Abstract
The human coxsackievirus and adenovirus receptor (CAR) represents the primary cellular site of adenovirus attachment during infection. An understanding of the mechanisms regulating its expression could contribute to improving efficacy and safety of adenovirus-based therapies. We characterized regulation of CAR expression in a 3D cell culture model of human breast cancer progression, which mimics aspects of the physiological tissue context in vitro. Phenotypically normal breast epithelial cells (S1) and their malignant derivative (T4-2 cells) were grown either on tissue culture plastic (2D) or 3D cultures in basement membrane matrix. S1 cells grown in 3D showed low levels of CAR, which was expressed mainly at cell–cell junctions. In contrast, T4-2 cells expressed high levels of CAR, which was mainly in the cytoplasm. When signaling through the epidermal growth factor receptor was inhibited in T4-2 cells, cells reverted to a normal phenotype, CAR protein expression was significantly reduced, and the protein relocalized to cell–cell junctions. Growth of S1 cells as 2D cultures or in 3D in collagen-I, a nonphysiological microenvironment for these cells, led to up-regulation of CAR to levels similar to those in T4-2 cells, independently of cellular growth rates. Thus, expression of CAR depends on the integrity and polarity of the 3D organization of epithelial cells. Disruption of this organization by changes in the microenvironment, including malignant transformation, leads to up-regulation of CAR, thus enhancing the cell's susceptibility to adenovirus infection.
The therapeutic use of recombinant adenoviruses represents a new chapter in the treatment of cancer. Two main strategies have been pursued: first, adenoviruses have been devised as vehicles for the delivery of therapeutic genes; second, replication-selective adenoviruses have been created that replicate in, and destroy, cells harboring certain mutations, such as cancer cells that have lost functional p53 (1). Studies evaluating such treatments in breast cancer have been initiated, in particular by using adenovirus expressing the tumor suppressor protein p53 or the cytokine granulocyte–macrophage colony-stimulating factor (2, 3). The efficacy of these viral agents critically depends on the expression of the human coxsackievirus and adenovirus receptor (CAR), which has been identified as the primary cellular receptor for adenovirus (4). This 46-kDa transmembrane protein mediates viral attachment through interaction with the adenovirus fiber-knob protein. Accordingly, loss of CAR expression on the cell surface leads to a significant reduction in susceptibility of cells to adenovirus infection, which can be rescued by ectopic expression of the protein (5). After attachment of the virus to CAR, internalization is mediated by binding of the adenovirus penton base to αvβ3 and αvβ5 integrin complexes (6).
To study the so-far unknown mechanisms of regulation of CAR expression, we took advantage of a 3D human breast cancer cell system that allows analysis of molecular events during tumor progression under conditions that model the physiological cellular context (7). The parental cell line, HMT-3522, was established from a biopsy of a nonmalignant breast lesion (7). Through continuous passaging in cell culture medium lacking epidermal growth factor (EGF), premalignant and malignant derivatives of these cells were established. Early passage cells (S1 cells) show a normal phenotype, undergo growth arrest, form phenotypically normal mammary tissue structures (acini) when grown in a basement membrane gel (Matrigel), and are nontumorigenic in athymic mice. In contrast, T4-2 cells grow as disorganized cell clusters, do not arrest under these conditions, and form xenograft tumors in nude mice (7, 9). Previously, we demonstrated that treatment of T4-2 cells with inhibitory antibodies to β1 integrins or EGF receptor (EGFR) (mAb225) leads to formation of growth-arrested, phenotypically normal acini-like structures, referred to as reverted T4-2 cells (T4-2rev) (9, 12). Therefore, this model allows the analysis of molecules and signaling events during tumor progression and in malignant cells that have undergone phenotypic differentiation.
We show here that signals important for maintaining the 3D integrity of mammary epithelial cells are crucial for the preservation of normal expression patterns of CAR. Disruption of the physiological interactions with the extracellular environment, either by alteration of the composition of the environment or by changes in signal transduction pathways during tumor progression, causes deregulation of CAR expression and, in turn, changes of the susceptibility of cells to adenovirus infection.
Materials and Methods
Cell Culture.
