Abstract
mtDNA mutations occur in a wide variety of degenerative diseases and cancer. mtDNA seems to be more susceptible to DNA damage and consequently sustains higher rates of mutation than does nuclear DNA (nDNA). Many of the somatic mtDNA mutations in human cancers are located in the displacement loop (D-loop) and in particular in a polycytidine stretch (C-tract) termed D310. The D310 region exhibits polymorphic length variation among individuals and has been described as a “hot spot” for somatic mutations in many cancer types. We used real-time quantitative PCR to analyze mtDNA integrity, damage repair, and induced mutations after exposure of human adult retinal pigment epithelial (ARPE)-19 cells to 4-nitroquinoline 1-oxide, a UV-mimetic and adduct-forming carcinogen, and tert-butyl hydroperoxide, an oxidant. The mtDNA-damage profile depended on the region. Thus, the tRNA coding for glycine (tRNAG) was the least affected region, whereas the D-loop, and especially its D310 region, were most sensitive to damage. The time course of repair of mutations of the D-loop and especially the D310 region after exposure to DNA-damaging agents was delayed when compared with other regions and gave rise to common D310 C-tract frame-shift mutations. The induced mutations in the D310 region were predominantly homoplasmic only 7 days after exposure to damage. Our results establish that the D-loop (especially its D310 region) is highly susceptible to mutations because of its vulnerability to DNA damage and inefficient repair mechanisms. Our findings may explain the high frequency of homoplasmic D310 somatic mutations in many tumor types.
Both inherited and somatic mitochondrial mutations cause severe degenerative disorders and occur in primary human cancers. The discovery that mutations in mitochondrial DNA (mtDNA) are causally related to a variety of inherited and acquired human diseases has stimulated interest in understanding how the integrity of this important component of the genome is maintained (1–4). This article describes the damaging effects of two chemical agents on mtDNA and the fate of the resultant mutations.
The human mitochondrial genome is a small (16,569-bp), closed-circular, duplex molecule that is present at a high copy number per cell, but this number varies widely among cell types. mtDNA contains 37 genes including the structural genes for 13 polypeptide components of the respiratory-chain enzyme complexes, two ribosomal RNAs, and a complete set of 22 tRNAs that are required for translation of the mtDNA-encoded mRNAs (5). In addition, the mtDNA molecule contains a noncoding region that includes a unique displacement loop (D-loop) that controls both replication and transcription (6). DNA replication and transcription are linked in vertebrate mitochondria because RNA transcripts initiated at the light-strand promoter are the primers for mtDNA replication at the heavy-strand origin. Mammalian mtDNA molecules are believed to replicate by an asynchronous displacement mechanism involving two unidirectional origins; however, recent data suggest that mtDNA may replicate through a strand-coupled mechanism (7). The genetic content and gene order in mtDNA are remarkably conserved among vertebrate species. In contrast, the nucleotide sequence and organization of the regulatory elements involved in transcription and replication of the mtDNA genome vary considerably.
mtDNA is believed to be more susceptible to DNA damage and consequently acquires mutations at a higher rate than does nuclear DNA (nDNA). Several possible factors may account for these differences in susceptibility including exposure to high levels of reactive oxygen species produced during oxidative phosphorylation, lack of protective histones, and limited capacity for repair of DNA damage (8–10). It is estimated that the mutational rate of mtDNA is at least 10 times higher than that of nDNA. Mitochondrial mutations are reported to occur in a wide variety of degenerative diseases and cancer (1, 2).
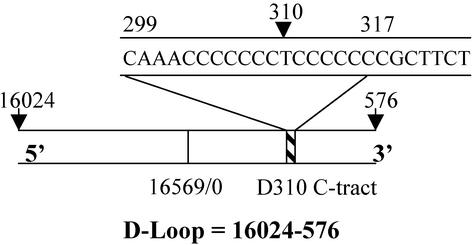
In many of the inherited mitochondrial degenerative diseases mtDNA mutations are located mainly in the tRNAs and coding regions (11). In contrast, many of the somatic mtDNA mutations in human cancers are located in the D-loop and in particular in a polycytidine stretch (C-tract) also termed the D310 region (12) (Fig. 1). The D310 region is composed of 12–18 cytidine bases that are interrupted by a thymidine base at position 310. The D310 region shows polymorphic length variation among individuals (13, 14) as well as heteroplasmic variation within an individual (15, 16). Moreover, the D310 D-loop region has been found to be a “hot spot” for somatic mutations in many cancer types (12). Severe mutations might be of biological significance, because the D310 region lies in a conserved sequence block that is hypothesized to be involved in some aspect of mtDNA replication and transcription (17). Maintenance of the integrity of mtDNA requires accurate replication and repair.
Figure 1.
