Abstract
Long interspersed nuclear elements 1 (L1) are active retrotransposons that reside in many species, including humans and rodents. L1 elements produce an RNA intermediate that is reverse transcribed to DNA and inserted in a new genomic location. We have tagged an active human L1 element (L1RP) with a gene encoding enhanced GFP (EGFP). Expression of GFP occurs only if L1-EGFP has undergone a cycle of transcription, reverse transcription, and integration into a transcriptionally permissive genomic region. We show here that L1-EGFP can undergo retrotransposition in vivo and produce fluorescence in mouse testis. The retrotransposition event characterized here has occurred at a very early stage in the development of an L1-EGFP transgenic founder mouse.
Keywords: L1 transgenic mice‖enhanced GFP
Long interspersed nuclear elements 1 (L1) are autonomous retrotransposons (1, 2). There are ≈40–80 active L1 elements in the human diploid genome (3, 4). Despite this rather unimpressive number, there are ≈500,000 inactive L1 elements, such that ≈17% of the mass of the genome is comprised of L1 sequences (5). The activity of L1s has contributed significantly to the shaping of the human genome. It is likely that L1 machinery has been “hijacked” by Alu elements, which are present in >1,000,000 copies per haploid genome (6, 7). L1s also contribute to processed pseudogene formation (8, 9). The demonstration that L1s can shuffle exons in cultured cells further highlights their evolutionary significance (10–12). Conversely, mammalian genomes may have co-opted the functions of transposable elements for their maintenance. For example, the enzyme telomerase has significant homology to the L1 reverse transcriptase (13).
Studying L1s is difficult because they are scattered in large numbers throughout the genome. To date, the activity of L1s in vivo has been inferred from the recovery of insertions that have been inherited and cause disease (14). Studies in the mouse have surveyed L1 RNA and ORF1 protein expression in various tissues (15, 16). However, until recently, a direct assay of L1 retrotransposition in mice has been lacking (17). We are interested in determining which tissues and cell types in the mouse are competent to support L1 retrotransposition. To approach this question, we took advantage of a cultured cell assay of retrotransposition (18). Instead of using neomycin phosphotransferase, we used enhanced GFP (EGFP) as a tag to track L1 insertions in vivo (17, 19). EGFP is expressed only when the L1 element undergoes a cycle of transcription, reverse transcription, and integration into a genomic site that permits EGFP transcription (Fig. 1a).
Figure 1.
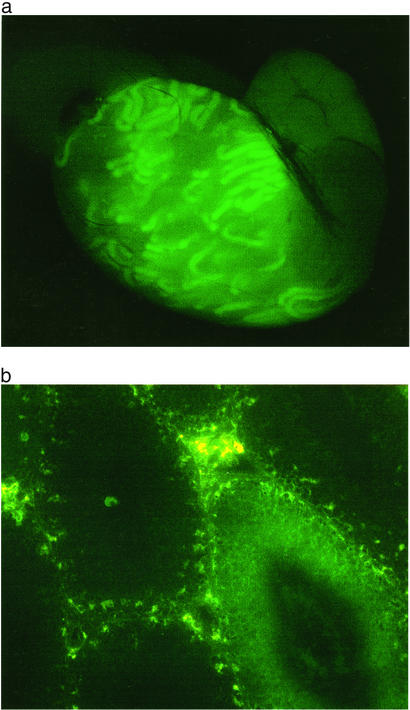
Retrotransposition of L1-EGFP. (a) Schematic of the L1-EGFP transgene and its retrotransposition. L1 transcription is driven by the mouse RNA pol II promoter in addition to the L1 5′ UTR (21). The EGFP gene is in the antisense orientation relative to L1. EGFP (green) is situated in the 3′ UTR (hatched) of L1 and is interrupted by the mouse γ-globin intron. The intron is in the same transcriptional orientation as L1 (18). Therefore, when the L1 sense-strand transcript is processed, the γ-globin intron is spliced out. The EGFP gene is driven by the human CMV MIE promoter (pCMV-MIE) and has an HSV thymidine kinase polyadenylation sequence (tkpA). pCMV-MIE, EGFP, and tkpA are all antisense relative to L1RP. At the very 3′ end of the L1-EGFP transgene is the SV40 late polyadenylation sequence (SV40pA) derived from the pCEP4 cloning vector (Invitrogen). 5′ truncation of the L1-EGFP insertion is depicted with a jagged line. Arrows depict the locations of the geno5 (left) and geno3 (right) genotyping primers used in the PCR assay shown in b (not drawn to scale). (b) The geno5 and geno3 primers flank the intron in EGFP and give rise to two products, a 1.5-kb amplicon (corresponding to the intron-containing transgene) and an ≈600-bp amplicon that lacks the 909-bp intron (corresponding to the insertion). Shown are the genotyping results on tail DNA from five offspring of founder 57 (lanes 1–5). dw, distilled water; neg, genomic DNA from the tail of a transgene negative mouse; tg, L1-EGFP transgene; XIV, 100-bp ladder with bright bands at 500 bp, 1,000 bp, and 2.6 kb (top band; Roche).
