Abstract
Although several genes that might mediate p53-induced apoptosis have been proposed, none have previously been shown to play an essential role in this process through a rigorous gene disruption approach. We used a gene-targeting approach to evaluate p53-mediated death in human colorectal cancer cells. Expression of p53 in these cells induces growth arrest through transcriptional activation of the cyclin-dependent kinase inhibitor p21. If p21 is disrupted via gene targeting, the cells die through apoptosis. If the PUMA gene is also disrupted in such cells, apoptosis is prevented. The effects of PUMA on apoptosis were observed after exogenous overexpression of p53 as well as after exposure to hypoxia, a physiologic activator of p53, and DNA damage. The PUMA protein interacts with Bcl-XL and promotes mitochondrial translocation and multimerization of Bax. Accordingly, genetic disruption of BAX makes cells resistant to the apoptosis resulting from PUMA expression. These results suggest that the balance between PUMA and p21 is pivotal in determining the responses to p53 activation and provide a model for understanding the basis of p53 mutations in human cancer.
That the p53 pathway is inactivated in the great majority of human cancers has stimulated an avalanche of research to understand the basis for its tumor suppressor activities (1–3). Of the many physiologic effects of p53 that have been described, current evidence suggests that apoptosis is critical for its tumor suppressor activities (4–6). At the biochemical level, it is known that p53 is a transcription factor that binds to specific sequences within the promoters of various genes and activates their expression (7). These genes include two major classes, one that controls the cell cycle and one that controls apoptosis. Although rigorous genetic evidence has supported the hypothesis that the first class of genes is required for p53-dependent growth arrest, no analogous evidence has definitively implicated any specific p53-regulated gene as an essential mediator of p53-dependent apoptosis (8, 9).
In mammalian cells, apoptosis is regulated by Bcl-2 family proteins (10, 11). This family includes multidomain members such as Bcl-2, Bcl-XL, and Bax and the “BH3-only” group of proteins, each of which contains a single amphipathic Bcl-2 homology (BH) domain (12). As sensors of discrete apoptotic stimuli, these BH3-only proteins interact with the multidomain Bcl-2 family proteins to either antagonize or activate their function (13, 14). As a result, a cascade of downstream events is triggered, including collapse of mitochondrial membrane potential, release of the apoptogenic mitochondrial proteins cytochrome c and SMAC, and activation of caspases (15).
In a systematic examination of the genes that were activated by p53 in apoptosis, we identified a BH3-only protein termed PUMA (p53 up-regulated modulator of apoptosis) that has an expression pattern consistent with a causative role in p53-dependent apoptosis (16). The same gene was independently identified by others (17, 18). PUMA can be directly activated by p53 through p53-responsive elements in its promoter region. Accordingly, PUMA is induced by the chemotherapeutic agents 5-fluorouracil and adriamycin in a p53-dependent fashion. The protein encoded by PUMA was exclusively localized to mitochondria where it interacted with Bcl-2 and Bcl-XL through its BH3 domain. Exogenous expression of PUMA resulted in rapid and complete apoptosis in a variety of cancer cell lines (16–18).
In this study, we demonstrate through gene-targeting experiments that PUMA plays an essential role in mediating the apoptotic responses to p53. We show that PUMA functions through Bax and propose a general model for the control of cell birth and death in tumors that reconciles much previous data on this topic.
Materials and Methods
Gene Targeting.
HCT116 cells were obtained from the American Type Culture Collection. HCT116 cells with two intact copies of PUMA were transfected with the bipartite targeting vectors shown in Fig. 1A by using Lipofectamine (Invitrogen) and selected with geneticin (0.4 mg/ml) for 3 weeks. The use of bipartite targeting vectors for homologous recombination in general is described in ref. 19. Geneticin-resistant clones were pooled and screened for targeting events by PCR with the primers P1 (5′-GGTGTGCAGGTCTTATCTCTG-3′) and P2 (5′-TTGTGCCCAGTCATAGCCG-3′). Two PCR-positive clones were recovered from ≈4,000 geneticin-resistant clones. The neomycin-resistance gene was excised from the heterozygous clones after infecting cells with an adenovirus (Ad) that expresses Cre recombinase (Ad-Cre) (19). The same targeting construct pair was then used to target the second allele of PUMA. Homologous recombination at the PUMA locus was verified by genomic Southern blotting and analysis of PUMA protein by immunoblotting.
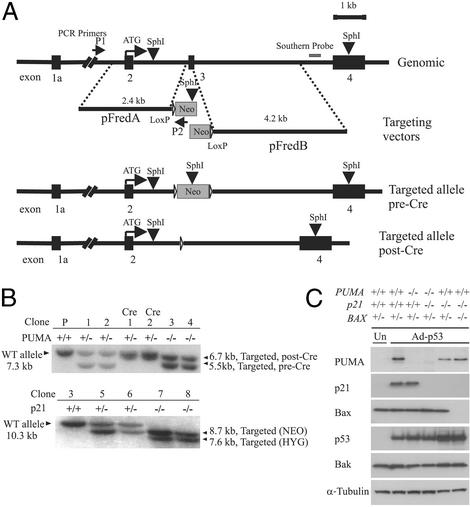
Figure 1.