Structure of human mitochondrial D-loop (1,122 bp) showing the location of the D310 cytidine-rich C-tract in relation to the origin of replication. Note that base 310 is thymine and is flanked by cytidine residues.
To our knowledge, there are no reports on the susceptibility of various mtDNA regions, and in particular the D-loop, to DNA-damaging agents. To address this question, we studied the extent of DNA damage in different mtDNA regions after exposure to DNA-damaging agents and the mutational consequences and fate of D-loop-induced damage. We used quantitative real-time PCR to determine the relative extent of regional mtDNA damage and in single sorted cells to identify C-tract frame-shift mutations. There are large differences in the susceptibility to mutation of various regions of mtDNA. Surprisingly, these mutations rapidly evolved into a homoplasmic state in which the mutations become identical in mitochondrial populations of cells.
The two damaging agents selected for these studies were 4-nitroquinoline 1-oxide (4NQO) and tert-butyl hydroperoxide (BHP). 4NQO is an electrophile and a powerful carcinogen and mutagen (18). It is a UV-mimetic agent and forms charge-transfer complexes with 5′-deoxyribonucleotides (19, 20). 4NQO forms DNA adducts and causes a wide range of DNA lesions including single-strand breaks, pyrimidine-dimer formation, abasic sites, and oxidized bases. Because earlier work showed that 4NQO treatment interrupted PCR amplification of human lymphocyte DNA, 4NQO seemed to be a promising agent for regional mapping of DNA damage (21). BHP is a powerful oxidant, and although its propensity for DNA-adduct formation is probably much lower than that of 4NQO, it generates free radicals and also causes DNA strand breaks and various types of base damage (22). The choice of human adult retinal pigment epithelial (ARPE)-19 cells was dictated by the great sensitivity of the retina to oxidative damage and the widespread use of these cells to assess oxidative damage and devise strategies for protection against such damage (23).
Experimental Procedures
Chemicals.
4NQO and BHP were purchased from Sigma.
Cell Culture and Drug Treatment.
Human ARPE-19 cells were obtained from the American Type Culture Collection. Cells were cultured in a mixture of equal volumes of DMEM and Ham's F-12 medium plus 10% heat-inactivated FBS and incubated at 37°C in an atmosphere of 5% CO2/95% relative humidity. Cells were exposed to either 10 μM 4NQO or 125 μM BHP and incubated at 37°C for 1 and 2 h, respectively. After exposure to the damaging agents, the cells were either harvested immediately or incubated for an additional 4 or 24 h in complete medium without the damaging agents to allow time for repair. Cells were trypsinized, washed with PBS, and incubated with 1% SDS and 1 mg/ml proteinase K for 24 h at 48°C. DNA was extracted by the standard phenol-chloroform procedure.
Quantitative Real-Time PCR.
The TaqMan 7900HT sequence-detection system was used to perform real-time PCR amplification for β-actin and mtDNA regions cytochrome c oxidase (COII), tRNA coding for glycine (tRNAG), D-loop (401–490), and D310. Table 1 lists the primers and probes used to amplify the respective DNA regions. All primers were obtained from Invitrogen. All TaqMan probes (Applied Biosystems) were labeled with 5′-FAM (6-carboxyfluorescein, fluorescent reporter) and 3′-TAMRA (6-carboxy-tetramethylrhodamine, fluorescence quencher). PCR amplifications were carried out in buffer containing 16.6 mM ammonium sulfate, 67 mM Tris base, 2.5 mM MgCl2, 10 mM 2-mercaptoethanol, 0.1% DMSO, 0.2 mM each of dATP, dCTP, dGTP, and dTTP, 600 nM each of forward and reverse primers, 200 nM TaqMan probe, 0.6 unit Platinum Taq polymerase, and 2% Rox reference dye (24). DNA (500 pg) was used to amplify mitochondrial regions, whereas 10 ng of DNA were used to amplify β-actin, a single-copy gene. The real-time PCR reactions were performed in triplicate for each gene. Standard curves were obtained by using ARPE-19 DNA from untreated cells. Data analysis was performed by using Microsoft EXCEL software. mtDNA/nDNA ratios were calculated by dividing the mtDNA signal for each gene by the β-actin signal and expressing the ratio as a percentage of the untreated control set at 100%.
Table 1.
