Abstract
α-N-Acetylglucosaminidase deficiency (mucopolysaccharidosis IIIB, MPS IIIB) and α-l-iduronidase deficiency (MPS I) are heritable lysosomal storage diseases; neurodegeneration is prominent in MPS IIIB and in severe cases of MPS I. We have obtained morphologic and molecular evidence for the involvement of microglia in brain pathology of mouse models of the two diseases. In the cortex, a subset of microglia (sometimes perineuronal) consists of cells that are probably phagocytic; they have large storage vacuoles, react with MOMA-2 (monoclonal antibody against macrophages) and Griffonia simplicifolia isolectin IB4, and stain intensely for the lysosomal proteins Lamp-1, Lamp-2, and cathepsin D as well as for GM3 ganglioside. MOMA-2-positive cells appear at 1 and 6 months in MPS IIIB and MPS I mice, respectively, but though their number increases with age, they remain sparse. However, a profusion of cells carrying the macrophage CD68/macrosialin antigen appear in the cortex of both mouse models at 1 month. mRNA encoding CD68/macrosialin also increases at that time, as shown by microarray and Northern blot analyses. Ten other transcripts elevated in both mouse models are associated with macrophage functions, including complement C4, the three subunits of complement C1q, lysozyme M, cathepsins S and Z, cytochrome b558 small subunit, macrophage-specific protein 1, and DAP12. An increase in IFN-γ and IFN-γ receptor was observed by immunohistochemistry. These functional increases may represent activation of resident microglia, an influx and activation of blood monocytes, or both. They show an inflammatory component of brain disease in the two MPS, as is known for many neurodegenerative disorders.
Mucopolysaccharidoses (MPS) type I and type IIIB are autosomal recessive disorders caused by mutations in the genes encoding α-l-iduronidase and α-N-acetylglucosaminidase, respectively. Both enzymes are required for degradation of glycosaminoglycans (GAG): α-l-iduronidase for dermatan sulfate and heparan sulfate, and α-N-acetylglucosaminidase for heparan sulfate. In the absence of either of these enzymes, the undegraded or partially degraded GAG accumulate in lysosomes, giving rise to cell and tissue pathology and to devastating clinical consequences (1). The clinical manifestations of MPS IIIB (Sanfilippo syndrome type B) are seen primarily in the central nervous system, with profound mental retardation, behavioral disturbances, and neurodegeneration; death usually occurs in the second decade. The manifestations of MPS I (Hurler, Hurler–Scheie, and Scheie syndromes) are more varied and affect many organs, including skeleton, joints, eyes, heart, liver, and spleen in addition to the central nervous system. Profound mental retardation and death in childhood are seen only in the most severe form of MPS I (Hurler syndrome). The less severely affected MPS I patients have normal intelligence and a lifespan ranging from two decades to near normal (Hurler–Scheie and Scheie).
The cellular and molecular mechanisms by which lysosomal storage of GAG can lead to neurodegeneration are not understood. The availability of mouse models of both MPS makes it possible to investigate such questions. Earlier studies have shown that neurons of mouse models of MPS I (2, 3) and MPS IIIB (4) contain storage vacuoles and that other cells in the brain are also affected. There is prominent storage in microglia (4), as well as a marked increase of reactive astrocytes in the MPS IIIB mice (5).
Microglia (cells of the monocyte/macrophage lineage in the brain) have been implicated in the pathogenesis of a number of neurodegenerative conditions, including Alzheimer's disease, HIV dementia, and multiple sclerosis (6). As part of the innate immune defense mechanism, microglia can defend the central nervous system against damage, but they can also produce neurotoxic substances (6–10). Involvement of microglia in the pathogenesis of a lysosomal storage disease was shown in the mouse model of Sandhoff disease (11). The appearance of activated microglia was found to precede the massive apoptosis that occurs in these mice. Bone marrow transplantation, and the resulting migration of healthy donor microglia to the Sandhoff mouse brain, led to suppression of activated microglia and to a delay of neuronal death, even though neuronal GM2 ganglioside storage was not decreased. It was suggested that the damage to neurons caused by the primary defect (lysosomal storage) was exacerbated by the inflammatory response of microglia. Involvement of microglia has also been suggested in mouse models of two other lysosomal storage diseases: metachromatic leukodystrophy (12) and Niemann–Pick disease type C (13).
In view of the implications of microglial involvement for the pathogenesis as well as the potential treatment of MPS, we have undertaken a study of these cells in the brain of mouse models of MPS I and MPS IIIB.
Methods
Animals.