Targeted deletion of PUMA. (A) Two targeting constructs were designed, each containing a homologous arm and an overlapping fragment of the neomycin-resistant gene. The SphI sites within the PUMA gene, the targeting construct, and the position of the probe used for Southern blotting are shown. PCR screening primers P1 and P2 were described in Materials and Methods. Homologous recombination results in the deletion of exon 3, which contains the BH3 domain. Filled boxes represent PUMA exons 1a–4. The same constructs were used in the second round of targeting after excision of the neomycin-resistance gene by Cre recombinase. (B Upper) A Southern blot obtained with a PUMA probe after SphI digestion of genomic DNA. (Lower) The blot obtained with a p21 probe after BglII digestion. Clone 3 (PUMA−/−, Upper) was used for targeting of the p21 locus, generating clones 5 and 6 (p21+/−). A second round of targeting generated clones 7 and 8, which were p21−/−. (C) Lysates from cells with the indicated genotypes were analyzed by immunoblotting with antibodies against PUMA, p21, p53, Bax, Bak, and α-tubulin. Cells were infected with Ad-p53 24 h before preparation of the lysates. Uninfected parental HCT116 cells (“Un”) show the basal level of p53-induced proteins in the absence of Ad-p53.
Cell lines with targeted disruptions of both alleles of the p21 gene or of the BAX gene have been described (20, 21).
To generate cell lines that were deficient in both PUMA and p21, the neomycin-resistance gene was excised from PUMA−/− cells with Ad-Cre. Targeting of the two alleles of p21 was as described (20). Homologous recombination at the p21 locus was verified by genomic Southern blotting and analysis of p21 by immunoblotting.
Cell lines that were deficient in both p21 and BAX were isolated by incubating p21−/− cells in the presence of 500 μM of indomethacin for 72 h, as described in ref. 21. Of 10 clones recovered after this procedure, eight were found to have deletions of both alleles of the G8 tract within the BAX coding region. The absence of Bax protein in all of these clones was confirmed by immunoblotting.
All lines were maintained in McCoy's 5A media (Invitrogen) supplemented with 10% FBS (HyClone), 100 units/ml penicillin, and 100 μg/ml streptomycin at 37°C.
PUMA- and p53-Inducible Cell Lines.
The generation of DLD-1 cells in which PUMA and p53 could be induced under control of a doxycycline-sensitive promoter has been described (16, 22). In these lines, doxycycline was removed from the media to induce expression of p53 or PUMA. In some cases, the caspase inhibitor Z-VAD (50 μM, Enzyme Systems Products, Livermore, CA) was added to the medium 8 h before doxycycline was removed. All inducible lines were maintained in McCoy's 5A medium (Invitrogen) supplemented with 10% FBS (HyClone), 100 units/ml penicillin, 100 μg/ml streptomycin, 0.4 mg/ml G418 (Invitrogen), 0.25 mg/ml hygromycin B (Calbiochem), and 20 ng/ml doxycycline at 37°C.
Ads.
Ads expressing the WT PUMA gene or a PUMA gene with a deletion of the BH3 domain were generated by using the AdEasy system (23). In brief, a fragment containing a human PUMA cDNA fused to a double hemagglutinin (HA)-epitope tag was excised from the constructs HA-PUMA or HA-PUMAΔBH3 (16), by using the restriction enzymes KpnI and BamHI, and inserted into the shuttle vector pAdTrack-CMV. After recombination with the pAdEasy-1 vector, high-titer viruses were generated in 293 cells. Viruses were purified by CsCl2 gradient centrifugation at 32,000 rpm (Sorvall SW41 rotor), and titers were assayed through measurement of the number of enhanced GFP-positive cells after infection of 293 cells. Ads expressing p53 (Ad-p53) or a mutant p53R175H (Ad-R175H) have been described (22).
Expression Constructs.
GFP-PUMA fusion expression vectors and their variants were constructed by cloning PCR products of pHA-PUMA or pHA-PUMAΔBH3 into the pEGFP-C1 vector (CLONTECH). In all cases, the expression vectors were verified to be free of unexpected mutations in PUMA through complete sequencing. These PUMA expression constructs were transfected into the retinal epithelial cell line 911 (gift of A. J. Van der Eb, University of Leiden, Leiden, The Netherlands) on glass chamber slides (Nalge Nunc/Lab-Tek). The expression of fusion proteins was verified by fluorescence microscopy and immunoblotting with a rabbit polyclonal antibody against GFP (CLONTECH). Twenty hours after transfection, the mitochondria were stained with MitoTracker red (0.1 μM, Molecular Probes). Slides were processed and analyzed by microscopy as described (16), except that nuclei were counterstained with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) instead of Hoechst dye.