Sequences of primers and probes used in the PCR and quantitative real-time PCR analyses
| Region | Forward primer (5′–3′) | Reverse primer (5′–3′) | TaqMan probe (5′–3′) |
|---|---|---|---|
| D-loop | tatcttttggcggtatgcacttttaacagt | tgatgagattagtagtatgggagtgg | caccccccaactaacacattattttcccc |
| (401–430) | (461–490) | (431–459) | |
| D310 | cacacagacatcataacaaaaaatttcc | ggtgttagggttctttgtttttgg | ccccccctcccccgcttct |
| (269–296) | (355–378) | (303–321) | |
| COII | ccccacattaggcttaaaaacagat | accgctacacgaccgggggtata | caattcccggacgtctaaaccaaaccactttc |
| (8,080–8,103) | (8,138–8,160) | (8,105–8,136) | |
| tRNAG | gccacctatcacaccccatc | tacccttcccgtactaattaatccc | gctatcgggcccataccccgaaaatgt |
| (4,378–4,397) | (4,454–4,478) | (4,420–4,446) | |
| β-Actin | tcacccacactgtgcccatctacga | cagcggaaccgctcattgccaatgg | atgccccccccatgccatcctgcgt |
| (2,141–2,165) | (2,411–2,435) | (2,171–2,196) | |
| Mt35A | ggagctctccatgcatttgg | gggtgatgtgagcccgtcta | |
| (35–53) Mt35F | (620–639) Mt635R | ||
| F57A | acaattgaatgtctgcacagccactt | ataccccaatcgtcgccacaca | |
| (241–266) F57A | (519–540) R15i2 |
Cloning of mtDNA C-Tract.
PCR amplification was performed by using 10 ng of total DNA. The amplification was performed in buffer containing 1.5 mM MgCl2, 16.6 mM (NH4)2SO4, 6.25% DMSO, 200 μM dNTPs, 250 nM each of Mt35F forward and Mt635R reverse primers, and 2.5 units of Platinum Taq polymerase. PCR cycling conditions were 1 cycle for 2 min at 94°C, 2 cycles for 30 s at 94°C, 1 min at 58°C, and 1 min at 72°C, followed by 25 cycles for 30 s at 94°C, 1 min at 56°C, and 1 min at 72°C, and a final extension for 5 min at 72°C. PCR products were separated on 1% agarose gels and purified by using the Qiagen (Valencia, CA) gel-purification kit according to the manufacturer's instructions. The PCR product was cloned into a Topo10TA vector (Invitrogen) according to the manufacturer's instructions. The resulting plasmid was used to transform TOPO10 bacterial cells that were subsequently plated on agar plates containing 50 μg/ml ampicillin. Colonies were picked and grown in 96-well plates. The plasmids were purified by using the Qiagen plasmid kit.
Analysis of Cloned C-Tract Variants.
Cloned D310 C-tract variants were analyzed by PCR amplification with use of a 32P end-labeled primer F57A under the following conditions: ≈10–20 ng of plasmid DNA, 200 μM dNTPs, and 1× buffer containing 0.5 μM each unlabeled forward (F57A) and reverse (R15i2) primers. Cycling conditions were 1 cycle for 2 min at 95°C, 35 cycles for 1 min at 95°C, 1 min at 60°C, and 1 min at 72°C, and a final extension for 4 min at 72°C. Stop buffer was added, and a fraction of the sample was loaded and separated on an 8% denaturing polyacrylamide gel.
Analysis of Mitochondria from Single Cells.
Single cells were sorted by flow cytometry into 96-well plates containing 5 μl of alkaline lysis solution (200 mM KOH/50 mM DTT) as described (25). After a 10-min incubation at 65°C, 5 μl of neutralization solution (900 mM Tris⋅HCl, pH 8.3/300 mM KCl/200 mM HCl) were added. To the lysed and neutralized sample we then added PCR buffer [20 mM Tris⋅HCl, pH 9.0/10 mM (NH4)2SO4/10 mM KCl/0.1% Triton X-100/2 mM MgCl2/0.1 mg/ml BSA/200 nM each of forward (Mt35F) and reverse (Mt635R) primers/0.75 mM dNTPs/2 units of Platinum Taq polymerase]. The cycling conditions of the first PCR stage were 1 cycle for 2 min at 94°C, 2 cycles for 30 s at 94°C, 1 min at 58°C, and 1 min at 72°C, followed by 35 cycles for 30 s at 94°C, 1 min at 56°C, and 1 min at 72°C, and a final extension for 5 min at 72°C. An aliquot of 2 μl was used as a template in the second PCR stage, which was performed in buffer containing a 32P end-labeled primer (F57A), 1.5 mM MgCl2, 16.6 mM (NH4)2SO4, 6.25% DMSO, 600 μM dNTPs, 200 nM each of forward (F57A) and reverse (R15i2) primer, and 0.5 unit of Platinum Taq polymerase. PCR cycling conditions were 1 cycle for 2 min at 95°C, 35 cycles for 1 min of 95°C, 1 min at 60°C, and 1 min at 72°C, followed by extension for 4 min at 72°C. The samples were separated on an 8% denaturing polyacrylamide gel.
