 5.
Overexpression of Sir3 concomitantly extends the telomeric domain of Sir3 and H3-K79 hypomethylation. (A) H3-K79 methylation at the indicated telomeric and control positions (see Fig. 1) in wild-type (white bars) and Sir3-overexpressed (black bars) strains. (B) Relative levels of Sir3 in wild-type (white bars) and Sir3-overexpressed (black bars) strains.
H3-K79 Methylation Is Associated with Active Chromatin Regions in Mammalian Cells.
We examined the pattern of H3-K79 methylation in mammalian cells at loci that are poised to undergo V(D)J recombination, the process by which antigen receptor genes are assembled (41), in a developmentally regulated manner. Specifically, we examined V, D, and J segments at the Ig heavy chain (IgH) and T cell receptor (TCRβ) loci in immortalized pro-B and pro-T cell lines that are developmentally arrested just before the first stage of rearrangement. In pro-B cell lines, D and J segments at the IgH locus are poised to undergo recombination, whereas the Ig VH segments and the TCRβ locus are not. Conversely, D and J segments of the TCRβ locus are recombinationally active in pro-T cell lines, whereas Ig segments are not. Detailed analysis indicates that recombinationally active loci contain high levels of acetylated histone H3 and Brg1, the catalytic subunit of the Swi/Snf nucleosome-remodeling complex.¶ In contrast, recombinationally inert loci show significant levels of histone H3-K9 methylation.¶ Thus, the active and inactive loci have properties consistent with euchromatin and heterochromatin, respectively.
As shown in Fig. 6, high levels of H3-K79 methylation are found exclusively at recombinationally active segments. Conversely, low levels of H3-K79 methylation are found at recombinationally inactive segments. Thus, H3-K79 methylation of Ig and TCR loci depends on the developmental status, and it tracks with euchromatic structure. It should be noted that the regions of euchromatin and heterochromatin at the Ig and TCR loci are large, spanning 200–300 kb, which is roughly the size of the smallest S. cerevisiae chromosome.
Figure 6.
H3-K79 methylation localizes to active gene segments and correlates with H3 acetylation at IgH and TCRβ loci. H3-K79 methylation (fold-enrichment in the immunoprecipitated samples relative to the input) at V, D, J, C (constant), and E (enhancer) regions of the IgH and TCRβ loci, and at CAD, a ubiquitously expressed gene involved in pyrimidine biosynthesis. The gene segments are arranged in 5′ to 3′ orientation as they appear at the endogenous IgH (VH15, VH81X, DQ52, JH1, JH4, Eμ, and Cγ3) and TCRβ (Vβ10, Dβ1, Jβ1.2, Jβ1.5, Cβ1, Eβ, and Vβ14).
Discussion
H3-K79 Methylation Is a Marker of Euchromatin in Yeast and Mammalian Cells.
Although euchromatic and heterochromatic regions of eukaryotic genomes were initially defined by cytological criteria, they can also be defined by their patterns of histone modifications, by accessibility of DNA to enzymatic probes, and by functional properties with respect to transcription and recombination (3, 4). The genome of S. cerevisiae is generally euchromatic, although telomeric, HM, and rDNA regions that are silenced by the Sir proteins are considered to be heterochromatic (2, 3). In S. pombe and mammalian cells, heterochromatin is characterized by low levels of histone acetylation, high levels of H3-K9 methylation, and the presence of nonhistone proteins such as HP1 that recognize methylated K9 (3, 4). Although euchromatin and heterochromatin represent general classes, rather than precise states, of chromatin structure, the distinction is useful.
Methylation of H3-K79 is found in a wide range of eukaryotic species, and Dot1 is conserved between S. cerevisiae and human (21, 24). Here, we show that the genomic pattern of H3-K79 methylation is remarkably similar in yeast and mouse cells. At the mouse IgH and TCRβ loci, methylated H3-K79 is associated with recombinationally active regions that have high levels of acetylated H3 and Brg1 and low levels of methylated H3-K9. In contrast, recombinationally silent regions that have high levels of H3-K9 methylation characteristic of heterochromatin are essentially unmethylated at H3-K79. In S. cerevisiae, H3-K79 methylation occurs at a relatively constant level over many types of genomic regions. However, H3-K79 methylation occurs at a very low but still detectable level at all heterochromatic loci that are silenced by the Sir proteins. Mass spectrometric analysis indicates that 10% of yeast histone H3 is nonmethylated at K79 (22), and it has been estimated that 7–10% of the yeast genome is heterochromatic (42) and associated with Sir proteins (43). These results are consistent with our finding that H3-K79 hypomethylation is restricted to silenced, heterochromatic regions in S. cerevisiae. Thus, in both yeast and mouse, H3-K79 methylation is associated with euchromatin and restricted from heterochromatin.