Hypoxia.
Hypoxia was generated through incubation of cells in a GasPak System (Becton Dickinson) maintained at 37°C in the presence of 25 mM Hepes.
Antibodies.
A PUMA-specific antibody was generated in rabbits by using the synthetic peptide RARQESSPEPVEGLA corresponding to amino acids 3–17. The p53, p21, HA, Cox IV, and α-tubulin proteins were detected by using mouse mAb DO1 (gift of David Lane, University of Dundee, Dundee, Scotland), OP64 (Oncogene Science), 12CA (Roche Applied Sciences), 20 E8-C12 (Molecular Probes), and OP06 (Oncogene Science), respectively. Bax and Bak were detected with rabbit polyclonal antibodies from Santa Cruz Biotechnology and Upstate Biotechnology (Lake Placid, NY), respectively.
Cellular Fractionation.
Two 75-cm2 flasks of cells were harvested after a PBS wash at 4°C. Whole-cell pellets were resuspended in homogenization buffer (0.25 M sucrose/10 mM Hepes, pH 7.4/1 mM EGTA) and subjected to 40 strokes in a 2-ml Dounce homogenizer at 4°C. The homogenates were subjected to centrifugation at 1,000 × g for 15 min at 4°C to pellet nuclei and unbroken cells. The supernatant was subsequently subjected to centrifugation at 10,000 × g for 15 min at 4°C to obtain the cytosolic fraction (supernatant) and mitochondrial fraction (pellet). In some experiments, cells were cross-linked with 1 mM dithiobis (DSP; Pierce) at 37°C for 30 min before subcellular fractionation, and fractions were subjected to SDS/PAGE under nondenaturing conditions followed by immunoblotting.
Apoptosis Assays.
Cells (attached cells plus those floating in the medium) were harvested at various time points and fixed in a solution containing a final concentration of 3.7% formaldehyde, 0.5% Nonidet P-40, and 10 μg/ml DAPI in PBS. Apoptosis was assessed through microscopic visualization of condensed chromatin and micronucleation. At least two independent experiments were carried out for each condition, and a minimum of 300 cells were counted in each measurement.
Colony Formation Assays.
Cells were infected with Ad-p53 or Ad-R175H virus for 48 h in 12-well plates and plated in six-well plates at dilutions of 1/50, 1/625, and 1/50, corresponding to ≈2,000–4,000 cells before treatment. Cells with different BAX genotypes were infected with Ad-PUMA or Ad-PUMAΔBH3 virus for 48 h and plated in 12-well plates at a dilution of 1/1,000, corresponding to ≈100–300 cells before treatment. To test the GFP-PUMA expression constructs, DLD-1 cells (≈2 × 106) were transfected in 25-cm2 (T25) flasks. Twenty-four hours after transfection, half of the cells were seeded in new T25 flasks under 1 mg/ml geneticin selection. Cells were allowed to grow for 10–14 days before staining with Crystal Violet (Sigma). All experiments were repeated at least twice, and similar results were obtained in each trial.
Results
Gene Targeting of PUMA.
The colorectal cancer cell line HCT116 was chosen for the current work because these cells have an intact p53 pathway and are susceptible to gene targeting through homologous recombination (20, 24). By using the bipartite targeting system depicted in Fig. 1A, we achieved a targeting frequency of ≈1 in 2,000 at the PUMA locus. A PCR-based strategy was used to screen for correctly targeted clones, and the results were confirmed through Southern blotting (Fig. 1B). Two sequential rounds of targeting resulted in the generation of two clones, each of which behaved identically in subsequent assays. We also generated specific antibodies to PUMA and used them to show that PUMA protein was absent in the PUMA−/− cells (Fig. 1C).
It has been shown that HCT116 cells undergo a growth arrest rather than apoptosis after exogenous p53 is expressed (25). Accordingly, we found that HCT116 cells arrested their growth after infection with an Ad expressing p53 (Ad-p53). As expected, disruption of the PUMA gene did not alter this response (data not shown). Previous experiments have shown that the choice between growth arrest and apoptosis in colorectal cancers can be modulated by p21WAF1/CIP1 (hereafter referred to as p21) (25, 26). Accordingly, when the p21 gene was disrupted in HCT116 cells, a robust apoptotic response was observed after infection with Ad-p53 (Fig. 2A). Because these p21-disrupted cells were more amenable to investigations of the apoptotic effects of p53, we analyzed the effects of PUMA disruption in p21−/− cells. Genetic disruptions of the two alleles of PUMA plus the two alleles of p21 were accomplished through four sequential rounds of gene targeting (Fig. 1 B and C). Four independent clones were obtained, each of which behaved identically in the assays described below.