Results
To assess mtDNA integrity we exposed ARPE-19 cells to 4NQO or BHP as DNA-damaging agents. ARPE-19 cells are immortalized but nonmalignant human ARPE cells and have been shown previously to be sensitive to the cytotoxicities of electrophiles and oxidants (23). Both 4NQO and BHP generate a wide variety of DNA damage. Specifically, 4NQO is a UV-mimetic agent that produces single DNA strand breaks and pyrimidine dimers, whereas BHP causes lipid peroxidation, induces single DNA strand breaks, and may cause some DNA base methylation (26).
Analysis of mtDNA Susceptibility to DNA-Damaging Agents by Quantitative PCR.
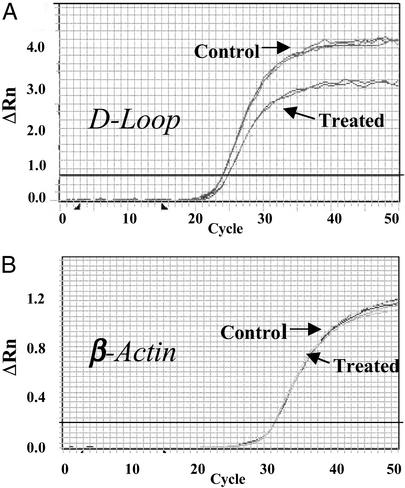
We hypothesized that damage to mtDNA would result in a reduction of the available template for PCR (decreased DNA integrity) in affected regions. In quantitative PCR this decrease in DNA integrity is reflected by a shift of the amplification time curve to the right. Fig. 2A is a representative amplification plot showing a shift to the right after damage in the D-loop. An undamaged template region would have produced overlapping curves before and after treatment as shown for the control nuclear β-actin gene (Fig. 2B). Because the damaging agents used in these studies did not affect the selected region of β-actin, we used this gene as a reference.
Figure 2.
Representative real-time PCR curves generated for the mitochondrial D-Loop (0.5 ng of DNA sample) (A) and β-actin gene (10 ng of DNA sample) (B) before and after treatment with the DNA-damaging agent 4NQO (2.5 μM, 1 h). Each experiment was performed in triplicate and is shown by overlapping amplification curves. ΔRn = (Rn+) − (Rn−), where Rn+ is the fluorescence emission intensity of reporter/emission intensity of quencher at any time point, and Rn− is the initial emission intensity of reporter/emission intensity of quencher in the same reaction vessel before PCR amplification is initiated.
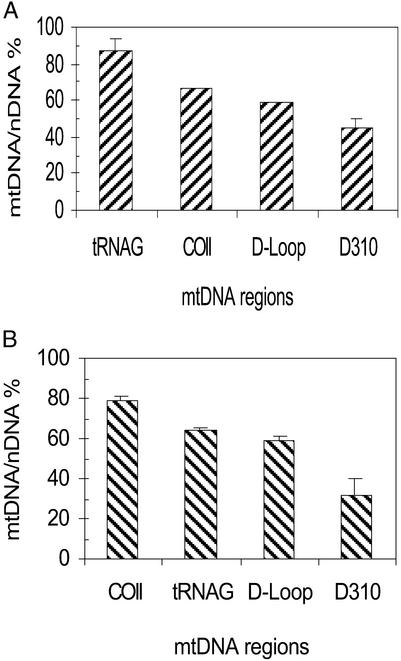
The extent of mtDNA damage was analyzed by calculating the mtDNA/nDNA ratio by normalizing (dividing) each mitochondrial fluorescence amplification measurement to the corresponding β-actin signal. The mtDNA/nDNA ratio of the untreated control was set at 100%, and mtDNA damage was expressed as a percentage of this control value. Thus, a lower mtDNA/nDNA ratio represents less initial template, denoting a decrease in the integrity of mtDNA. A quantitative analysis of induced damage in tRNAG, COII, D-loop, and D310 is shown in Fig. 3A. This analysis revealed that the tRNAG region was the least affected region, whereas the COII region was moderately damaged. The D-loop was more sensitive to damage than the tRNAG and COII regions. Furthermore, the D310 region, postulated to be a hot spot for mutation, was also hypersensitive to 4NQO damage when compared with the other mtDNA regions. Similarly, treatment with BHP resulted in more damage in the D310 area as compared with the other mtDNA regions analyzed here (Fig. 3B). A comparison of Fig. 3 A and B shows that the profile of DNA damage depended on the nature of the damaging agent. Damage by 4NQO decreased in the order D310 > D-loop > COII > tRNAG, whereas the damage profile for BHP was D310 > D-loop > tRNAG > COII. Interestingly, the D310 region was the most susceptible to damage by both agents.