The mammary epithelial cell line HMT-3522 was maintained in H14 medium [DMEM/F12 (Invitrogen) with 250 ng/ml insulin, 10 μg/ml transferrin, 2.6 ng/ml sodium selenite, 10−10 M estradiol, 1.4 × 10−6 M hydrocortisone, and 5 μg/ml prolactin]. Nonmalignant S1 cells were grown on plastic in the presence of 10 ng/ml EGF; malignant T4-2 cells were cultured on collagen type I-coated dishes (Vitrogen 100, Celtrix Laboratories, Palo Alto, CA) in the absence of EGF. 3D cultures were prepared by growing S1 and T4-2 cells to confluence as monolayers, followed by trypsin treatment and embedding (8.5 × 105/ml) into a commercially prepared reconstituted basement membrane from Englebreth-Holm-Swarm tumors (Matrigel, Collaborative Research). Chinese hamster ovary (CHO) cells and CHO cells expressing hCAR (a kind gift of J. M. Bergelson, Children's Hospital of Philadelphia, Philadelphia) were cultured in Iscove's medium (University of California, San Francisco Cell Culture Facility) containing 10% FBS and 1% streptomycin/penicillin.
In Vitro Transduction Assay.
A nonreplicating, E1A-deleted adenovirus (a kind gift from D. Stokoe, University of California, San Francisco) containing an expression cassette for enhanced GFP was used to assess susceptibility to adenovirus infection of cells in 2D and 3D cultures. The fraction of infected cells grown in 2D that showed green fluorescence was measured by fluorescence-activated cell sorting analysis. For infection of cells in 3D culture, virus was suspended in PBS, then added to the surface of the basement membrane gel at a multiplicity of infection of 10 plaque-forming units per cell and allowed to enter the gel for 90 min. Subsequently, fresh medium was added. Green fluorescence was assessed by fluorescence microscopy or by fluorescence-activated cell sorting 48 h postinfection. The percentage of cells demonstrating green fluorescence was estimated for 10 microscopic fields at 400-fold magnification.
Antibodies.
A polyclonal anti-CAR antibody (Ab72) was a kind gift of L. Post (Onyx Pharmaceuticals, Richmond, CA). Antibodies against αv-integrin, E-cadherin, and β-actin were obtained from Chemicon, Transduction Laboratories (Lexington, KY), and Sigma, respectively. The human EGFR-blocking mAb225 (Oncogene) was used at a concentration of 4 μg/ml.
Immunoblotting.
Cells grown as monolayers were lysed in RIPA buffer (1% Nonidet P-40/0.5% deoxycholate/0.2% SDS/150 mM sodium chloride/50 mM Tris⋅HCl, pH 7.4 containing 2 mM sodium fluoride, 1 mM sodium orthovanadate, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 10 μg/ml aprotinin, 10 μg/ml E 64, and 1 mM Pefabloc). Cells grown in 3D basement membrane cultures for 10 days were isolated as colonies by using ice-cold PBS/EDTA (0.01 M sodium phosphate, pH 7.2 containing 138 mM sodium chloride and 5 mM EDTA) as described and thereafter lysed in RIPA buffer. Equal amounts of protein lysates were loaded on reducing Laemmli gels, immunoblotted, and detected with an ECL system (Amersham Pharmacia).
Immunofluorescence.
Cells or fresh frozen tissue sections were fixed with 1% paraformaldehyde for 2 min. After blocking with 2% BSA, sections were incubated with primary antibodies against CAR or E-cadherin. Nonimmune Igs from rabbits or mice was used as control. Primary antibodies were detected with a FITC-conjugated anti-rabbit antibody or a Cy-3-conjugated anti-mouse antibody, respectively (Molecular Probes). Multicolor fluorescence microscopy was performed by using a DMRCA fluorescence microscope (Leica) and a Leica confocal microscope, respectively. To control for bleed-through artifacts, single-color staining was performed. No significant signal was detected in channels other than the one corresponding to the secondary antibody.
Immunohistochemistry.
Tissue sections were deparaffinized in xylene and dehydrated in ethanol. For CAR staining standard indirect immunoperoxidase methods were used for immunostaining with rabbit polyclonal anti-CAR antibody Ab72 as described (10). In brief, the slides were baked for 30 min at 60°C and standard antigen retrieval methods including trypsinization and microwave treatment in 10 mM citrate buffer were performed. The tissue was blocked in 10% goat serum/PBS and incubated in primary polyclonal antibody CAR Ab72 diluted 1:7,000 for 8–16 h at 4°C. The tissue sections were incubated in secondary biotinylated goat anti-rabbit Ig diluted 1:200 (Vector Laboratories) and then treated with steptavidin-biotinylated horseradish peroxidase complex (Vectastain Elite ABC kit, Vector Laboratories). The sections were subsequently developed by using diaminobenzidine tetrahydrochloride (Sigma) in hydrogen peroxide/PBS and counterstained with hematoxylin.