In S. cerevisiae, hypomethylation of H3-K79 is not simply a consequence of transcriptional repression but rather is restricted to heterochromatin. First, transcriptionally repressed loci, including those that contain very low levels of histone acetylation (e.g., INO1, IME2, and SPO11), show normal levels of H3-K79 methylation. Thus, even though histones within the silenced telomeric, HM, and rDNA regions are essentially deacetylated, histone deacetylation per se is not sufficient for H3-K79 hypomethylation. Second, all mutations tested that decrease Sir protein association and hence disrupt repressive heterochromatin lead to increased levels of H3-K79 methylation. Third, creation of extended telomeric heterochromatin by overexpression of Sir3 results in a corresponding extension of the domain of H3-K79 hypomethylation.
Dot1-mediated methylation of H3-K79 is strongly influenced by chromatin structure. In vitro, Dot1 methylates H3-K79 only in the context of nucleosomes (21–24). In vivo, Dot1 methylation of H3-K79 strongly depends on ubiquitination of H2B-K123 (25, 26), possibly because these two lysine residues on different histones lie in close proximity in the nucleosomal context (26). The relatively constant levels of H3-K79 methylation levels throughout the yeast genome suggest that Dot1 functions in a nontargeted manner to methylate H3-K79 at nucleosomes that have a typical structure. Further, we suggest that nucleosomes containing Sir proteins are poor substrates for Dot1, presumably because the Sir proteins block (directly or indirectly) the nucleosome surface recognized by Dot1. Although the basis of H3-K79 hypomethylation in mammalian cells is unknown, we presume that silencing proteins (e.g., HP1 and Sir protein homologs) create a specialized heterochromatin structure that is refractory to H3-K79 methylation by Dot1 homologs.
Role of H3-K79 Methylation for Association of Sir Proteins.
Methylation of H3-K79 is not just a structural probe that distinguishes euchromatin from heterochromatin but rather plays an important role in Sir protein association and heterochromatic silencing. In S. cerevisiae, loss of Dot1 or alteration of H3-K79 significantly impairs telomeric and HM silencing and Sir protein association at these loci (21, 22). Paradoxically, overexpression of Dot1 also impairs silencing (27) and Sir protein association (22) at these loci. These results are equally consistent with models in which Sir proteins associate preferentially with histone H3 containing methylated (21) or nonmethylated K79 (22). However, the high level of H3-K79 methylation in bulk chromatin is more consistent with association of Sir proteins to nonmethylated K79 (22). To accommodate the similar silencing phenotypes of Dot1 deletion and overexpression, both models propose (in opposite manners) that Sir proteins delocalize from the silenced regions and redistribute around the genome in response to alteration of H3-K79 methylation.
Our observation that H3-K79 hypomethylation at telomeric and HM regions is Sir dependent is inconsistent with the model that Sir preferentially associates with H3 that is methylated at K79. Instead, it suggests that Sir proteins prefer to interact with H3 molecules that are unmethylated (or perhaps monomethylated) at K79. It is likely that the K79 moiety itself contributes to Sir protein association, because alteration of K79 has a more significant effect on Sir protein occupancy than loss of Dot1 (21, 22). Interestingly, alteration of H3 residues in close proximity to K79 also results in silencing defects at all heterochromatic loci (44), and it is likely (although untested) that such mutations reduce Sir protein occupancy. From these observations, we hypothesize that Sir proteins directly interact with a core nucleosomal surface that includes unmethylated K79. Sir3 and Sir4 are likely to play an important role in this interaction at telomeric and HM loci, given their ability to interact with histones H3 and H4 in vitro (10). At the rDNA locus, we suspect that Net1 and/or Cdc14 play an analogous role to Sir3 and Sir4, and indeed, Net1 and Cdc14 should be considered as silencing proteins in the context of the rDNA locus. Given the importance of chromatin structure for Dot1-mediated methylation, this hypothesis provides a simple explanation for why H3-K79 is hypomethylated at heterochromatic loci that are silenced by Sir protein complexes.
H3-K79 Methylation as a Mechanism for Position-Effect Variegation.