Figure 3.
mtDNA-damage profile after exposure to the DNA-damaging agents 4NQO (2.5 μM, 1 h) (A) and BHP (125 μM, 3 h) (B). Analysis was performed immediately after exposure to damage. Error bars represent the SD of two experiments, each done in triplicate.
Assessment of mtDNA Repair by Quantitative PCR.
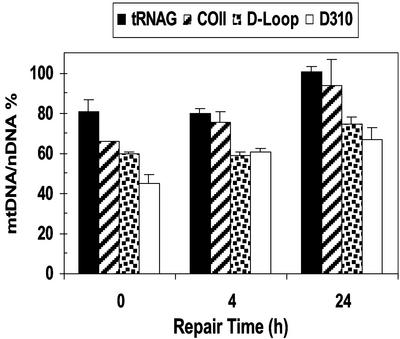
We examined the temporal repair of mtDNA by using quantitative PCR. The different regions of DNA isolated from ARPE-19 cells at 0.5, 4, and 24 h after exposure to 4NQO and BHP were subjected to quantitative PCR. Four hours after exposure to 4NQO, the mtDNA/nDNA ratios for D-loop and the D310 region were both lower than those of COII and tRNAG (Fig. 4), indicating decreased mtDNA integrity of the D-loop. Moreover, at 24 h after exposure, the mtDNA/nDNA ratios for COII and tRNAG returned to >95% of control values, whereas both D-loop and D310 lagged behind at 74% and 66%, respectively. These results indicate that damage induced by 4NQO was repaired more rapidly in the COII and tRNAG regions compared with the D-loop, and in particular the D310 region. Similar results were observed in BHP-treated cells (data not shown). Together these findings confirm that D310 is highly prone to damage by quite different chemical agents and is repaired more slowly in comparison to the other mtDNA regions examined. These findings led us to analyze the nature and frequency of induced mutations in the D310 region.
Figure 4.
Repair of induced damage in mitochondrial regions. The mtDNA/nDNA ratio after exposure to the DNA-damaging agent 4NQO (10 μM, 1 h) is shown. Repair analysis was performed at the indicated times after exposure to DNA damage. The time “0 h” represents DNA isolated immediately after exposure to 4NQO.
Mutational Analysis of the D310 Region.
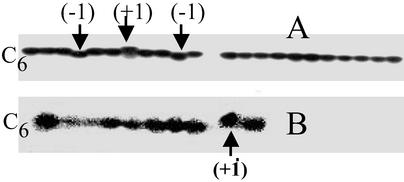
To assess mutations in the D310 region, we first performed direct manual DNA sequencing from pooled cell DNA but were unable to detect any mutations. In general, DNA mutations cannot be detected by direct sequencing analysis unless they represent >10% of the starting template. We therefore searched for rare mutations in the D310 region by cloning PCR fragments encompassing this region. Before exposure to damaging agents, <2% of the mitochondria harbored D310 C-tract variants (heteroplasmy) in the parental ARPE-19 DNA (Table 2). At 4 h after exposure to damage, D310 C-tract heteroplasmy increased to 6.5% (P = 0.018). The distribution of frame-shift variants also widened, indicating the presence of new mutations. Interestingly, 7 days after exposure to damage D310 C-tract heteroplasmy fell to 4.1% (P = 0.02) and the distribution narrowed again (Table 2). Fig. 5A is a representative of the cloned PCR products of the D310 region showing the wild-type C6 D310 C-tract and the +1 and −1 variants. These data suggest that the variant forms of D310 C-tract were present in mitochondria from the cell but could also reflect imprecise PCR due to unrepaired template. To determine the origin of the observed heteroplasmy we exposed cells to 10 μM 4NQO for 1 h, incubated the treated cells for 7 days in fresh medium, and analyzed the DNA of single cells by PCR. Fig. 5B shows representative results of single-cell PCR showing that +1 variants were present before treatment. Furthermore, the results also revealed that each cell contained only one D310 variant (single-cell homoplasmy). These results suggest that fixed mtDNA mutations evolve rapidly to a homoplasmic state.
Table 2.
Analysis of D310 C-tract variants by sequencing of cloned PCR products
| Control | Exposed to 4NQO
|
||
|---|---|---|---|
| 4 h | 7 days | ||
| Total clones examined | 372 | 438 | 456 |
| +1 variants | 3 (0.81%) | 8 (1.83%) | 3 (0.66%) |
| −1 variants | 4 (1.08%) | 16 (3.65%) | 16 (3.5%) |
| +2 variants | 0 | 1 (0.23%) | 0 |
| −2 variants | 0 | 3 (0.68%) | 0 |
| % variants | 1.88 | 6.39 | 4.17 |
Control represents untreated cells, and 4 h and 7 days represent time after treatment with 4NQO (10 μM, 1 h). +1 and −1 variants represent the insertion and deletion, respectively, of one cytosine; +2 and −2 variants represent the insertion and deletion, respectively, of two cytosines.