Here we document retrotransposition of the L1-EGFP transgene in vivo. On retrotransposition, the EGFP reporter is activated: green fluorescence is observed in the testes of mice that inherit the insertion. Genetic and phenotypic analysis of the mouse in which the insertion arose demonstrates that the insertion occurred at an early stage of development. These and other studies (17) indicate that tagged L1 transgenes may be used to study L1 biology in vivo. These L1-EGFP mice also illustrate an application for tagged L1 transgenes as cell lineage markers.
Materials and Methods
Transgene Construction.
A fragment containing the entire L1RP retrotransposon (20) up to the unique BstZ17I site at position 5964 in the 3′ UTR was cloned into a pBS (KS-) shuttle vector containing a genetically engineered L1.2 3′ UTR (18, 20). A linker (AAAAAAGTATACGTAAAAAAACCCGGGGGGA, gift from Eric Ostertag, University of Pennsylvania, Philadelphia) was cloned into the BstZ17i and XmaI sites (which were preserved in the linker) with a unique SnaB1 site corresponding to position 5967 in the L1 3′ UTR (18). The EGFP marker [cytomegalovirus (CMV)-EGFPint-tkpA] was inserted via the SnaB1 and XmaI sites of the linker in JCC5-RPs-SnaB1. The RP-EGFP fragment was directionally cloned into a pCEP4 derivative (pol II-RJD99, a gift from Ralph DeBerardinis, University of Pennsylvania) that contained the mouse RNA polymerase II (large subunit) promoter (pol II) (21). A negative control construct was cloned by swapping part of RP with JM111. JM111 contains a human L1 element with mutations in ORF1 that abrogate retrotransposition activity (18). To clone JM111-RP-EGFP, JM111 and CMV-RP-EGFP (19) plasmids were isolated from dam-dcm- bacteria. This allowed us to use the (normally methylated) unique BclI site in L1 to swap the downstream portion of L1 and the EGFP marker from CMV-RP-EGFP into pJM111, creating a hybrid but nonfunctional L1 element. The transgene and negative control constructs were tested in a transient cell culture assay for EGFP expression (19). A similar construct has been used to make transgenic mice (JM111-EGFP) that lack genetically detectable retrotransposition events (17). The L1-EGFP transgene was liberated from the pCEP4 backbone by combined SalI + MluI digestion and agarose gel electrophoresis, band purified (Gene Clean, Bio 101) followed by an Elutip column (Schleicher & Schuell). Three lines of L1-EGFP transgenic mice were produced (lines 57, 59, and 63). These transgenic mouse studies were approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
PCR Primers.
The primers, sequences, and uses are as follows: 6896rev, TATATCTCCCAATGCTATCC, inverse PCR (IPCR); SV40for, ATGATAAGATACATTGATGAGTTTGGA, IPCR; SV40rev, TCCAAACTCATCAATGTATCTTATCAT, RT-PCR; HSVtkrev, AAGGCGATGCGCTGCGAATCGG, RT-PCR; geno5, TTTATTGCCGATCCCCTCAGAAGAA, genotyping; geno3, TTCAAGGACGACGGCAACTACAAGA, genotyping; 6032for, AACACCCGTGCGTTTTATTC, intron PCR; and 6543rev, CAGCCCAGTTAGTCCTCTGC, intron PCR.
IPCR.
Five to 10 μg of genomic DNA were digested for 4 h with SspI, extracted with phenol then chloroform, and subjected to overnight ligation in a 0.6-ml volume. The ligation products were reextracted, ethanol precipitated, and subjected to PCR amplification by using inverse IPCR primers situated just upstream of the SV40 polyA signal (SV40for) and just downstream of the EGFP cassette (6896rev). IPCR was carried out with the Expand Long kit using buffer system 1 following the manufacturer's directions (Roche, Nutley, NJ). The 4.3-kb amplicon was gel purified (Bio101; Qbiogene, Montreal) and cloned into pCR2.1 (Invitrogen). Portions of the cloned amplicon were sequenced (Fig. 2).
Figure 2.