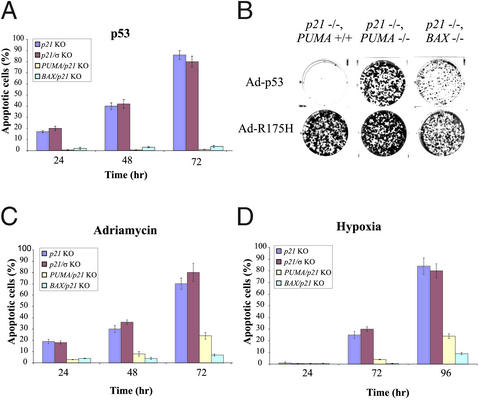
Figure 2.
Apoptosis induced by p53 and hypoxia. (A) Cells of the indicated genotypes were infected with Ad-p53 for the times shown on the x axis. The fraction of apoptotic cells was assessed by fluorescence microscopy of DAPI-stained cells. The means and standard errors of the mean are illustrated. (B) Cells were infected with Ads encoding WT (Ad-p53) or mutant p53 (Ad-R175H) and then seeded in six-well plates. Attached cells were stained with crystal violet 14 days later. (C) Cells of the indicated genotypes were exposed to adriamycin for various periods and scored as in A. (D) Cells of the indicated genotypes were exposed to hypoxic conditions for various periods and scored as in A.
PUMA Is Required for the Apoptosis Induced by Overexpression of p53.
The PUMA−/−, p21−/− cells proved completely resistant to the apoptosis induced by Ad-p53 (Fig. 2A). Moreover, as these cells neither underwent growth arrest (because of the absence of p21) nor cell death (because of the absence of PUMA), they remained viable after p53 infection. Colony formation assays showed that p21−/− cells failed to form colonies after p53 infection, whereas numerous colonies were formed after p53 infection of PUMA−/−, p21−/− cells. In contrast, an Ad expressing a mutant p53 commonly found in tumors (p53R175, Ad-R175H) did not induce apoptosis or growth arrest in any HCT116 cell derivative (Fig. 2B). The GFP marker present in the Ad-p53 constructs was used to demonstrate that all of the clonal cell lines used in this study could be infected with Ads to similar extents.
The BAX–PUMA Connection.
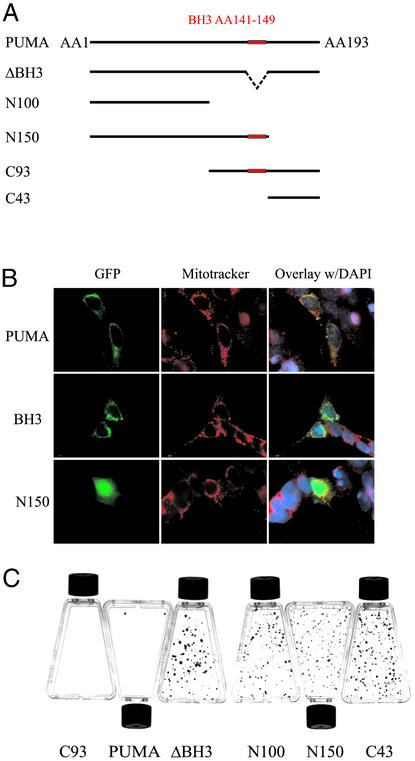
We next attempted to determine the mechanisms through which PUMA induced apoptosis. As PUMA is a BH3 domain-containing protein found predominantly in mitochondria, we expected that its apoptotic effects would be related to the mitochondrial pathway of cell death (16–18). We first tested various deletion constructs of PUMA to see which domains were required for mitochondrial localization and apoptotic induction (Fig. 3A). We found that the C-terminal 43 residues were required for mitochondrial localization and apoptosis induction, whereas the BH3 domain was required for apoptosis but not mitochondrial localization (Fig. 3 B and C). Accordingly, a PUMA expression construct containing only the C-terminal 93 residues (C93, containing amino acids 101–193) was able to induce apoptosis at a level comparable to the full-length PUMA protein (Fig. 3C). In contrast, PUMA constructs that did not contain both the BH3 domain and the C-terminal mitochondrial localization domain had no apoptotic effects (Fig. 3C).
Figure 3.
Protein domains required for PUMA function. (A) GFP-PUMA expression constructs. GFP was fused to the N terminus of the indicated fragments of PUMA cDNA. (B) GFP-PUMA constructs were transfected into 911 cells and visualized through the fluorescence of GFP. MitoTracker red dye was used to visualize mitochondria. The N150 construct devoid of the C-terminal 43 residues had a diffuse staining pattern that was distinct from GFP-PUMA or GFP-PUMAΔBH3. (C) DLD-1 cells were transfected with the indicated constructs and selected with geneticin for 2 weeks before staining with crystal violet. There was no difference in colony formation after transfection with PUMA-ΔBH3 compared with transfection with the empty vector (data not shown).