Figure 5.
Detection of the D310 C-tract variants. (A) Cloned PCR products of the D310 region from untreated cells showing C-tract, the +1 and −1 variants. (B) Single-cell PCR amplification of the D310 region showing homoplasmic variants of C-tract. The wild-type D310 C-tract has a stretch of six polycytidine (C6) separated by a thymine. −1 and +1 represent deletion and insertion of one cytidine into the wild-type (C6) D310, respectively.
Discussion
To understand the nature of mtDNA and the process of chemical mutations, we analyzed mtDNA integrity, the time course of repair, and damage-induced mutations after exposure to DNA-damaging agents. Our results showed that the damage profile of the mtDNA was region-dependent. The damage to the D-loop region was higher than that of tRNAG and the region encoding COII. Interestingly, within the D-loop the D310 region was the most vulnerable to damage. Our observation that the D-loop was prone to DNA-damaging agents raised the question of the effectiveness of DNA repair. Our results indicate that the absolute level of repair of the D-loop region was lower than that of the COII and tRNAG regions. This finding clearly indicates the presence of differential repair capacity within mtDNA. Moreover, the D310 region was repaired more slowly when compared with other regions within the D-loop. The possibility that damaged mtDNA molecules are discarded and replaced by replication of undamaged mtDNA molecules is unlikely because of the high mutational rate observed after treatment, suggesting inefficient repair. Although the reasons for this differential mtDNA damage and repair are unknown, it is possible that the unique D-loop structure predisposes this region to excessive DNA damage. In addition, this unique D-loop structure coupled with the polycytidine stretch and the proximity of the D-loop to the mitochondrial inner membrane may hinder access to repair proteins.
Several enzymatic activities involved in mtDNA repair have been identified in mammalian cells (27–30). In this study we used 4NQO, a UV-mimetic agent that induces a wide range of lesions including DNA adducts, single-strand breaks, pyrimidine dimers, abasic sites, and oxidized bases. These lesions are repaired by nucleotide and base excision-repair pathways. Although nucleotide excision-repair mechanisms have not been fully established yet in mitochondria, our finding that most mtDNA regions with the exception of the D-loop were repaired provides support for previous suggestions that mitochondria possess DNA-repair systems. Further characterization of the induced lesions in mtDNA is required to identify the different repair mechanisms in detail.
The notion that less efficiently repaired regions are expected to harbor higher mutation rates prompted us to analyze mutation induction in the D310 region. We found that 4 h after the D310 region was exposed to the DNA-damaging agents, there was a significant increase in the quantity of D310 C-tract variants. This coexistence of mutant and wild-type mtDNA is termed heteroplasmy. Seven days after DNA damage, the frequency of C-tract heteroplasmy was still higher than in the control, but the differences were narrowed. The concomitant increase in the frequency of C-tract variants coupled with the reduction in the observed heteroplasmy after 7 days could be attributed either to the fixation of scattered mtDNA mutations throughout the cell population (single-cell heteroplasmy) or the emergence of mutant homoplasmic cells (cell-population heteroplasmy). To test the latter possibility we isolated single cells by flow cytometry and subjected the D310 region to PCR. The results showed a homoplasmic population of mtDNA per cell, supporting the rapid emergence of clonal mutant mitochondria. Our findings help to explain the high frequency of D310 homoplasmic mtDNA somatic mutations detected in human tumors (31, 32). It is possible that certain mtDNA mutations may confer a selective advantage or disadvantage for the carrier mtDNA copy or the host cell. This hypothesis of clonal selection is supported by several studies that showed a nonrandom segregation of mtDNA in cybrids (33–36).
Although the mechanisms of mitochondrial replication and segregation are still unknown, several theories have been proposed: (i) the bottle-neck, (ii) clonal-selection, and (iii) random-segregation theories. The bottle-neck theory proposes that only a small fraction of mtDNA molecules are sampled from a larger population for transmission and amplification. The bottle-neck fails to explain the expansion of somatic mutations, because it is thought to occur only during the early stages of oogenesis (10). The clonal-selection theory proposes that certain mtDNA mutations may confer a selective advantage or disadvantage for the carrier mtDNA copy or the host cell. mtDNA mutants that confer disadvantages to the cell would be eliminated, whereas those that possess survival advantage are propagated. Our results showed propagation of D310 C-tract frame shift of one cytosine insertion after 7 days, whereas the other induced variants were eliminated. These findings are consistent with the selection theory or nonrandom segregation and further show that this process may be very rapid.