The mouse model of MPS IIIB, in which the Naglu gene is disrupted in exon 6, has been described (4). The mouse model of MPS I was generated by disruption of the Idua gene by insertion of the neomycin resistance gene on a tk (herpes simplex virus thymidine kinase) promoter into the unique BstEII site (blunt-ended) in exon 6, in the opposite orientation. Both mutant genes were placed onto a C57BL/6 inbred background by repeated back-crossing. The phenotype of our MPS I mice is similar, though not identical, to that described earlier (2, 3), perhaps because of the greater inbreeding. The animal studies were approved by the University of California, Los Angeles, Animal Research Committee.
Tissue Preparation for Light Microscopy.
Before dissecting the brain, mice were deeply anesthetized with 100 mg/kg pentobarbital, then perfused through the left ventricle with PBS, pH 7.4. For frozen sections, the brains were submerged in Tissue-Tek Optimal Temperature Cutting Compound (Sakura Finetek, Torrance, CA) for freezing and kept at −80°C before sectioning in a cryostat. For vibratome sectioning, the brains were postfixed in 4% formaldehyde overnight and stored at 4°C in 70% ethanol. Frozen and vibratome sections were 7 and 40 μm thick, respectively.
Electron Microscopy.
Mice anesthetized as above were perfused with PBS, followed by PBS containing 4% formaldehyde and 0.1% glutaraldehyde. The brain was rapidly removed, and 500 μm-thick slices in the coronal plane were fixed overnight at 4°C in 4% paraformaldehyde/2% glutaraldehyde in PBS, and then for 2 h on ice in 1% OsO4. The slices were then processed, embedded in Spurr resin by standard protocols, and cut to 30–40 nm. Sections were stained with uranyl acetate and lead citrate by standard procedures and analyzed in a JEM-1200EX electron microscope (JEOL) at ×3–6,000 magnification.
Reagents for Staining.
A rat monoclonal antibody against mouse macrophages/monocytes (MOMA-2) and a rat monoclonal antibody against mouse CD68 (macrosialin) were purchased from Serotec. Goat polyclonal antibodies against Lamp-1 and Lamp-2 were purchased from Santa Cruz Biotechnology. A mouse monoclonal antibody against GM3 ganglioside was purchased from Seikagaku America (Falmouth, MA). Rabbit anti-mouse IFN-γ and rabbit anti-mouse IFN-γ receptor chain 1 were from PBL Biomedical Laboratories (New Brunswick, NJ). These reagents were used at the concentration recommended by the supplier. For secondary antibodies, biotin-labeled F(ab′)2 for staining and FITC and TRITC-labeled F(ab′)2 for fluorescence were obtained from Jackson ImmunoResearch. Goat Fab anti-mouse IgG and goat Fab anti-mouse IgM were also obtained from Jackson ImmunoResearch. FL (bodipy)-pepstatin A, a fluorescent probe for cathepsin D, was purchased from Molecular Probes. Griffonia (Bandeireae) simplicifolia isolectin IB4, a probe for α-galactosyl groups and a macrophage marker, was purchased from Sigma.
Immunostaining.
Sections were preincubated sequentially for 30 min with PBS, 5 min with PBS containing 0.1% H2O2, and 5 min with PBS. To minimize nonspecific staining, the sections were incubated for 30 min with a mixture of goat Fab anti-mouse IgG and anti-mouse IgM in PBS, followed by 30 min in blocking solution of PBS containing 2% BSA for anti-GM3, or 2% BSA plus 0.1% Triton X-100 for other antibodies. The sections were then reacted overnight with primary antibody in blocking solution and washed for 30 min with PBS. For visualization by light microscopy, the sections were reacted for 30 min with secondary antibody conjugated to biotin. Color was developed by the avidin-biotin-peroxidase method (ABC Elite kit, Vector Laboratories) with diaminobenzidine (DAB) or VIP (Vector Laboratories) as chromogen. For visualization by fluorescence, the sections were reacted with secondary antibody conjugated to fluorescent dye, FITC or tetramethylrhodamine B isothiocyanate (sequentially for double staining). Staining was observed by fluorescence microcopy (DIA PHOTO, Nikon Instruments) or by confocal laser scanning microscopy (Leica). Some sections were counterstained with Hoechst dye 33342 (Molecular Probes). Staining for G. simplicifolia isolectin IB4 and for cathepsin D was performed as per instructions of the suppliers.
Microarray Analysis.
After death, the brain was removed, and cortex was dissected. The cortexes of three +/+ or of three −/− mice, age 3 months, were pooled. Total RNA was isolated with Trizol Reagent (Invitrogen) by the supplier's instructions. Synthesis of cDNA and biotin-labeled cRNA, fragmentation, and hybridization were performed according to the Affimetrix Genechip Expression Analysis Technical Manual (Affimetrix, Santa Clara, CA). Briefly, 20 μg of total RNA was used for cDNA synthesis using the Superscript II cDNA synthesis kit (Invitrogen); the cDNA was then cleaned by Phase Lock Gel Centrifugation (Eppendorf). Biotin-labeled cRNA was synthesized by in vitro transcription of the cDNA with the Enzo BioArray High Yield RNA transcript labeling kit (Enzo Diagnostics), fragmented, and then hybridized to duplicate Affimetrix Murine Genome U74v2, GeneChip A. The chip contains probe sets for 12,000 full-length mouse genes and EST clusters. Hybridization and scanning were done by the University of California, Los Angeles, Microarray Core facility, with the Affimetrix Fluidic Station 400 and Gene Array scanner. Analysis was performed by using the Affimetrix Microarray Suite version 4.0. Only genes with a >1.7-fold increase and flagged as Present were considered significantly increased over the wild type.