Position-effect variegation in flies (1), centromeric position effect in S. pombe (45), and telomeric position effect in S. cerevisiae (8) are phenomena in which a gene in the vicinity of heterochromatin is either expressed or silenced in individual cells. Both the expressed and silenced states are epigenetically inherited through many cell divisions, although switching between the two states occurs at low frequency. Sir proteins and H3-K79 methylation are critical for telomeric silencing, but this is not sufficient to explain how two distinct epigenetic states can be stably maintained in genetically identical cells.
Sir proteins are recruited to telomeres by Rap1 and associated proteins, whereupon they spread away from the telomere by means of interactions among Sir3, Sir4, and histones H3 and H4 tails (11). We propose that Sir proteins maintain an epigenetic state by preferentially binding chromatin containing undermethylated H3-K79 and then blocking H3-K79 methylation (Fig. 7). These two biochemical properties essentially constitute a positive-feedback loop that mutually reinforces the stability of Sir proteins on chromatin with hypomethylated H3-K79. Conversely, the nonsilenced state is governed by a positive-feedback loop in which methylation of H3-K79 weakens Sir protein binding, which subsequently leads to increased H3-K79 methylation and even weaker Sir protein binding. Thus, stochastic or regulated (e.g., by DNA replication or Rad6-mediated ubiquitination of H2B) effects on either Dot1-mediated methylation or Sir protein occupancy will drive individual cells to exist in stable repressed or expressed states. Furthermore, this “mutual reinforcement” hypothesis explains why the normal physiological level of H3-K79 methylation is required for restricting Sir proteins and heterochromatin to specific regions.
Figure 7.
Model for maintaining stable on–off states. Positive-feedback loops for the on and off states involve the preferential recognition of chromatin with hypomethylated H3-K79 by Sir proteins and inhibition of Dot1-mediated methylation of H3-K79 by bound Sir proteins. Stochastic events that affect either Sir protein association or H3-K79 methylation can lead to switching between the two states.
In principle, preferential binding of silencing proteins to specifically modified histones and subsequent inhibition of those histone modifications represent a general mechanism for maintaining stable on or off epigenetic states in genetically identical cells. A similar mechanism for maintaining on–off epigenetic states in S. cerevisiae might also apply to Set1-mediated methylation of histone H3-K4, which is low at telomeres (33, 40), and histone acetylation (10). In addition, H3-K4 is hypomethylated at heterochromatic loci in S. pombe, in a manner that depends on and spreads in conjunction with factors involved in heterochromatin assembly (5). The stability of the epigenetic state would be enhanced considerably if multiple histone modifications were subject to positive-feedback loops. It is tempting to speculate that epigenetic states in mammalian cells involve a similar mechanism, although the silencing proteins might differ. In this regard, H3-K79 methylation is particularly attractive for maintaining epigenetic states, because this enzymatic process is particularly sensitive to nucleosome structure.
Acknowledgments
We thank Danesh Moazed for yeast strains and antibodies, and Masayasu Nomura and David Gross for plasmids. This work was supported by a postdoctoral fellowship from the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation (to H.H.N.) and research grants from the National Institutes of Health to M.A.O. (GM 48026) and K.S. (GM 53720). H.H.N. is on leave from the Department of Biological Sciences, National University of Singapore.
Abbreviations
- HM
mating type
- rDNA
ribosomal DNA
- IgH
Ig heavy chain
- TCR
T cell receptor
Footnotes
K.B.M., D.N.C., S. D. Taverna, C. D. Allis, and M.A.O., unpublished work.