Because the D310 area lies in a conserved sequence block that presumably controls mtDNA replication and transcription, it is possible that severe D310 C-tract length variations may have an important role in the regulation of mtDNA metabolism. Minimal D310 C-tract frame shifts of one or two base pairs are common polymorphisms in the population; however, they are present in low-level heteroplasmy in normal tissue, suggesting that such minimal changes are unlikely to cause disease. Studies of mitochondrial degenerative diseases showed that mutant mtDNA must represent >80–90% of cellular mitochondria content to cause a clinical phenotype (37–39). This threshold is likely to differ between mutations in the control and the coding regions.
We have shown that the D-loop, and in particular the D310 region, is highly susceptible to mutations, possibly because of a combination of vulnerability to damage and inefficient repair. To our knowledge, complete analysis of the DNA damage, repair, and mutation of the D310 region has not been described previously. The susceptibility of the D310 region to mutations may explain the high frequency of common polymorphisms and the high frequency of somatic D310 mutations detected in many tumor types (31, 32). Furthermore, our findings strongly suggest that emergence of homoplasmic mitochondrial variants is a very rapid event that occurs within days after damage. Understanding how the mitochondrial genome is controlled will increase the opportunities for clinical intervention against the pathogenic effects of defective mitochondria.
Acknowledgments
We thank Dr. Susan V. Harden for helping with the TaqMan experiments. This work was supported by National Institutes of Health Grant 5 U01 CA84986-04 titled Early Detection Research Network–Integrated Development of Novel Molecular Markers. This work was also supported by a collaborative research agreement with Virco, Inc. Under a licensing agreement between The Johns Hopkins University (JHU) and Virco, D.S. is entitled to a share of royalty received by JHU. D.S. is a paid consultant to Virco. The terms of this arrangement are being managed by JHU in accordance with its conflict-of-interest policies. This study was also supported by a grant from the American Institute for Cancer Research (Washington, DC) and generous gifts from the Lewis B. and Dorothy Cullman Foundation, the McMullan Family Fund, and the Barbara Lubin Goldsmith Foundation.
Abbreviations
- D-loop
displacement loop
- nDNA
nuclear DNA
- 4NQO
4-nitroquinoline 1-oxide
- BHP
tert-butyl hydroperoxide
- ARPE
adult retinal pigment epithelial
- COII
cytochrome c oxidase
- tRNAG
tRNA coding for glycine
References
- 1.Wallace D C, Lott M T, Shoffner J M, Ballinger S. Epilepsia. 1994;35, Suppl. 1:S43–S50. doi: 10.1111/j.1528-1157.1994.tb05928.x. [DOI] [PubMed] [Google Scholar]
- 2.Wallace D C. Am J Med Genet. 2001;106:71–93. doi: 10.1002/ajmg.1393. [DOI] [PubMed] [Google Scholar]
- 3.Orth M, Schapira A H V. Am J Med Genet. 2001;106:27–36. doi: 10.1002/ajmg.1425. [DOI] [PubMed] [Google Scholar]
- 4.DiMauro S, Schon E A. Am J Med Genet. 2001;106:18–26. doi: 10.1002/ajmg.1392. [DOI] [PubMed] [Google Scholar]
- 5.Shadel G S, Clayton D A. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 6.Clayton D A. Cell. 1982;28:693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 7.Yang M Y, Bowmaker M, Reyes A, Vergani L, Angeli P, Gringeri E, Jacobs H T, Holt I J. Cell. 2002;111:495–505. doi: 10.1016/s0092-8674(02)01075-9. [DOI] [PubMed] [Google Scholar]
- 8.Croteau D L, Bohr V A. J Biol Chem. 1997;272:25409–25412. doi: 10.1074/jbc.272.41.25409. [DOI] [PubMed] [Google Scholar]
- 9.Beal M F. Curr Opin Neurobiol. 1996;6:661–666. doi: 10.1016/s0959-4388(96)80100-0. [DOI] [PubMed] [Google Scholar]
- 10.Lightowlers R N, Chinnery P F, Turnbull D M, Howell N. Trends Genet. 1997;13:450–455. doi: 10.1016/s0168-9525(97)01266-3. [DOI] [PubMed] [Google Scholar]
- 11.Kogelnik A M, Lott M T, Brown M D, Navathe S B, Wallace D C. Nucleic Acids Res. 1996;24:177–179. doi: 10.1093/nar/24.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez-Cespedes M, Parrella P, Nomoto S, Cohen D, Xiao Y, Esteller M, Jeronimo C, Jordan R C K, Nicol T, Koch W M, et al. Cancer Res. 2001;61:7015–7019. [PubMed] [Google Scholar]