Northern Blot Analysis.
Aliquots (5 μg) of total RNA (obtained from the cortex of individual mice) were used for Northern blot analysis. The RNA was subjected to electrophoresis on a 1% agarose-formaldehyde gel and blotted onto a nylon membrane. Templates for the synthesis of riboprobes, 250–300 bp long, were generated by PCR with oligonucleotide primers from the coding region of each gene. Reverse primers contained a T7 promoter sequence. 32P-labeled riboprobes were prepared with Maxiscript T7 labeling kit (Ambion, Austin, TX). Radioactivity was visualized and quantitated with a PhophorImager (Molecular Dynamics) and normalized to β-actin.
Results
Morphological Evidence of Microglia.
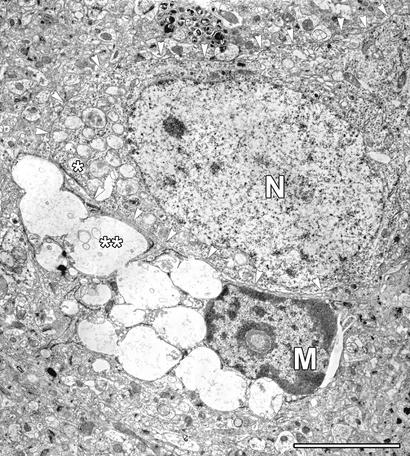
Electron microscopic examination of the cortex of an MPS IIIB mouse shows microglia that are sometimes apposed to neurons (Fig. 1). The microglial cell, identifiable in the electron micrograph by the shape of the nucleus, has enormous vacuoles that appear empty or contain some flocculent material and have the characteristic appearance of lysosomes engorged with GAG. The adjoining neuron, on the other hand, has very small vacuoles filled with denser material. Unattached microglia have similar morphology. Similar images were seen in the cortex of MPS I mice (data not shown).
Figure 1.
A microglial cell with large inclusions, juxtaposed to a neuron in the cortex of a 6-month-old MPS III B mouse. M and N identify the nucleus of the microglia and of the neuron, respectively. Note the large, nearly empty inclusions in the microglia (**) and the much smaller and denser inclusions in the neuron (*). Arrowheads trace the membrane of the neuron. (Scale bar = 5 μm.)
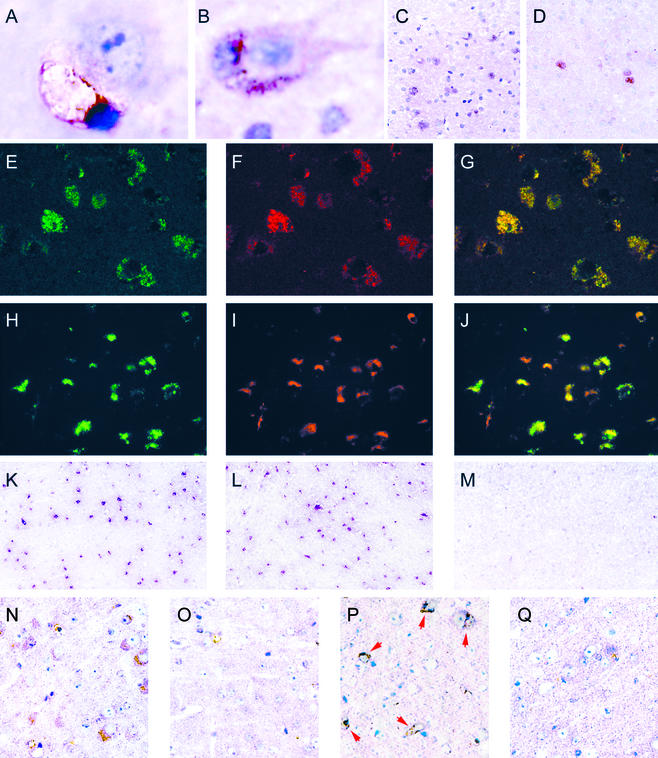
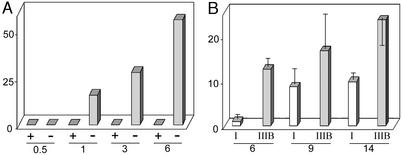
The identification of the perineuronal cells as microglia was confirmed by staining with G. simplicifolia isolectin IB4 (Fig. 2A) or MOMA-2 (Fig. 2B), reagents that specifically recognize cells of the macrophage/monocyte lineage (14–16). MOMA-2-positive microglia appeared in the cortex of MPS IIIB mice between 2 weeks and 1 month, and increased over time (Fig. 3A); they were not seen in the cortex of wild-type mice at any time (Fig. 3A) or of heterozygous mice up to 3 months, the only ages examined (not shown). MOMA-2-positive cells were also seen in other parts of the brain, but in lesser number (not shown). The MOMA-2 cells were also present in the brain of MPS I mice, but in lower number and with later appearance (Fig. 3B). However, they remained sparse in both mouse models (Figs. 2 C and D and 3B).