Nucleotide sequence of the L1-EGFP insertion flanks. The genomic sequences flanking the L1-EGFP insertion are lowercase. L1-EGFP sequences adjoining the flanks are given in uppercase. The 5′ flanking sequence is shown in the top portion of the figure, followed by the 3′ flanking sequence in the bottom. Each flank is numbered separately. The complete L1-EGFP sequence (between the 5′ and the 3′ flanks) is not shown. The discontinuity corresponding to the L1-EGFP sequence is denoted by three asterisks at the end of the 5′ flank. Target site duplications (TSD) are in bold. The thymine at position 422, just downstream of the 5′ TSD, is not present in the L1RP sequence at that position. A nucleotide blast search of the mouse genome database using sequences flanking the L1-EGFP insertion suggests that the insertion is on Mus musculus chromosome 4 (http://www.ncbi.nlm.nih.gov).
RT-PCR.
RNA was extracted from mouse tissues using an RNAeasy midi kit (Qiagen, Chatsworth, CA) and digested with RNase-free DNAse1 (Roche). cDNA synthesis was performed with AMV reverse transcriptase following the manufacturer's instructions (Roche). The SV40rev oligonucleotide was used to produce L1 sense-strand cDNA (from the transgene), whereas the HSVtkrev oligonucleotide was used to produce EGFP cDNA (from the insertion). Each 50-μl PCR contained 5 μl of 10× PCR buffer (Roche buffer 1 with 15 mM MgCl2), 1 μl of 10 mM dNTPs, 200 ng of each oligonucleotide, 0.3 μl of AmpliTaq Gold, and 5 μl of reverse transcriptase extension product. Amplifications were performed on a Peltier thermal cycler (Hybaid, Asford Middlesex, U.K.) by using the following program: 95°C for 15 min, followed by 40 cycles of 95°C for 25 sec, 62°C for 30 sec, and 72°C for 90 sec. This was followed by a final extension at 72°C for 10 min and a 4°C hold. The following tissues were surveyed for L1 sense-strand transcripts in L1-EGFP line 57 transgenic mice: testis, pooled ovaries, spleen, liver, and brain. Testis, pooled ovary, liver, and lung were surveyed in L1-EGFP line 59 transgenics. Testis and lung were surveyed in line 63 L1-EGFP transgenics. EGFP transcripts were surveyed in testis, spleen, liver, and brain of mice with the insertion.
Genotyping PCR.
Genotyping PCR was carried out with the geno5 and geno3 primers, 250 ng of tail DNA, and a mix like the one described for RT-PCR. Amplification conditions were 94°C for 15 min, followed by 35–40 cycles of 94°C for 30 sec, 58°C for 30 sec, 72°C for 90 sec. This was followed by a final extension at 72°C for 5 min and a 4°C hold. Tail DNA from Fo57 offspring with the insertion band by genotyping PCR were separately amplified by using intron primers (and the same amplification conditions) to confirm the presence (or absence) of the transgene.
Primary Cell Cultures.
A 5 × 5-mm portion of the inner aspect of the earlobe was harvested, washed three times in PBS, cut into smaller pieces, and compressed with the ribbed plunger of a syringe. Macerated tissue fragments were transferred to a sterile 15-ml conical tube, pelleted by centrifugation, and digested in 0.25% trypsin at 37°C for 1.5 h. The trypsin was neutralized with FCS, and the cells were pelleted and resuspended in 10 ml of fibroblast medium (DMEM with 4.5 g of glucose per liter and 10% FCS supplemented with penicillin and streptomycin). Cells were cultured in T25 flasks at 37°C in a humidified atmosphere with 7.5% CO2. Tumor cells were isolated for culture by using a similar method, except that there were no washing steps before trypsin digestion, and trypsin digestion was carried out for ≈30 min.
Production of Ear Fibroblast Subclones.
Fibroblasts were harvested by brief trypsinization, neutralized with serum, pelleted, and resuspended in fibroblast medium. The cell suspension was serially diluted and cultured in 96-well flat-bottom plates (Nunc). Cultures were evaluated for cell growth by light microscopy. Two groups of clones were used for analysis. One group consisted of eight subclones plated at a dilution at which at least two-thirds of the wells did not contain viable cells. A second group consisted of 11 wells that contained only one visible focus of cells. Lysates that typed negative by L1-EGFP genotyping PCR had amplifiable DNA by PCR for glyceraldehyde phosphate dehydrogenase.
Lysates for PCR Analysis.
Ear fibroblast cultures and blastocysts were subjected to hypotonic lysis for 10 min at 80°C and overnight digestion with proteinase K at 55°C. Digested lysates were boiled for 15 min to inactivate the proteinase K and were stored at –20°C until PCR was performed. Five microliters of lysate were used in 25–50 μl of PCR.