To evaluate further the apoptosis resulting from PUMA expression, we generated Ads expressing WT PUMA (Ad-PUMA) or a mutant PUMA gene missing the BH3 domain (Ad-PUMAΔBH3). The proteins encoded by both Ads were localized to mitochondria but only the WT gene induced apoptosis (16). This apoptosis was associated with depolarization of the outer mitochondrial membrane (Fig. 4A) and activation of caspases (Fig. 4B). The apoptosis induced by Ad-PUMA or Ad-p53 could be completely inhibited by Z-VAD, a pan-caspase inhibitor (Fig. 4C).
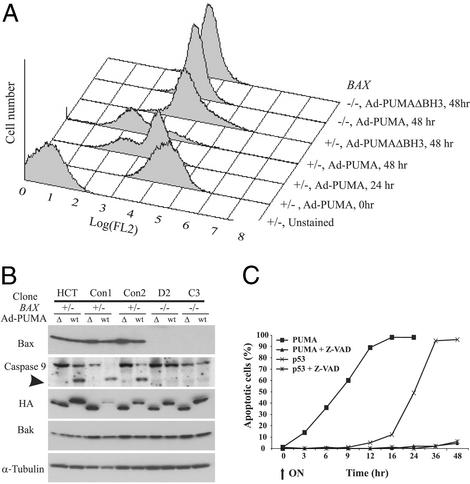
Figure 4.
Biochemical changes during PUMA-induced apoptosis are Bax-dependent. (A) Cells with the indicated genotype were infected with Ad-PUMA or Ad-PUMAΔBH3 viruses, harvested at the indicated times, stained with CMXRos, and analyzed by flow cytometry to measure changes in mitochondrial membrane potential (Δψ). FL2 on the x axis reflects the intensity of the CMXRos signal. (B) Lysates from cells indicated genotypes infected with Ad-PUMA (wt) or Ad-PUMAΔBH3 (Δ) were analyzed by immunoblotting, using antibodies specific for the indicated proteins. The intact caspase 9 polypeptide is 46 kDa, whereas a degraded fragment of 37 kDa is detected in cells undergoing apoptosis. Tagged PUMA and PUMAΔBH3 proteins were detected by an antibody to HA. (C) DLD-1 cells were induced to express either PUMA or p53 in the presence or absence of the caspase inhibitor Z-VAD. At the indicated times, cells were assayed for apoptosis by fluorescence microscopy after staining with DAPI.
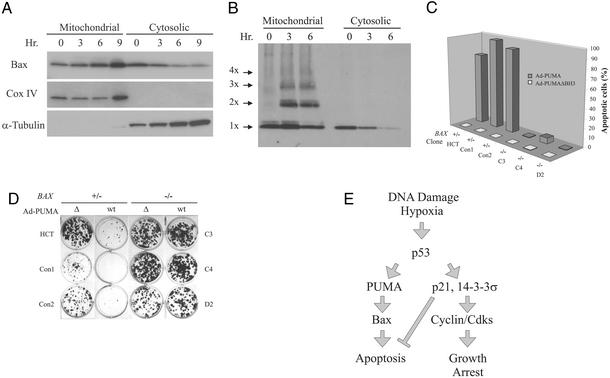
Previous experiments have shown that BH3 domain-containing proteins such as BIK and BID induce apoptosis in a Bax- or Bax/Bak-dependent manner (13, 27, 28). We found that the apoptosis induced by PUMA completely depended on the presence of intact Bax. After PUMA expression, Bax translocated to the mitochondria (Fig. 5A) and multimerized (Fig. 5B). The extent of Bax multimerization was compatible with that demonstrated to cause pore formation in isolated mitochondria and artificial liposomes (29). HCT116 cells in which the BAX genes had been disrupted through homologous recombination were completely resistant to the apoptotic effects of PUMA (Fig. 5C), even though they still expressed Bak (Figs. 1C and 4B). As expected, biochemical changes associated with apoptosis, such as depolarization of the outer mitochondrial membrane and activation of caspases, were completely absent in BAX−/− cells (Fig. 4 A and B). The BAX−/− cells also formed many more colonies after Ad-PUMA infection than did parental cells (Fig. 5D). It was of interest to test the colonies that did form after Ad-PUMA infection of parental HCT116 cells. The parental HCT116 cells are heterozygous for BAX, containing one WT and one mutant allele (21, 30). We found that 10 of 28 colonies formed after infection with Ad-PUMA contained homozygous mutants of BAX, whereas only 1 of 28 colonies formed after infection with Ad-PUMAΔBH3 contained homozygous BAX mutations (P < 0.02, χ2 test). These spontaneously arising mutations strongly confirmed the importance of BAX in determining cell viability after PUMA expression.