Figure 2.
Histochemical and immunohistochemical characterization of microglia in cortex. (A) G. simplicifolia isolectin IB4-positive microglia apposed to neuron; MPS IIIB, 7 months (×1,750). (B) MOMA-2-positive microglia apposed to neuron; MPS IIIB, 3 months (×1,150). (C and D) MOMA-2-positive cells seen at low power; MPS IIIB 3 months and MPS I, 14 months, respectively (×140). (E–G) Confocal images of sections stained with antibody against Lamp-1 (green), MOMA-2 (red), and merged; MPS IIIB, 6 months (×350). (H–J) Fluorescent images using antibody against ganglioside GM3 (green), MOMA-2 (red), and merged; MPS IIIB, 6 months (×260). (K–M) Images of sections stained with antibody against CD68/macrosialin, MPS IIIB, MPS I, and control, respectively, 3 months (×65). (N and O) Images of sections stained with antibody against IFN-γ; MPS IIIB, and control, respectively, 3 months (×250). (P and Q) Images of sections stained with antibody against IFN-γ receptor; MPS IIIB and control, respectively, 7 months (×250).
Figure 3.
Increase of MOMA-2-positive microglia with age. (A) Comparison of the number of MOMA-2-positive cells in cortex of MPS IIIB (−) and wild-type control (+) mice at 0.5, 1, 3, and 6 months. Each bar represents the number of MOMA-2-positive cells in a low-power field of the cortex of one mouse. (B) Comparison of density of MOMA-2-positive cells in a low-power field in the cortex of MPS I and MPS IIIB mice of different ages; each bar represents one mouse, with SD derived from counting cells in three low-power fields.
The MOMA-2-positive cells in both mouse models stained intensely for lysosomal markers. Staining by MOMA-2 coincided with immunohistochemical staining of Lamp-1 (Fig. 2 E–G) as well as of Lamp-2 (data not shown), both lysosomal membrane proteins. The colocalization observed by confocal microscopy suggests that the MOMA-2 antigen and Lamp-1 are present in the same intracellular organelles. The MOMA-2-positive cells also stained with pepstatin-A, an inhibitory peptide that can be used as a probe for the soluble lysosomal enzyme cathepsin D (data not shown). Such strong immunostaining of lysosomal membrane and soluble proteins is in agreement with the presence of the very large storage vacuoles (lysosomes) seen in the microglia (Fig. 1).
The MOMA-2-positive cells also showed immunostaining for GM3 ganglioside (Fig. 2 H–J). The colocalization was not absolute in that not all MOMA-2 cells stained for GM3, and not all cells staining for GM3 were microglia. Some neurons were also stained by the antibody (not shown).
Although MOMA-2 staining was seen in relatively few cells of younger mice, particularly of the MPS I model, immunostaining of macrosialin, the murine homolog of CD68 and a marker of cells of the macrophage lineage, showed a profusion of positive cells in mutant mice of both genotypes at 3 months (Fig. 2 K and L) and even as early as 3 weeks (data not shown). No CD68/macrosialin-positive cells were seen in the parenchyma of the cortex of normal mice (Fig. 2M). However, CD68/macrosialin-positive staining was seen in both normal and mutant mice in cells lining the capillaries and in the corpus callosum (not shown).
Preliminary experiments showed that immunostaining for IFN-γ was increased in the cortex of MPS IIIB mice relative to control mice (Fig. 2 N and O), as was immunostaining for IFN-γ receptor 1 (Fig. 2 P and Q). The staining was present in microglia and perhaps also in neurons, but because of the close apposition of microglia and neurons, the identification of the cells is not definitive.
Molecular Evidence for Microglial Involvement.
Microarray analysis showed that a number of transcripts were present at a level significantly higher in the cortex of MPS IIIB and MPS I mice than in the cortex of control mice (Table 1). Of the 39 transcripts increased in MPS IIIB mice and 17 transcripts increased in MPS I mice, 13 were increased in both mouse models (Table 1, shown in bold). One of the transcripts increased in both is glial fibrillary acid protein, a marker of activated astrocytes, whereas another, granulin, is of unknown cellular origin. The others are transcripts produced by, or enriched in, cells of the macrophage/monocyte lineage.