Fluorescence Microscopy.
Stereofluorescence microscopy was performed with a Leica MZFLIII stereofluorescent dissecting microscope, using a 100-W mercury bulb and the following filters: exciter HQ470/40 and emitter 515 nm LP (Chroma Technology, Brattleboro, VT). Images were captured by using a MagnaFire charge-coupled device (CCD) camera (Optronics International, Goleta, CA) and formatted in photoshop (Adobe Systems, Mountain View, CA). Blastocysts were imaged for fluorescence at ×200–400 with the MagnaFire CCD camera mounted on an inverted fluorescence microscope (DM-IL, Leica, Deerfield, IL). Long exposures revealed a consistent pattern of autofluorescence among all of the blastocysts at a given developmental stage. The following tissues were surveyed by stereofluorescence in all three lines of L1-EGFP transgenic mice: liver, lung, heart, kidney, spleen, testis, ovary, uterus, skeletal muscle, and brain. In addition, thymus, bone marrow, eye, skin, and stomach were evaluated in line 57 L1-EGFP transgenics. The following organs were evaluated by fluorescence from mice with the L1-EGFP insertion: liver, lung, heart, kidney, thymus, spleen, testis, ovary, uterus, brain, eye, and skin.
Results
We chose a human L1 element, L1RP, to study retrotransposition in mice, because L1RP is highly active, undergoing retrotransposition in approximately 1 of 30 cultured cells (20). Transgenes consisting of L1RP driven by its endogenous promoter, the 5′ untranslated region (5′ UTR), produced detectable transcripts only in the testis (E.T.L.P., unpublished data; ref. 17). Therefore, the mouse RNA polymerase II large subunit promoter (pol II) was added upstream of L1RP to achieve a broader tissue distribution of L1 expression (17, 21). The pol II promoter was added upstream of the 5′ UTR because constructs in which the 5′ UTR had been deleted were less active in the cell culture based retrotransposition assay (data not shown). The CMV-major immediate early (MIE) promoter was chosen to drive expression of EGFP because it was effective in producing fluorescent signals in the cultured cell assay and was known to be expressed in a wide range of tissues (19, 22). Transgene constructs were tested for retrotransposition activity in cultured HeLa cells before pronuclear injection. No EGFP-expressing cells were observed with a negative control construct that contained a nonfunctional L1 element (JM111-EGFP). To control for integration site effects on transgene expression and other unknown position effects on retrotransposition, we generated three independent lines of L1-EGFP transgenic mice.
PCR genotyping of offspring from an L1-EGFP transgenic founder (founder number 57, Fo57) revealed a low molecular-weight amplicon that was consistent in size with a spliced L1-EGFP fusion and a high molecular-weight amplicon that was consistent with the original transgene (Fig. 1b). DNA sequence analysis confirmed the identities of both bands.
To verify that the spliced L1-EGFP sequence arose via retrotransposition, we cloned the genomic flanks of the L1 insertion by inverse PCR. The insertion is a truncated L1, consisting of 208 base pairs of the L1RP ORF2 and the 3′ UTR containing the entire EGFP cassette lacking the intron (Fig. 2). L1 insertions often truncate within the most downstream kilobase of the L1 sequence (23). This particular insertion is flanked by target site duplications having the sequence AAAAATAAATTGTTCT. Genomic L1 target sites are characteristically 7–20 nt in length and AT rich (2). The presence of target site duplications is consistent with an endonuclease-dependent retrotransposition event. In the 3′ flank, the target site is preceded by a polyadenylate tail, which originates from the SV40pA signal. The 5′ genomic flank contains part of a mouse B2 element, whereas the 3′ genomic flank contains 506 bases of the 3′ end of a mouse L1 element (Fig. 2).
Of 42 offspring from Fo57, 13 inherited the transgene, 8 inherited the insertion, 4 inherited both, and 17 inherited neither (Table 1). Thus, the insertion segregates from the transgene in offspring of Fo57, indicating that it is likely to be at a significant physical distance from the transgene. Frequent transmission of the insertion could be due to multiple independent insertions in the germ cells of the founder or to a single insertion that took place in a cell that ultimately gave rise to ≈30% of germ cell precursors. To distinguish between these two alternatives, we performed Southern analysis of genomic DNA from tail biopsies of four different offspring harboring the insertion-sized PCR amplicon. When probed with EGFP exonic sequence, all of the mice with the insertion shared the same sized band (data not shown.) This result was confirmed with a second restriction enzyme and implies a single retrotransposition event.
Table 1.