RNA Extraction and cDNA Synthesis.
Total RNA was extracted from cells and tissues by using TRIzol reagent (Life Technologies, Grand Island, NY). cDNA was synthesized by using 2.5 units/μl Moloney murine leukemia virus reverse transcriptase (Invitrogen). Reverse transcription was performed in a 100 μl final volume containing 10 μl of 10× PCR buffer, 30 μl MgCl2, 4 μl dNTP mix (25 mM each; all Roche Molecular Systems, Branchburg, NJ), 5 μl of random primers (100 μM; Invitrogen), 1 μl RNase inhibitor (Roche Molecular Systems), and 43.75 μl RNase free water. Reverse transcriptase reactions were incubated at room temperature for 10 min and 48°C for 40 min, followed by 5 min at 95°C. cDNA was stored at −20°C until use.
Real-Time PCR.
For real-time PCR detection of CAR mRNA expression, oligonucleotide primers and TaqMan probe with the following sequences were used: forward primer, GGCG-CTCCTGCTGTGC; reverse primer, CTTTGGCTTTTTCAATCATCTCTTC; probe, 5′-(6FAM)-TGCGGAGTAGTGGATTTCGCCAGAAG-(TAMRA)-3′. PCR was conducted in triplicate with 50-μl reaction volumes of 1× PCR buffer A (Applied Biosystems), 2.5 mM MgCl2, 0.4 μM each primer, 200 μM each dNTP, 100 nM probe, and 0.025 units/μl Taq Gold (Applied Biosystems). After addition of 10 μl primer/probe and 10 μl cDNA the PCR was performed by using the following parameters: 95°C 12 min × 1 cycle, 95°C 20 sec, 60°C 1 min × 40 cycles. Analysis was carried out by using the sequence detection software supplied with the ABI 7700 (Applied Biosystems). Expression was quantified based on the δ-CT for CAR expression relative to expression of β-glucuronidase (11).
Results
CAR Expression in HMT-3522 Cells.
To investigate the mechanisms regulating CAR expression during tumor progression and to examine the consequences of this regulation for adenovirus infection, we chose the HMT-3522 breast cancer progression model. This system has the advantage that the nonmalignant S1 cells and the malignant T4-2 cells are derived from the same parental cells, yet demonstrate dramatic genotypic and phenotypic differences (8, 9). In addition, sublines of premalignant cells have been isolated where the cells exhibit loss of structural integrity but are not yet malignant (unpublished work). S1 and T4-2 cells can be grown inside a reconstituted basement membrane (9). Under these conditions, nonmalignant S1 cells form organized, acinus-like structures, whereas T4-2 cells grow as disorganized, loosely adherent cell clusters. Interestingly, T4-2 cells assume a normal phenotype on treatment with inhibitors of EGFR (mAb225 and tyrophostin; ref. 12) and β1 integrin (mAb AIIB2) signaling (9).
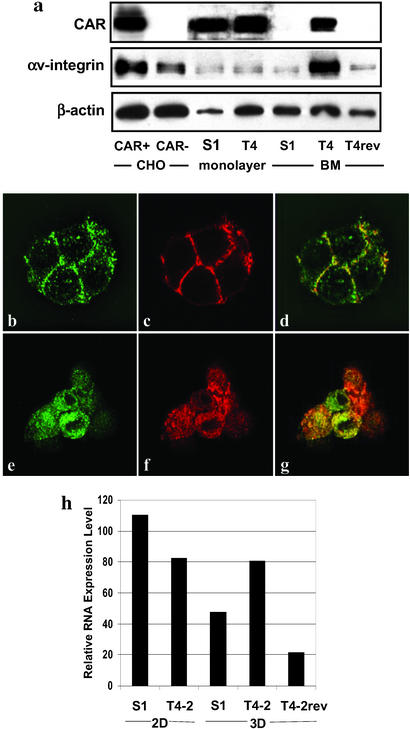
To ask whether the progression to malignancy is associated with changes in protein levels of CAR, we first assessed CAR protein expression in S1 and T4-2 cells by Western blotting. No significant differences in CAR expression levels were found between S1 and T4-2 cells grown in 2D cultures. In contrast, the expression level of CAR was strikingly reduced in S1 cells compared with T4-2 cells when these cells were grown in 3D (Fig. 1a). Overall αv-integrin levels were also dramatically increased when T4-2 cells were cultured in 3D. In T4-2 cells that had been treated with mAb225, resulting in phenotypic reversion (T4-2rev cells), CAR and αv-integrin levels were reduced to levels similar to those found in S1 cells in 3D (Fig. 1a).