Table 1.
Transcripts increased in cortex of mouse models of MPS IIIB and MPS I, determined by microarray analysis
| GenBank accession no. | Fold* | Transcripts |
|---|---|---|
| MPS III B mice | ||
| M21050 | 6.0 | Lysozyme M |
| M64086 | 5.8 | Mouse spi2 proteinase inhibitor (spi2/eb4) |
| X02801 | 4.1 | Glial fibrillary acidic protein |
| X06454 | 3.8 | Complement C4 |
| AW123191 | 3.5 | Similar to interferon-inducible protein |
| AF024637 | 3.3 | DAP 12 |
| AV00854 | 3.0 | Glyceraldehyde-3-phosphate dehydrogenase |
| X68273 | 2.9 | CD68/macrosialin |
| M22531 | 2.5 | Complement C1q B chain |
| X61800 | 2.5 | CCAAT/enhancer binding protein (C/EBP) δ |
| U22033 | 2.3 | Large multifunctional protease 7 |
| AJ242663 | 2.3 | Cathepsin Z precursor |
| L20315 | 2.3 | Macrophage-specific gene 1 |
| U74683 | 2.2 | Cathepsin C |
| X60980 | 2.2 | Thymidine kinase |
| AB007599 | 2.2 | MD-1 |
| X53929 | 2.2 | Decorin |
| X00496 | 2.2 | Mouse Ia-associated invariant chain (Ii) |
| AF010254 | 2.1 | Complement component 1 inhibitor |
| X58861 | 2.1 | Complement C1q, A chain |
| AJ223208 | 2.1 | Cathepsin S |
| AI430879 | 2.1 | Mus musculus cDNA |
| AW124151 | 2.1 | Mus musculus cDNA |
| U06119 | 2.0 | Cathepsin H |
| X66295 | 2.0 | Complement C1q, C chain |
| AV356071 | 2.0 | Lysosomal transmembrane protein |
| AF053943 | 2.0 | Aortic carboxypeptidase-like protein ACLP |
| U29539 | 2.0 | Retinoic acid-inducible E3 protein |
| X98113 | 1.9 | Lymphocyte-activation gene 3 |
| X57796 | 1.9 | Tumor necrosis factor receptor superfamily, member 1a |
| D16195 | 1.8 | Granulin |
| U14419 | 1.8 | GABA-benzodiazepine receptor β-2 subunit |
| AV242495 | 1.8 | Mus musculus cDNA |
| AF069051 | 1.8 | Pituitary tumor transforming gene protein |
| U60020 | 1.8 | Transporter 1, ABC |
| AF013486 | 1.8 | Type I interferon receptor soluble isoform |
| M31775 | 1.7 | Cytochrome β-558, small subunit |
| AI642048 | 1.7 | Mus musculus cDNA |
| MPS I mice | ||
| M21050 | 4.4 | Lysozyme M |
| X02801 | 4.4 | Glial fibrillary acid protein |
| R75193 | 3.9 | Mus musculus cDNA |
| L20315 | 3.2 | Macrophage-specific gene 1 |
| X68273 | 3.2 | CD68/macrosialin |
| AF024637 | 3.1 | DAP 12 |
| M22531 | 2.4 | Complement C1q, B chain |
| X58861 | 2.3 | Complement C1q, A chain |
| AJ223208 | 2.3 | Cathepsin S |
| X06454 | 2.2 | Complement C4 |
| X66295 | 2.1 | Complement C1q, C chain |
| X87142 | 2.0 | Ca/calmodulin-dependent protein kinase II α |
| AJ242663 | 1.9 | Cathepsin Z precursor |
| D16195 | 1.8 | Granulin |
| AA815845 | 1.8 | Mus musculus cDNA |
| M31775 | 1.8 | Cytochrome β-558, small subunit |
| AJ007909 | 1.7 | Erythroid differentiation regulator (activin A) |
The transcripts shown in bold are those increased in both mouse models.
Average fold increase over normal.
Northern blot analyses of selected transcripts confirmed the increase observed by microarray and showed that the mRNA levels were significantly elevated in the cortex of MPS I and MPS IIIB mice as young as 1 month of age (Table 2). The elevated level persisted for up to 9 months for those transcripts that were examined in older mice.
Table 2.