References
- 1.Weiler K S, Wakimoto B T. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- 2.Grunstein M. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- 3.Moazed D. Mol Cell. 2001;8:489–498. doi: 10.1016/s1097-2765(01)00340-9. [DOI] [PubMed] [Google Scholar]
- 4.Richards E, Elgin S C. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- 5.Noma K, Allis C D, Grewal S I. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 6.Braunstein M, Sobel R E, Allis C D, Turner B M, Broach J R. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suka N, Suka Y, Carmen A A, Wu J, Grunstein M. Mol Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- 8.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 9.Grewal S I, Klar A J. Cell. 1996;86:95–101. doi: 10.1016/s0092-8674(00)80080-x. [DOI] [PubMed] [Google Scholar]
- 10.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 11.Hecht A, Strahl-Bolsinger S, Grunstein M. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 12.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 13.Straight A F, Shou W, Dowd G J, Turck C W, Deshaies R J, Johnson A D, Moazed D. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- 14.Shou W, Seol J H, Shevchenko A, Baskerville C, Moazed D, Chen Z W, Jang J, Shevchenko A, Charbonneau H, Deshaies R J. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 15.Hoppe G J, Tanny J C, Rudner A D, Gerber S A, Danaie S, Gygi S P, Moazed D. Mol Cell Biol. 2002;22:4167–4180. doi: 10.1128/MCB.22.12.4167-4180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo K, Vega-Palas M A, Grunstein M. Genes Dev. 2002;16:1528–1539. doi: 10.1101/gad.988802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imai S-I, Armstrong C M, Kaeberlein M, Guarente L. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 18.Landry J, Sutton A, Tafrov S T, Heller R C, Stebbins J, Pillus L, Sternglanz R. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith J S, Brachmann C B, Celic I, Kenna M A, Muhammad S, Starai V J, Avalos J L, Escalante-Semerena J C, Grubmeyer C, Wolberger C, Boeke J D. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanny J C, Moazed D. Proc Natl Acad Sci USA. 2001;98:415–420. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng H H, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Leeuwen F, Gafken P R, Gottschling D E. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 23.Lacoste N, Utley R T, Hunter J, Poirier G G, Cote J. J Biol Chem. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- 24.Feng Q, Wang H, Ng H H, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Curr Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 25.Briggs S D, Xiao T, Sun Z-W, Caldwell J A, Shabanowitz J, Hunt D F, Allis C D, Strahl B D. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- 26.Ng H H, Xu R M, Zhang Y, Struhl K. J Biol Chem. 2002;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- 27.Singer M S, Kahana A, Wolf A J, Meisinger L L, Peterson S E, Goggin C, Mahowald M, Gottschling D E. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keener J, Dodd J A, Lalo D, Nomura M. Proc Natl Acad Sci USA. 1997;94:13458–13462. doi: 10.1073/pnas.94.25.13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longtine M S, McKenzie A, Demarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 30.Kuras L, Struhl K. Nature. 1999;399:609–612. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 31.Kirch S A, Rathbun G A, Oettinger M A. EMBO J. 1998;17:4881–4886. doi: 10.1093/emboj/17.16.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mombaerts P, Terhorst C, Jacks T, Tonegawa S, Sanch J. Proc Natl Acad Sci USA. 1995;92:7420–7424. doi: 10.1073/pnas.92.16.7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernstein B E, Humphrey E L, Erlich R L, Schneider R, Bouman P, Liu J S, Kouzarides T, Schreiber S L. Proc Natl Acad Sci USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mencia M, Moqtaderi Z, Geisberg J V, Kuras L, Struhl K. Mol Cell. 2002;9:823–833. doi: 10.1016/s1097-2765(02)00490-2. [DOI] [PubMed] [Google Scholar]
- 35.Gottschling D E. Proc Natl Acad Sci USA. 1992;89:4062–4065. doi: 10.1073/pnas.89.9.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rundlett S E, Carmen A A, Suka N, Turner B M, Grunstein M. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 37.Deckert J, Struhl K. Mol Cell Biol. 2001;21:2726–2735. doi: 10.1128/MCB.21.8.2726-2735.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu J, Suka N, Carlson M, Grunstein M. Mol Cell. 2001;7:117–126. doi: 10.1016/s1097-2765(01)00160-5. [DOI] [PubMed] [Google Scholar]
- 39.Armstrong C M, Kaeberlein M, Imai S I, Guarente L. Mol Biol Cell. 2002;13:1427–1438. doi: 10.1091/mbc.01-10-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bryk M, Briggs S D, Strahl B D, Curcio M J, Allis C D, Winston F. Curr Biol. 2002;12:165–170. doi: 10.1016/s0960-9822(01)00652-2. [DOI] [PubMed] [Google Scholar]
- 41.Bassing C H, Swat W, Alt F W. Cell. 2002;109:S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 42.Laurenson P, Rine J. Microbiol Rev. 1992;56:543–560. doi: 10.1128/mr.56.4.543-560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lieb J D, Liu X L, Botstein D, Brown P O. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 44.Park J-H, Cosgrove M S, Youngman E, Wolberger C, Boeke J D. Nat Genet. 2002;32:273–279. doi: 10.1038/ng982. [DOI] [PubMed] [Google Scholar]
- 45.Allshire R C, Javerzat J P, Redhead N J, Cranston G. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]