L1-EGFP genotyping data
| Breeding surveys | ||||
| Cross | tg+ | I+ | tg+, I+ | Negative |
| Fo57 × WT | 13 | 8 | 4 | 17 |
| 57tg+/− × WT (or tg+) | 64 | 0 | 0 | 41 |
| 59tg+/− × WT (or tg+) | 54 | 0 | 0 | 26 |
| 63tg+/− (or 63Fo) × WT | 13 | 0 | 0 | 24 |
| Blastocyst (B) and morulae (M) surveys | ||||
| Fluorescence | Yes | No | ||
| 57tg+/− × WT | 0 | 198 | ||
| Single B or M PCR | tg+ | I+ | tg+, I+ | Negative |
| 57tg+/− × WT | 17 | 0 | 0 | 18 |
| Ear fibroblast subclones from Fo57 | ||||
| Clones | tg+ | I+ | tg+, I+ | Negative |
| 19 | 9 | 0 | 3 | 7 |
Three independent lines of L1-EGFP mice (57, 59, 63) were bred to screen for germ-line insertion events by genotyping PCR of tail DNA. All transgenes were maintained on the male line beyond the first generation. Fo57, line 57 founder; 129/SvEmsJ, 57tg+/−, heterozygous for the line 57 L1-EGFP transgene; tg+, transgene positive; l+, has the L1-EGFP insertion.
At the age of 11.5 mo, Fo57 was killed because of a rapidly growing 9-g tumor originating from the right leg/hip. On the basis of histology, we believe that the tumor is a high-grade sarcoma, possibly a rhabdomyosarcoma (data not shown). Very few tumors and no sarcomas have been reported in the strain of Fo57, (SJL/B6)F1 (http://tumor.informatics.jax.org). The freshly harvested tumor was not fluorescent, nor was GFP fluorescence detected in frozen sections of the tumor. However, tumor DNA contained both the transgene and the insertion by PCR (data not shown). We have intercrossed mice carrying the line 57 transgene and have not seen any tumors in offspring (now up to 6 mo of age), including those that are homozygous for the transgene. We have also intercrossed mice carrying the insertion and have not observed any tumors to date in the homozygous offspring (now 6 mo of age). Given the late age of tumor onset in the founder and the absence of similar tumors in other line 57 mice with the transgene or the insertion, the development of this tumor is likely unrelated to the transgene or the L1 insertion.
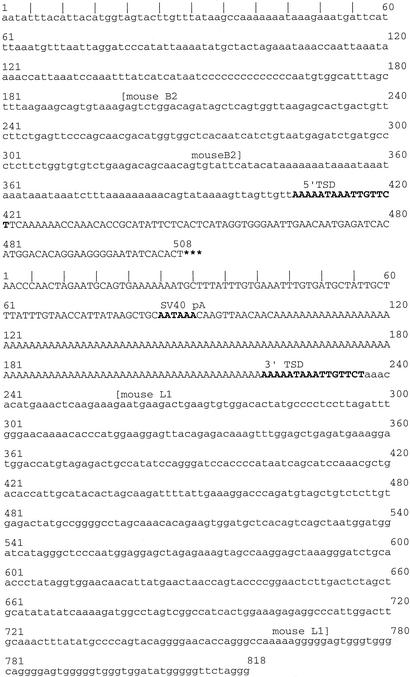
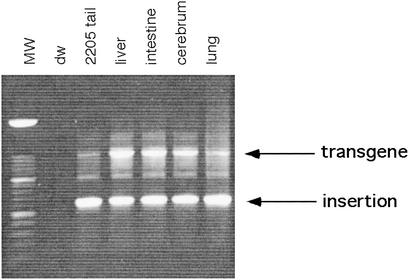
The only organ with demonstrable fluorescence in Fo57 was the testis (Fig. 3a). On frozen section, GFP was discernible in individual seminiferous tubules (Fig. 3b). This pattern of uniform fluorescence across longitudinal aspects of single tubules suggests that cells giving rise to segments within the tubules already contained the L1-EGFP insertion (24). Given the evidence for a single insertion event among offspring inheriting the insertion, this pattern of fluorescence suggests that these segment-producing cells were derived from an earlier precursor cell. To determine whether the precursor cell gave rise to any tissues other than the testis, Fo57 genomic DNA was extracted from the liver, small intestine, cerebrum, and lung and amplified for L1-EGFP. Amplicons corresponding to the transgene and the insertion were obtained in all of these tissues (Fig. 4). It is possible that only a few cells or even a single lineage of cells harboring the insertion could account for these results. Alternatively, metastatic spread of the tumor could have seeded these different organs. However, we disfavor the theory that the insertion arose in a tumor cell, because it is inconsistent with the pattern of fluorescence seen in the testis, which also shows no gross or microscopic evidence of tumor (Fig. 3).