Increase in transcripts in cortex of mice at different ages determined by Northern blot analysis
| MPS III B, mo.
|
MPS I, mo.
|
||||
|---|---|---|---|---|---|
| 1 | 3 | 9 | 1 | 3 | |
| Complement C1q A chain | 2.3 | 2.0 | 1.9 | 2.5 | 2.2 |
| CD68/macrosialin | 1.7 | 2.1 | 2.0 | 2.2 | 2.6 |
| Lysozyme M | 13 | 15 | – | 3.0 | 9.0 |
| Macrophage-specific gene 1 | 3.6 | 1.9 | 2.4 | 3.3 | 3.2 |
| DAP12 | 1.7 | 2.3 | 1.7 | 1.7 | 2.0 |
| Cathepsin S | 2.2 | 2.2 | 1.8 | – | – |
| Glial fibrillary acid protein | 2.0 | 2.9 | 3.5 | – | – |
The numbers represent the fold-increase of the transcripts, determined by Northern blot quantitation as described in Methods.
Discussion
Our interest in microglia in the brain of the mouse models of MPS I and IIIB was prompted by the remarkable appearance of perineuronal microglia in electron micrographs. Similar pictures had been shown for the brain of a canine model of MPS I (17) and of a mouse model of MPS IIIA, a deficiency of heparan N-sulfatase (18). The enormous and nearly empty inclusions in the microglial cell, characteristic of lysosomes loaded with GAG, contrast with the small inclusions in the neuron to which it is apposed. Many microglia not apposed to neurons have similarly large inclusions. The basis of the difference in storage between neurons and microglia is not known, but we surmise that the large amount of undegraded GAG stored in microglia is not just of endogenous origin, but is also acquired by scavenging from the environment or by phagocytosis of dead or damaged neurons.
Identification of the perineuronal cell as a microglia was confirmed by its reactivity with G. simplicifolia isolectin IB4 and with MOMA-2. The isolectin has long been used to identify macrophages (14) and microglia (15), and is greatly increased on activation of these cells. It reacts with α-galactoside residues on an unidentified membrane glycoprotein(s). MOMA-2 is a monoclonal antibody that had been selected to react with mononuclear phagocytes (16). The original study showed that it reacted with monocytes and most tissue macrophages (although not microglia) of normal mice, and that it was localized to the same cells as acid phosphatase. It was later shown to stain only a subset of macrophages (19). The only non-macrophage cells reported to react with MOMA-2 are 3T3-L1 cells during their phagocytic preadipocyte stage (20). In the brains of MPS IIIB and MPS I mice, MOMA-2 reactivity was found on a relatively small number of cells that also stained strongly for lysosomal proteins (Lamp-1, Lamp-2, and cathepsin D). Confocal microscopy indicated localization of MOMA-2 in the same subcellular organelles as Lamp-1, a lysosomal membrane protein. Taken together, the data suggest that MOMA-2 reactive cells in the mouse models represent a subset of microglia that are phagocytic and have a strongly developed lysosomal system, and that the unknown antigen of MOMA-2 is probably a lysosomal protein.
The phagocytic nature of the MOMA-2 microglia may explain the enrichment of GM3 ganglioside in these cells. Ganglioside GM3, and to a lesser extent ganglioside GM2, are present in excess amount in the brain of animal models (3, 4, 21) and of humans patients affected by MPS I and MPS III B (22, 23). Although GM3 ganglioside is ubiquitously made, it is both a precursor and a degradation product of the more complex gangliosides (such as GM2, GM1, and the di- and tri-sialylated gangliosides) that are especially abundant on neuronal membranes (24). The microglia may have acquired the excess ganglioside GM3 by phagocytosis of neurons that had undergone normal apoptosis during development or of neurons that had been damaged by the disease.
Although MOMA-2-positive cells are limited in number, increase slowly over the lifetime of the mouse, and are more numerous in MPS IIIB than in MPS I mice, CD68/macrosialin-positive cells appear in large number at 1 month or even earlier (data not shown), and in roughly equal numbers in the two mouse models. Macrosialin, the murine homolog of CD68, is related to the lysosome-associated membrane glycoprotein (Lamp) family of endosomal-lysosomal proteins, and is restricted to cells of the macrophage lineage (25). The immunohistochemical appearance of CD68/macrosialin corresponds temporally with a doubling of its transcript, as seen on microarray and Northern blots.
Coinciding with an increase in the transcripts for CD68/macrosialin, there is an increase in both mouse models of other transcripts associated with cells of the macrophage/monocyte lineage. Cathepsin S is preferentially expressed in such cells (26), as is lysozyme M (27) and macrophage-specific gene 1 (28). Cytochrome b558 is part of the NADPH oxidase complex, which is responsible for the oxidative burst of phagocytes, including microglia (29). DAP12, a tyrosine protein kinase binding protein, was originally described in natural killer cells (30) but later was also found in monocytes and microglia (31). Cathepsin Z has been found in phagosomes of macrophages (32). Complement C1q is specifically and dramatically increased in microglia in transient global cerebral ischemia (33). C1q and complement C4 are produced by microglia (34) but also by other brain cells in Alzheimer's disease (35). It is noteworthy that the increase in transcripts observed in the cortex of MPS I and MPS IIIB mice is an early event, seen by 1 month of age. It precedes the behavioral abnormality in an open field test, which was observed in MPS IIIB mice at 4.5 months but not at 2.5 months (4). Finally, the preliminary evidence for increase in IFN-γ and its receptor indicate an inflammatory response in the brain; IFN-γ is produced by microglia and astrocytes (36) as well by activated macrophages (37).