Figure 5.
BAX deficiency rescues PUMA-induced apoptosis. (A) Mitochondrial and cytosolic fractions were prepared from DLD-1 cells in which PUMA had been induced by the removal of doxycycline. The fractions were analyzed by immunoblotting with antibodies to the indicated proteins. Cox IV and α-tubulin provided controls for the fractionation. (B) DLD-1 cells in which PUMA had been induced were cross-linked before subcellular fractionation and subjected to immunoblotting with an antibody to Bax. Bax monomers (1X) and multimers (2X–4X) are indicated. (C) Clones with various BAX genotypes were infected with Ad-PUMA (wt) or Ad-PUMAΔBH3 (Δ) for 48 h and the fraction of apoptosis was assessed by fluorescence microscopy of DAPI-stained cells. (D) HCT116 clones with the indicated BAX genotypes were infected with Ad-PUMA (wt) or Ad-PUMAΔBH3 (Δ) for 48 h then diluted into 12-well plates. Colonies were stained with crystal violet 10–14 days later. (E) Model of p53 function during tumorigenesis. As tumors progress to malignancy, they accumulate DNA damage (1). They also outgrow their blood supply, resulting in hypoxia of large regions of the tumors (31). The DNA damage and/or hypoxia activate p53, which transcriptionally activates several genes, including p21, 14-3-3σ, PUMA, and, in some cases, BAX. The cells either arrest or enter into apoptosis, depending on which of the two arms of the pathway is predominant in the particular cell type and microenvironment. Cells with a mutant p53 gene would have a selective growth advantage in either case, because both growth arrest and apoptosis would be diminished in such mutant cells. Although many other genes are induced by p53 (7), the ones included in the model have all been shown to be essential for the indicated downstream effects through gene-targeting experiments in mammalian cells.
PUMA Is Required for the Apoptosis Induced by Hypoxia and DNA-Damaging Drugs.
We also wanted to test the effects of disruption of PUMA on cellular responses to signals that elicit a p53 response. Treatment with the DNA-damaging drug adriamycin resulted in significant apoptosis of p21−/− cells, and this response was attenuated in cells in which the PUMA gene had also been disrupted (Fig. 2C). Although p53 can be activated by drugs like adriamycin, cells in vivo are generally not exposed to such agents, especially at the high concentrations used in vitro. We therefore chose to examine the responses of HCT116 cells to hypoxia, a more physiologic form of stress that is often naturally encountered during tumorigenesis. Previous experiments have shown that hypoxia activates p53 in vitro and within the cancer cell environment in vivo (31). Exposure of p21−/− cells to low concentrations of oxygen resulted in apoptosis. Isogenic p21−/− cells that were also PUMA-deficient were highly resistant to such apoptosis, especially at early time periods (Fig. 2D). As a control for this experiment, we also tested cells in which both p21 and the 14-3-3σ genes had been targeted through homologous recombination (32); such cells were as susceptible to apoptosis induced by p53, DNA damage, or hypoxia because cells with only the p21 gene disrupted (Fig. 2 A, C, and D). Finally, if BAX was epistatic to PUMA, as suggested previously, cells with disruptions of both p21 and BAX should be as resistant to apoptosis as cells with both p21 and PUMA disrupted. Two p21−/−, BAX−/− clones were generated to test this hypothesis (Fig. 1D). As shown in Fig. 2, the absence of BAX, like the absence of PUMA, made p21−/− cells relatively resistant to apoptosis.
Discussion
The results described above show that PUMA is essential for the apoptosis induced by exogenous or endogenous p53. Furthermore, they demonstrate that PUMA-induced apoptosis depends on the presence of Bax. These results provide the basis for a molecular model of the effects of p53 on cell growth and death (Fig. 5E). As tumors progress, angiogenesis fails to keep pace and tumor cells become hypoxic (31, 33). The hypoxia and resultant metabolic abnormalities induce p53, which in turn transcriptionally activates genes, including p21 and PUMA. The p21 induction results in inhibition of cyclin-dependent kinases and growth arrest. The PUMA induction results in Bax multimerization and mitochondria-dependent death. Cells that are mutant for p53 will not properly activate any of these pathways and will have a selective advantage in such microenvironments. Whether growth arrest vs. apoptosis, predominates will depend on the interaction with other signaling pathways (34). For example, p21 can be induced by oxidative stress independently of p53 (35–37).