Figure 3.
EGFP fluorescence in the seminiferous tubules of Fo57. (a) Stereofluorescent image (×20), testis (fresh tissue) from Fo57. (b) Fluorescent light image (×200, frozen section from Fo57 testis. GFP fluorescence (dark green) is restricted to the seminiferous tubule in the lower right. Interstitial areas are autofluorescent (yellow–green speckled pattern).
Figure 4.
Tissue DNA PCR surveys in Fo57. Genomic DNA from Fo57 tissues (liver, intestine, cerebrum, and lung) was amplified with geno5 and geno3 primers. A short extension time was used to favor the smaller amplicon (corresponding to the L1-EGFP insertion) relative to the larger amplicon (corresponding to the transgene). MW, 100-bp molecular weight ladder; dw, water; 2205, tail DNA from a line 57 mouse that has the transgene (present in one to five copies) and the single copy insertion.
To assess mosaicism in Fo57 directly, we cultured ear fibroblasts and produced monoclonal subclones by limiting dilution. Of 19 subclones, 7 were negative for both the transgene and the insertion (but contained amplifiable DNA), 9 had the transgene, and 3 had both the transgene and the insertion (Table 1). These results confirm that Fo57 is mosaic for the transgene and the insertion. They also indicate that the L1-EGFP retrotransposition event occurred at an early stage of development.
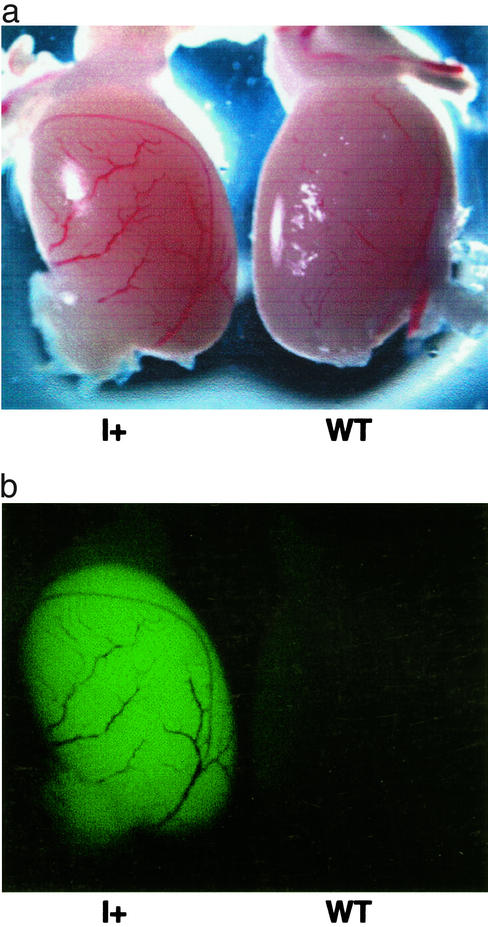
The L1-EGFP insertion resulted in heritable fluorescence that was restricted to the testis. Fig. 5 shows that the testis from a mouse that inherited the insertion from Fo57 is fluorescent, whereas testes from WT littermates are negative. Sperm were not fluorescent, possibly due to cytoplasmic loss during spermatogenesis or lack of EGFP expression in this cell type (25). We were surprised that fluorescence was limited to the testis in mice that had inherited the insertion, given the broad pattern of expression of the CMV-MIE promoter in transgenic mice (22). EGFP sense-strand RNA was readily detected by RT-PCR in the testis and was present at lower levels in multiple organs outside of the testis (data not shown). Lower levels of EGFP transcripts in organs outside of the testis suggest that decreased or undetectable fluorescence could be explained by decreased transcript abundance. DNA sequence analysis revealed no mutations in the CMV promoter in the insertion.
Figure 5.
EGFP fluorescence in the testes of mice inheriting the L1-EGFP insertion. (a) Transmitted light image, testis from an F1 mouse with only the insertion (I+, son of Fo57), and a WT littermate. (b) Stereofluorescent light image, same testis pair as in a.
We searched for somatic retrotransposition events by surveying numerous organs from all three lines of L1-EGFP mice for EGFP fluorescence. All organs surveyed were negative (see Materials and Methods). These findings suggest that the frequency of somatic retrotransposition of the L1-EGFP transgene is low. One possible explanation for the low frequency is that the transgene or the marker gene is rarely expressed or not readily detected in somatic tissues. Consistent with this explanation, sense-strand L1 RNA was low or undetectable in organs other than the testis in several independent RT-PCR experiments (data not shown).