Our histological and molecular results do not discriminate between an expansion and activation of microglia already present in the brain, an influx and activation of monocytes from blood, or a combination thereof. However, the distinction bears primarily on the mechanism underlying the appearance of antigens and the increase in certain transcripts. Regardless of whether the mechanism involves activation of preexisting microglia and up-regulation of gene expression in these cells and/or an admixture and activation of blood monocytes displaying the antigens and transcripts, the effect is the same. The cortexes of the MPS IIIB and the MPS I mice show a marked increase in functions associated with cells of the macrophage/monocyte lineage. An important question, relevant to development of therapy, is whether this increase results in an exacerbation of the disease process. At the very least, microglia should be considered a target cell in therapeutic strategies.
Acknowledgments
We thank Dr. Alexei Slesarev for preparing the constructs used in making the MPS I mouse model, and Hui-Zhi Zhao for excellent technical assistance. This work was supported in part by National Institutes of Health Grants NS22376 and DK38857, a fellowship from the National MPS Society, Inc., and a grant from the Children's Medical Research Foundation.
Abbreviations
- MPS
mucopolysaccharidosis
- GAG
glycosaminoglycan
References
- 1.Neufeld E F, Muenzer J. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 2001. pp. 3421–3452. [Google Scholar]
- 2.Clarke L A, Russell C S, Pownall S, Warrington C L, Borowski A, Dimmick J E, Toone J, Jirik F R. Hum Mol Genet. 1997;6:503–511. doi: 10.1093/hmg/6.4.503. [DOI] [PubMed] [Google Scholar]
- 3.Russell C, Hendson G, Jevon G, Matlock T, Yu J, Aklujkar M, Ng K Y, Clarke L A. Clin Genet. 1998;53:349–361. doi: 10.1111/j.1399-0004.1998.tb02745.x. [DOI] [PubMed] [Google Scholar]
- 4.Li H H, Yu W H, Rozengurt N, Zhao H Z, Lyons K M, Anagnostaras S, Fanselow M S, Suzuki K, Vanier M T, Neufeld E F. Proc Natl Acad Sci USA. 1999;96:14505–14510. doi: 10.1073/pnas.96.25.14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H H, Zhao H Z, Neufeld E F, Cai Y, Gomez-Pinilla F. J Neurosci Res. 2002;69:30–38. doi: 10.1002/jnr.10278. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez-Scarano F, Baltuch G. Annu Rev Neurosci. 1999;22:219–240. doi: 10.1146/annurev.neuro.22.1.219. [DOI] [PubMed] [Google Scholar]
- 7.Streit W J, Walter S A, Pennell N A. Prog Neurobiol. 1999;57:563–581. doi: 10.1016/s0301-0082(98)00069-0. [DOI] [PubMed] [Google Scholar]
- 8.Smits H A, Boven L A, Pereira C F, Verhoef J, Nottet H S. Eur J Clin Invest. 2000;30:526–535. doi: 10.1046/j.1365-2362.2000.00661.x. [DOI] [PubMed] [Google Scholar]
- 9.Gebicke-Haerter P J. Microsc Res Tech. 2001;54:47–58. doi: 10.1002/jemt.1120. [DOI] [PubMed] [Google Scholar]
- 10.Hanisch U K. Glia. 2002;40:140–155. doi: 10.1002/glia.10161. [DOI] [PubMed] [Google Scholar]
- 11.Wada R, Tifft C J, Proia R L. Proc Natl Acad Sci USA. 2000;97:10954–10959. doi: 10.1073/pnas.97.20.10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hess B, Saftig P, Hartmann D, Coenen R, Lullmann-Rauch R, Goebel H H, Evers M, von Figura K, D'Hooge R, Nagels G, et al. Proc Natl Acad Sci USA. 1996;93:14821–14826. doi: 10.1073/pnas.93.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.German D C, Liang C L, Song T, Yazdani U, Xie C, Dietschy J M. Neuroscience. 2002;109:437–450. doi: 10.1016/s0306-4522(01)00517-6. [DOI] [PubMed] [Google Scholar]