This model explains some previously confusing observations. For example, it explains why p53 mutations arise only late in colorectal tumorigenesis, near the transition from benign tumor growth to malignancy (38). It is only at this stage that significant hypoxia occurs, as indicated by the multiple necrotic regions commonly found within cancers but rarely found in benign precursor lesions. It also explains the interesting but curious relationship between Bax and p53. In this regard, it has been shown that BAX disruption can partially recapitulate the effects of p53 disruption in certain cellular contexts (21, 39, 40). Yet in many cell types, including the HCT116 cells used here (Fig. 1C), p53 expression does not result in a significant induction of Bax compared with PUMA or p21, excluding BAX as a direct target of p53 in these cases (16, 41–45). Such results are consistent with the fact that the BAX gene promoter is only weakly bound to p53 in cells compared with the promoters of p21 or PUMA (46). Our results reconcile these observations by demonstrating that BAX can function as an indirect target of p53 through the PUMA intermediary. They also support the idea that the BAX mutations found in cancers without p53 mutations create an apoptotic phenotype that is similar to that produced by p53 mutations (47). Finally, it is notable that each of the steps of the model depicted in Fig. 5E has been rigorously tested through genetic disruption experiments in either mouse or human cells. It will be of interest to determine whether this model applies to other mammalian cell types as well as to simpler eukaryotes with ancestral p53 genes.
Acknowledgments
We thank L. Meszler for help with imaging and flow cytometry and the members of the Kinzler/Vogelstein laboratories for advice and discussion. This work was supported by the Clayton Fund and National Institutes of Health Grants CA 43460, CA 62924, and CA 06973. Under agreements between Genzyme Molecular Oncology, CalBiochem, Cyclis Pharmaceuticals, and Johns Hopkins University, the authors and university are entitled to a share of the sales royalty for reagents related to p21, p53, or PUMA. K.W.K. is also a consultant to Genzyme Molecular Oncology.
Abbreviations
- HA
hemagglutinin
- BH
Bcl-2 homology
- Ad
adenovirus
- DAPI
4′,6-diamidino-2-phenylindole
References
- 1.Vogelstein B, Lane D, Levine A J. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Hollstein M, Hergenhahn M, Yang Q, Bartsch H, Wang Z Q, Hainaut P. Mutat Res. 1999;431:199–209. doi: 10.1016/s0027-5107(99)00162-1. [DOI] [PubMed] [Google Scholar]
- 3.Hussain S P, Harris C C. Mutat Res. 1999;428:23–32. doi: 10.1016/s1383-5742(99)00028-9. [DOI] [PubMed] [Google Scholar]
- 4.Prives C, Hall P A. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 5.Oren M. J Biol Chem. 1999;274:36031–36034. doi: 10.1074/jbc.274.51.36031. [DOI] [PubMed] [Google Scholar]
- 6.Schmitt C A, Fridman J S, Yang M, Baranov E, Hoffman R M, Lowe S W. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 7.El-Deiry W S. Semin Cancer Biol. 1998;8:345–357. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 8.Vousden K H. Cell. 2000;103:691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 9.Gottlieb T M, Oren M. Semin Cancer Biol. 1998;8:359–368. doi: 10.1006/scbi.1998.0098. [DOI] [PubMed] [Google Scholar]
- 10.Gross A, McDonnell J M, Korsmeyer S J. Genes Dev. 1999;13:1899–1911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- 11.Cory S, Adams J M. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 12.Huang D C, Strasser A. Cell. 2000;103:839–842. doi: 10.1016/s0092-8674(00)00187-2. [DOI] [PubMed] [Google Scholar]
- 13.Zong W X, Lindsten T, Ross A J, MacGregor G R, Thompson C B. Genes Dev. 2001;15:1481–1486. doi: 10.1101/gad.897601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Letai A, Bassik M C, Walensky L D, Sorcinelli M D, Weiler S, Korsmeyer S J. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 15.Wang X. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- 16.Yu J, Zhang L, Hwang P M, Kinzler K W, Vogelstein B. Mol Cell. 2001;7:673–682. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- 17.Nakano K, Vousden K H. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 18.Han J, Flemington C, Houghton A B, Gu Z, Zambetti G P, Lutz R J, Zhu L, Chittenden T. Proc Natl Acad Sci USA. 2001;98:11318–11323. doi: 10.1073/pnas.201208798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jallepalli P V, Waizenegger I C, Bunz F, Langer S, Speicher M R, Peters J, Kinzler K W, Vogelstein B, Lengauer C. Cell. 2001;105:445–457. doi: 10.1016/s0092-8674(01)00340-3. [DOI] [PubMed] [Google Scholar]