Figure 1.
Analysis of CAR expression in S1 and T4-2 cells. (a) Western blot analysis of expression of CAR, αv-integrin, and β-actin in S1, T4-2, and phenotypically reverted T4-2 (T4-2rev) cells. Cells were grown as monolayer or 3D cultures in basement membrane (BM). Chinese hamster ovary cells (CHO) with or without stable expression of CAR were used as a control. CAR (b) and E-cadherin (c) expression in S1 cells grown in 3D as detected by confocal fluorescence microscopy. (d) The overlay of images b and c. Staining of T4-2 cells grown in 3D for CAR and E-cadherin (e and f) and the overlay image (g) is shown. (Magnification: ×630.) (h) CAR mRNA expression levels in S1 and T4 cells grown in 2D and 3D were measured relative to expression of β-glucuronidase by using a real-time PCR assay.
As the level of cellular organization differs distinctly among S1, T4-2, and T4-2rev cells, we tested whether the differences in CAR protein levels were associated with changes in the spatial distribution of the protein. Cells grown on conventional 2D cultures and fresh frozen sections from 3D cultures containing these cells were stained by using the rabbit polyclonal anti-CAR antibody, Ab72, and a mouse mAb against E-cadherin as primary antibodies and analyzed by confocal immunofluorescence microscopy. S1 cells grown in 2D demonstrated consistent CAR staining at the cell–cell junctions sites as well as in the perinuclear cytoplasm (data not shown). A similar pattern was seen in T4-2 cells, with the difference that these cells exhibited a less consistent staining at the cell–cell interaction sites and less protein in the perinuclear cytoplamatic compartment (data not shown). Frozen sections of S1 cells grown in 3D revealed prominent staining of CAR and E-cadherin at sites of cell–cell contact, with no staining at the basal portions of these cells (Fig. 1 b–d) and only a weak cytoplasmic staining as seen by confocal microscopy. In contrast, T4-2 cells demonstrated a strong, diffuse cytoplasmatic localization of CAR without enhanced staining at cell–cell junctions (Fig. 1 e–g). Phenotypic reversion of these cells (described above) reestablished a staining pattern that was identical to that of S1 cells in 3D (data not shown). These findings indicate that the phenotypic differences between S1 and T4-2 cells are accompanied by changes in subcellular distribution as well as in the levels of CAR protein. Moreover, structural changes induced by pharmacological inhibition of the EGFR in tumor cells were associated with reestablishment of a CAR expression pattern found in normal cells.
Real-time RT-PCR showed changes in CAR mRNA expression that resembled those observed for the protein. S1 and T4-2 cells grown on 2D cultures expressed high levels of CAR (Fig. 1h). In contrast, CAR mRNA levels were markedly lower in 3D cultures of S1 cells compared with T4-2 cells. Reverted T4-2 cells were similar to S1 cells in that CAR expression was reduced to normal, nonmalignant levels (Fig. 1h). These data suggest that CAR expression level is regulated by microenvironmental factors that control the structural organization of the tissue by both transcriptional and translational mechanisms.
Correlation of Susceptibility to Infection with Adenovirus and CAR Expression.
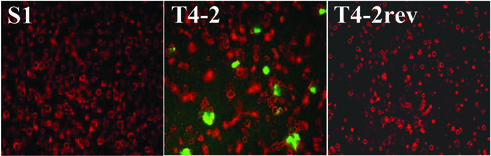
To assess whether the differences in CAR and αv-integrin protein levels were associated with differences in susceptibility to infection with adenovirus, 2D and 3D cultures of S1 and T4-2 cells were infected, at a multiplicity of infection of 10 plaque-forming units per cell, with a nonreplicating adenovirus construct encoding GFP (Fig. 2). S1 and T4-2 cells grown in 2D showed similar infection rates, as determined by fluorescence-activated cell sorting analysis, resulting in GFP expression in ≈40% of cells (data not shown) whereas S1 cells grown in 3D were hardly infectable at this multiplicity of infection. Only a few cells that were not part of the organized, acini-like structures showed a fluorescent signal. In contrast, strong green fluorescence was seen in ≈15% of T4-2 cells grown in 3D (Fig. 2). We also analyzed susceptibility to adenovirus infection of cells that demonstrated phenotypic reversion after treatment with mAb225 or tyrphostin. As in S1 cells, only very few infected cells (<1%) were found in T4-2rev cells (Fig. 2). These data demonstrate that susceptibility to infection with adenovirus changes dramatically in parallel to altered expression levels of CAR and integrins in S1, T4-2, and T4-2rev cells. Thus, the organizational integrity of the normal or malignant cells overrides the genotype of the cells in terms of susceptibility to adenovirus infection.