- 13.Anderson S, Bankier A T, Barrell B G, de Bruijn M H, Coulson A R, Drouin J, Eperon I C, Nierlich D P, Roe B A, Sanger F, et al. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 14.Greenberg B D, Newbold J E, Sugino A. Gene. 1983;21:33–49. doi: 10.1016/0378-1119(83)90145-2. [DOI] [PubMed] [Google Scholar]
- 15.Jazin E E, Cavelier L, Eriksson I, Oreland L, Gyllensten U. Proc Natl Acad Sci USA. 1996;93:12382–12387. doi: 10.1073/pnas.93.22.12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchington D R, Hartshorne G M, Barlow D, Poulton J. Am J Hum Genet. 1997;60:408–416. [PMC free article] [PubMed] [Google Scholar]
- 17.Ghivizzani S C, Madsen C S, Nelen M R, Ammini C V, Hauswirth W W. Mol Cell Biol. 1994;14:7717–7730. doi: 10.1128/mcb.14.12.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugimura T. Carcinogenesis: A Comprehensive Survey. Vol. 6. New York: Raven; 1981. [Google Scholar]
- 19.Kondo S. Br J Cancer. 1977;35:595–601. doi: 10.1038/bjc.1977.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winkle S A, Tinoco I., Jr Biochemistry. 1979;18:3833–3839. doi: 10.1021/bi00585a001. [DOI] [PubMed] [Google Scholar]
- 21.Govan H L, III, Valles-Ayoub Y, Braun J. Nucleic Acids Res. 1990;18:3823–3830. doi: 10.1093/nar/18.13.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochi T, Miyaura S. Toxicology. 1989;55:69–82. doi: 10.1016/0300-483x(89)90175-3. [DOI] [PubMed] [Google Scholar]
- 23.Gao X, Dinkova-Kostova A T, Talalay P. Proc Natl Acad Sci USA. 2001;98:15221–15226. doi: 10.1073/pnas.261572998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heid C A, Stevens J, Livak K J, Williams P M. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Cui X, Arnheim N. Proc Natl Acad Sci USA. 1990;87:4580–4584. doi: 10.1073/pnas.87.12.4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hix S, Augusto O. Chem Biol Interact. 1999;118:141–149. doi: 10.1016/s0009-2797(99)00079-4. [DOI] [PubMed] [Google Scholar]
- 27.LeDoux S P, Wilson G L, Beecham E J, Stevnsner T, Wassermann K, Bohr V A. Carcinogenesis. 1992;13:1967–1973. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- 28.Myers K A, Saffhill R, O'Connor P J. Carcinogenesis. 1988;9:285–292. doi: 10.1093/carcin/9.2.285. [DOI] [PubMed] [Google Scholar]
- 29.Pettepher C C, LeDoux S P, Bohr V A, Wilson G L. J Biol Chem. 1991;266:3113–3117. [PubMed] [Google Scholar]
- 30.Satoh M S, Huh N, Rajewsky M F, Kuroki T. J Biol Chem. 1988;263:6854–6856. [PubMed] [Google Scholar]
- 31.Polyak K, Li Y, Zhu H, Lengauer C, Willson J K, Markowitz S D, Trush M A, Kinzler K W, Vogelstein B. Nat Genet. 1998;20:291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 32.Fliss M S, Usadel H, Caballero O L, Wu L, Buta M R, Eleff S M, Jen J, Sidransky D. Science. 2000;287:2017–2019. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 33.Danielson S R, Wong A, Carelli V, Martinuzzi A, Schapira A H, Cortopassi G A. J Biol Chem. 2002;277:5810–5815. doi: 10.1074/jbc.M110119200. [DOI] [PubMed] [Google Scholar]
- 34.Brown M D, Trounce I A, Jun A S, Allen J C, Wallace D C. J Biol Chem. 2000;275:39831–39836. doi: 10.1074/jbc.M006476200. [DOI] [PubMed] [Google Scholar]
- 35.Dunbar D R, Moonie P A, Jacobs H T, Holt I J. Proc Natl Acad Sci USA. 1995;92:6562–6566. doi: 10.1073/pnas.92.14.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoneda M, Chomyn A, Martinuzzi A, Hurko O, Attardi G. Proc Natl Acad Sci USA. 1992;89:11164–11168. doi: 10.1073/pnas.89.23.11164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chomyn A, Martinuzzi A, Yoneda M, Daga A, Hurko O, Johns D, Lai S T, Nonaka I, Angelini C, Attardi G. Proc Natl Acad Sci USA. 1992;89:4221–4225. doi: 10.1073/pnas.89.10.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Attardi G, Yoneda M, Chomyn A. Biochim Biophys Acta. 1995;1271:241–248. doi: 10.1016/0925-4439(95)00034-2. [DOI] [PubMed] [Google Scholar]
- 39.Boulet L, Karpati G, Shoubridge E A. Am J Hum Genet. 1992;51:1187–1200. [PMC free article] [PubMed] [Google Scholar]