- 14.Maddox D E, Shibata S, Goldstein I J. Proc Natl Acad Sci USA. 1982;79:166–170. doi: 10.1073/pnas.79.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Streit W J, Kreutzberg G W. J Neurocytol. 1987;16:249–260. doi: 10.1007/BF01795308. [DOI] [PubMed] [Google Scholar]
- 16.Kraal G, Rep M, Janse M. Scand J Immunol. 1987;26:653–661. doi: 10.1111/j.1365-3083.1987.tb02301.x. [DOI] [PubMed] [Google Scholar]
- 17.Shull R M, Breider M A, Constantopoulos G C. Pediatr Res. 1988;24:347–352. doi: 10.1203/00006450-198809000-00015. [DOI] [PubMed] [Google Scholar]
- 18.Bhaumik M, Muller V J, Rozaklis T, Johnson L, Dobrenis K, Bhattacharyya R, Wurzelmann S, Finamore P, Hopwood J J, Walkley S U, Stanley P. Glycobiology. 1999;9:1389–1396. doi: 10.1093/glycob/9.12.1389. [DOI] [PubMed] [Google Scholar]
- 19.Leenen P J, de Bruijn M F, Voerman J S, Campbell P A, van Ewijk W. J Immunol Methods. 1994;174:5–19. doi: 10.1016/0022-1759(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 20.Cousin B, Munoz O, Andre M, Fontanilles A M, Dani C, Cousin J L, Laharrague P, Casteilla L, Penicaud L. FASEB J. 1999;13:305–312. doi: 10.1096/fasebj.13.2.305. [DOI] [PubMed] [Google Scholar]
- 21.Constantopoulos G, Shull R M, Hastings N, Neufeld E F. J Neurochem. 1985;45:1213–1217. doi: 10.1111/j.1471-4159.1985.tb05544.x. [DOI] [PubMed] [Google Scholar]
- 22.Constantopoulos G, Eiben R M, Schafer I A. J Neurochem. 1978;31:1215–1222. doi: 10.1111/j.1471-4159.1978.tb06245.x. [DOI] [PubMed] [Google Scholar]
- 23.Hara A, Kitazawa N, Taketomi T. J Lipid Res. 1984;25:175–184. [PubMed] [Google Scholar]
- 24.Gravel R A, Kaback M M, Proia R L, Sandhoff K, Suzuki K, Suzuki K. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 2001. pp. 3827–3876. [Google Scholar]
- 25.Holness C L, da Silva R P, Fawcett J, Gordon S, Simmons D L. J Biol Chem. 1993;268:9661–9666. [PubMed] [Google Scholar]
- 26.Petanceska S, Canoll P, Devi L A. J Biol Chem. 1996;271:4403–4409. doi: 10.1074/jbc.271.8.4403. [DOI] [PubMed] [Google Scholar]
- 27.Cross M, Mangelsdorf I, Wedel A, Renkawitz R. Proc Natl Acad Sci USA. 1988;85:6232–6236. doi: 10.1073/pnas.85.17.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spilsbury K, O'Mara M A, Wu W M, Rowe P B, Symonds G, Takayama Y. Blood. 1995;85:1620–1629. [PubMed] [Google Scholar]
- 29.Klegeris A, McGeer P L. J Neuroimmunol. 1994;53:83–90. doi: 10.1016/0165-5728(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 30.Lanier L L, Corliss B C, Wu J, Leong C, Phillips J H. Nature. 1998;391:703–707. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- 31.Bakker A B, Wu J, Phillips J H, Lanier L L. Hum Immunol. 2000;61:18–27. doi: 10.1016/s0198-8859(99)00160-3. [DOI] [PubMed] [Google Scholar]
- 32.Lennon-Dumenil A M, Bakker A H, Maehr R, Fiebiger E, Overkleeft H S, Rosemblatt M, Ploegh H L, Lagaudriere-Gesbert C. J Exp Med. 2002;196:529–540. doi: 10.1084/jem.20020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schafer M K, Schwaeble W J, Post C, Salvati P, Calabresi M, Sim R B, Petry F, Loos M, Weihe E. J Immunol. 2000;164:5446–5452. doi: 10.4049/jimmunol.164.10.5446. [DOI] [PubMed] [Google Scholar]
- 34.Eikelenboom P, Veerhuis R. Neurobiol Aging. 1996;17:673–680. doi: 10.1016/0197-4580(96)00108-x. [DOI] [PubMed] [Google Scholar]
- 35.Emmerling M R, Watson M D, Raby C A, Spiegel K. Biochim Biophys Acta. 2000;1502:158–171. doi: 10.1016/s0925-4439(00)00042-9. [DOI] [PubMed] [Google Scholar]
- 36.De Simone R, Levi G, Aloisi F. Cytokine. 1998;10:418–422. doi: 10.1006/cyto.1997.0314. [DOI] [PubMed] [Google Scholar]
- 37.Munder M, Mallo M, Eichmann K, Modolell M. J Exp Med. 1998;187:2103–2108. doi: 10.1084/jem.187.12.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]