- 20.Waldman T, Kinzler K W, Vogelstein B. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 21.Zhang L, Yu J, Park B-H, Kinzler K W, Vogelstein B. Science. 2000;290:989–992. doi: 10.1126/science.290.5493.989. [DOI] [PubMed] [Google Scholar]
- 22.Yu J, Zhang L, Hwang P M, Rago C, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1999;96:14517–14522. doi: 10.1073/pnas.96.25.14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He T C, Zhou S, da Costa L T, Yu J, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown J P, Sedivy J M, Kinzler K W, Vogelstein B. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 25.Polyak K, Waldman T, He T-C, Kinzler K W, Vogelstein B. Genes Dev. 1996;10:1945–1952. doi: 10.1101/gad.10.15.1945. [DOI] [PubMed] [Google Scholar]
- 26.Gartel A L, Tyner A L. Mol Cancer Ther. 2002;1:639–649. [PubMed] [Google Scholar]
- 27.Cheng E H, Wei M C, Weiler S, Flavell R A, Mak T W, Lindsten T, Korsmeyer S J. Mol Cell. 2001;8:705–711. doi: 10.1016/s1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- 28.Bouillet P, Strasser A. J Cell Sci. 2002;115:1567–1574. doi: 10.1242/jcs.115.8.1567. [DOI] [PubMed] [Google Scholar]
- 29.Waterhouse N J, Ricci J E, Green D R. Biochimie. 2002;84:113–121. doi: 10.1016/s0300-9084(02)01379-2. [DOI] [PubMed] [Google Scholar]
- 30.Ionov Y, Yamamoto H, Krajewski S, Reed J C, Perucho M. Proc Natl Acad Sci USA. 2000;97:10872–10877. doi: 10.1073/pnas.190210897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graeber T G, Osmanian C, Jacks T, Housman D E, Koch C J, Lowe S W, Giaccia A J. Nature. 1996;379:88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- 32.Chan T A, Hwang P M, Hermeking H, Kinzler K W, Vogelstein B. Genes Dev. 2000;14:1584–1588. [PMC free article] [PubMed] [Google Scholar]
- 33.Kinzler K W, Vogelstein B. Nature. 1996;379:19–20. doi: 10.1038/379019a0. [DOI] [PubMed] [Google Scholar]
- 34.Seoane J, Le H V, Massague J. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 35.Russo T, Zambrano N, Esposito F, Ammendola R, Cimino F, Fiscella M, Jackman J, O'Connor P M, Anderson C W, Appella E. J Biol Chem. 1995;270:29386–29391. doi: 10.1074/jbc.270.49.29386. [DOI] [PubMed] [Google Scholar]
- 36.O'Reilly M A, Staversky R J, Watkins R H, Reed C K, de Mesy Jensen K L, Finkelstein J N, Keng P C. Am J Respir Cell Mol Biol. 2001;24:703–710. doi: 10.1165/ajrcmb.24.6.4355. [DOI] [PubMed] [Google Scholar]
- 37.Esposito F, Cuccovillo F, Russo L, Casella F, Russo T, Cimino F. Cell Death Differ. 1998;5:940–945. doi: 10.1038/sj.cdd.4400427. [DOI] [PubMed] [Google Scholar]
- 38.Baker S J, Preisinger A C, Jessup J M, Paraskeva C, Markowitz S, Willson J K, Hamilton S, Vogelstein B. Cancer Res. 1990;50:7717–7722. [PubMed] [Google Scholar]
- 39.Yin C, Knudson C M, Korsmeyer S J, Van Dyke T. Nature. 1997;385:637–640. doi: 10.1038/385637a0. [DOI] [PubMed] [Google Scholar]
- 40.McCurrach M E, Connor T M, Knudson C M, Korsmeyer S J, Lowe S W. Proc Natl Acad Sci USA. 1997;94:2345–2349. doi: 10.1073/pnas.94.6.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strobel T, Swanson L, Korsmeyer S, Cannistra S A. Oncogene. 1997;14:2753–2758. doi: 10.1038/sj.onc.1201132. [DOI] [PubMed] [Google Scholar]
- 42.Shu H K, Kim M M, Chen P, Furman F, Julin C M, Israel M A. Proc Natl Acad Sci USA. 1998;95:14453–14458. doi: 10.1073/pnas.95.24.14453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adachi J, Ookawa K, Kohno T, Tomizawa Y, Tsuchida S, Yokota J. Cell Death Differ. 1998;5:148–155. doi: 10.1038/sj.cdd.4400325. [DOI] [PubMed] [Google Scholar]
- 44.Tatebe S, Matsuura T, Endo K, Teramachi K, Nakamura T, Sato K, Ito H. Int J Oncol. 1999;15:229–235. doi: 10.3892/ijo.15.2.229. [DOI] [PubMed] [Google Scholar]
- 45.Huang T G, Ip S M, Yeung W S, Ngan H Y. Eur J Cancer. 2000;36:249–256. doi: 10.1016/s0959-8049(99)00247-6. [DOI] [PubMed] [Google Scholar]
- 46.Kaeser M D, Iggo R D. Proc Natl Acad Sci USA. 2002;99:95–100. doi: 10.1073/pnas.012283399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed J C, Perucho M. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]