Given the low levels of L1 transcripts in organs other than the testis, we focused on looking for germ-line retrotransposition events in transgenic animals. Of 222 offspring from three independent lines of L1-EGFP transgenic mice, 131 were transgene positive, 91 were transgene negative, and none exhibited a spliced L1-EGFP product by PCR (Table 1). We also assayed 56 morulae and 142 blastocysts derived from mating a heterozygous transgene positive line 57 male to superovulated WT females. None of the blastocysts or morulae was fluorescent. It is possible that the EGFP marker is not expressed in blastocysts. Therefore, we looked for retrotransposition events by PCR of single blastocyst lysates with primers flanking the EGFP intron. Of 35 lysates surveyed, approximately half were transgene positive, and none contained the smaller spliced product (Table 1). Taken together, these findings lead us to a conservative estimate of an L1-EGFP germ-line retrotransposition frequency of <1 in 100.
Discussion
We have traced the origins of an L1 retrotransposition event to an early stage in the embryonic development of a mouse, Fo57. Fo57 is mosaic for the insertion as well as the L1-EGFP transgene. This is demonstrated visually for the L1 insertion by expression of EGFP in some but not all tubules within the Fo57 testis (Fig. 3a). Mosaicism is also supported by our limiting dilution analysis of skin fibroblasts of Fo57, in which we recover cells that are either transgene positive, transgene and insertion positive, or negative for both (Table 1). These three different genotypes indicate that the insertion took place after the two-cell stage, possibly as early as the four-cell stage.
Little is known about the activity of L1 elements during the early stages of mouse development. Others have shown that full-length L1 transcripts can be found in murine blastocysts (26). L1 ORF1 protein and sense-strand L1 RNA are expressed in male and female murine germ cells and embryos (15, 16). Recently L1s have been demonstrated to retrotranspose in male germ cells (17). Retrotransposition in germ cells is an attractive strategy for a genomic parasite. In theory, germ cells could tolerate a high rate of retrotransposition without significantly reducing the reproductive fitness of the host, but the chances of transmitting a particular insertion to the offspring are low. Here we provide evidence for another approach for L1 to colonize the genome: embryonic retrotransposition. An L1 retrotransposon that inserts in an embryonic cell (or germ cell precursor) has greatly increased its chances of propagating that insertion to the next generation, unless the insertion is deleterious to the host.
It is possible that the L1-EGFP insertion observed here originated from an extrachromosomal source. Clearly it is mechanistically allowable for retrotransposition to proceed from an extrachromosomal substrate, such as a plasmid in the cultured cell assay (18). Here we show that the cellular machinery exists for an L1 of unknown structure (extrachromosomal or chromosomal) to retrotranspose during early development.
We have been unable to identify any germ-line L1 retrotransposition events in any of the 222 progeny and 198 blastocysts derived from our L1-EGFP transgenic mice. These results differ significantly from the retrotransposition frequency of 1 in 70 sperm observed by Ostertag et al. (17) in one of their L1-EGFP transgenic lines. Chromosomally based L1s or multicopy transgene arrays may be subjected to a variety of constraints that could limit L1 mobility, including transcriptional inhibition by methylation or posttranscriptional silencing (27). It is possible, for example, that the integration sites of our transgenes were unfavorable compared with the most active line in Ostertag et al. Alternatively, the difference in rate could be due to differences in the transgenes themselves, such as the promoters driving the EGFP marker gene. EGFP is likely to be transcribed in both the sense and antisense directions (relative to L1) in our L1-EGFP transgenic mice. The EGFP transcript, originating from the CMV-MIE promoter, is expressed in multiple tissues and may be active at a very early stage of development, unlike the acrosin promoter used by Ostertag et al. (17), which appears to be restricted to the male germ line (22, 28). The EGFP transcript (which is antisense relative to L1) may inhibit L1 transcription or form dsRNA that targets other L1-EGFP transcripts for destruction.
L1-EGFP transgenics may prove useful in determining whether altered host cell characteristics (such as tissue injury, neoplastic transformation, defective DNA repair machinery, or altered RNA regulation) result in increased L1 mobility. If L1s can be induced to retrotranspose at high frequencies, they could also be used as cell lineage markers. The L1 insertion permanently tags the cell in which it arose and all of the descendants of that cell. As L1 insertions accumulate, a dendogram of lineal descendants could be created.
Acknowledgments
We thank Julia Sarino and Stephanie Johnson for technical assistance; Paula Stein for the blastocyst harvests; the University of Pennsylvania Transgenic facility; and the Institute for Human Gene Therapy morphology core facility. We thank George Gerton, Brad Johnson, Shane Horman, James Wheeler, Paul Zhang, Richard Schultz, and members of the Kazazian laboratory for helpful discussions and cloning reagents. E.T.L.P. is supported by the National Cancer Institute (5-K08-CA83977), the University of Pennsylvania Research Foundation, and the McCabe Foundation. A.W.D. was supported by a summer fellowship from Biomedical Graduate Studies at the University of Pennsylvania. E.A.F. is supported by the National Institutes of Health (NIH) (T32 H10791) and H.H.K. is also supported by the NIH.
Abbreviations
- L1
long interspersed nucleotide element 1
- EGFP
enhanced GFP
- IPCR
inverse PCR
- CMV
cytomegalovirus
- MIE
major immediate early
- SV
simian virus
- pol II
polymerase II large subunit promoter
References
- 1.Skowronski J, Fanning T G, Singer M F. Mol Cell Biol. 1988;8:1385–1397. doi: 10.1128/mcb.8.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kazazian H H, Moran J V. Nat Genet. 1998;19:19–23. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- 3.Ostertag E, Kazazian H H., Jr Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 4.Sassaman D M, Dombroski B A, Moran J V, Kimberland M L, Naas T P, DeBerardinis R J, Gabriel A, Swergold G D, Kazazian H H., Jr Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 5.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.Boeke J D. Nat Genet. 1997;16:6–7. doi: 10.1038/ng0597-6. [DOI] [PubMed] [Google Scholar]
- 7.Smit A F A. Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 8.Esnault C, Maestre J, Heidmann T. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 9.Wei W, Gilbert N, Ooi S L, Lawler J F, Ostertag E M, Kazazian H H, Boeke J D, Moran J V. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moran J V, DeBerardinis R J, Kazazian H H., Jr Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 11.Pickeral O K, Makalowski W, Boguski M S, Boeke J D. Genome Res. 2000;10:411–415. doi: 10.1101/gr.10.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodier J L, Ostertag E M, Kazazian H H., Jr Hum Mol Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- 13.Eickbush T H. Science. 1997;277:911–912. doi: 10.1126/science.277.5328.911. [DOI] [PubMed] [Google Scholar]
- 14.Kazazian H H., Jr Curr Opin Genet Dev. 1998;8:343–350. doi: 10.1016/s0959-437x(98)80092-0. [DOI] [PubMed] [Google Scholar]
- 15.Trelogan S A, Martin S. Proc Natl Acad Sci USA. 1995;92:1520–1524. doi: 10.1073/pnas.92.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brancifort D, Martin S L. Mol Cell Biol. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ostertag E, DeBerardinis R J, Goodier J L, Zhang Y, Yang N, Gerton G, Kazazian H H. Nat Genet. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- 18.Moran J V, Holmes S E, Naas T, DeBerardinis R J, Boeke J D, Kazazian H H. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 19.Ostertag E M, Luning Prak E T, DeBerardinis R J, Moran J V, Kazazian H H. Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimberland M L, Dovoky V, Prchal J, Schwahn U, Berger W, Kazazian H H. Hum Mol Genet. 1999;8:1557–1560. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- 21.Ahearn J M, Bartolomei M S, West M L, Cisek L J, Corden J L. J Biol Chem. 1987;262:10695–10705. [PubMed] [Google Scholar]
- 22.Baskar J F, Smith P P, Ciment G S, Hoffman S, Tucker C, Tenney D J, Colberg-Poley A M, Nelson J A, Ghazal P. J Virol. 1996;70:3207–3214. doi: 10.1128/jvi.70.5.3207-3214.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boissinot S, Chevret P, Furano A V. Mol Biol Evol. 2000;6:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- 24. Histological and Histopathological Evaluation of the Testis (1990), eds. Russell, L. D., Ettlin, R. A., Hikim, S. & Clegg, E. D. (Cache River Press, Clearwater, FL), pp. 41–58.
- 25.Villuendas G, Gutierrez-Adan A, Jimenez A, Rojo C, Roldan E R, Pintado B. Int J Androl. 2001;24:300–305. doi: 10.1046/j.1365-2605.2001.00302.x. [DOI] [PubMed] [Google Scholar]
- 26.Packer A I, Manova K, Bachvarova R F. Dev Biol. 1993;157:281–283. doi: 10.1006/dbio.1993.1133. [DOI] [PubMed] [Google Scholar]
- 27.Bestor T H, Tycko B. Nat Genet. 1996;12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe K, Baba T, Kashiwabara S, Okamoto A, Arai Y. J Biochem (Tokyo) 1991;109:828–833. doi: 10.1093/oxfordjournals.jbchem.a123466. [DOI] [PubMed] [Google Scholar]