Figure 2.
Assessment of susceptibility to infection with adenovirus of S1 and T4-2 cells grown in basement membrane matrix in 3D. 3D cultures of S1, T4-2, and T4-2rev cells were grown in basement membrane gel and infected with a GFP-expressing, nonreplicating adenovirus (Ad-GFP) at a multiplicity of infection of 10 plaque-forming units per cell. Microscopic images (magnification: ×50) represent overlays of phase-contrast (red, false-color) and fluorescence microscopy (green) after infection. Each equatorial section of S1 cell spheroids contains an average of eight cells (8).
Deregulation of CAR Expression in Normal Cells Grown in Nonphysiologic, 3D Microenvironments.
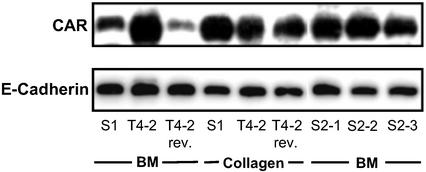
In contrast to S1 cells grown in 3D basement membrane cultures, expression levels of S1 cells grown as conventional monolayer cultures on plastic (2D cultures) showed high CAR expression levels similar to the levels observed in T4-2 cells grown in 2D or 3D basement membrane cultures (Fig. 1a). Levels of αv-integrins were also equal in S1 and T4-2 cells grown in 2D. This finding suggested that extracellular signals are necessary to maintain physiologic CAR levels. Because S1 cells undergo growth arrest in 3D, we tested whether CAR levels are regulated by growth rate. In addition, it was necessary to differentiate whether three-dimensionality per se was sufficient to maintain low levels of CAR expression or whether basement membrane signaling and correct polarity was important. To answer both of these questions, we examined CAR protein levels in S1 cells that were grown in 3D collagen I gels, a nonphysiological microenvironment for mammary epithelial cells. In this matrix, S1 cells undergo growth arrest and form organized colonies that are growth arrested but reversely polarized, as determined by sialomucin, epithelial specific antigen, or occludin (14, 15). Western blot analysis showed a significantly higher expression of CAR in these cells compared with S1 cells grown in 3D basement membrane cultures whereas CAR levels in T4-2 cells in basement membrane culture were not different from those grown in collagen (Fig. 3). These findings demonstrate that changes in CAR expression are not caused by growth rate or cultivation in 3D cultures but correlate with correct polarity (15). The results further demonstrate that normal cells require basement membrane signals to keep CAR levels in check and respond with CAR up-regulation under conditions where these signals are missing or aberrant. Although malignant T4-2 cells did not respond to the basement membrane in 3D, they became responsive when receptors were manipulated to down-modulate signaling and restore polarity (Fig. 1). This experiment also demonstrates that CAR levels do not correlate with the cell cycle status of S1 cells as these cells have undergone cell cycle arrest in both collagen I and basement membrane.
Figure 3.
Western blot analysis of CAR protein expression in S1, T4-2, and S2 (clones 1–3) cells grown in basement membrane matrix (BM) or collagen I. Protein levels are also shown for T4-2rev cells after treatment with the anti-EGFR antibody mAb225. E-cadherin expression was shown previously not to change and is used as a control.
Up-Regulation of CAR in Premalignant Cells with Perturbed 3D Organization.
The results with S1 cells alone indicated that even cells with relatively normal genotype express high levels of CAR when their polarity is compromised. To investigate whether the relative up-regulation of CAR expression in 3D basement membrane cultures in T4-2 cells was a consequence of the malignant phenotype or whether it was caused by loss of polarity and tissue organization, we analyzed 3D basement membrane cultures of premalignant cells. These cells, designated S2, were also derived from HMT-3522 cells by continuous cell passaging in EGF-depleted medium (8). They resemble T4-2 cells in that they also display disorganized cell growth in 3D basement membrane matrix where they form cell clusters of different sizes. In contrast to T4-2 cells, these cells are not tumorigenic in nude mice. We selected and propagated three of these clusters that exhibited different sizes but were all disorganized in 3D basement membrane matrix (designated S2-1, S2-2, and S2-3). In contrast to T4-2 cells, these cells do not form xenograft tumors when transplanted into mice (unpublished work). As such they can be considered as equivalents of early precursors of breast cancer. CAR levels in all three sublines were significantly up-regulated (Fig. 3), suggesting that it is the disturbance in the 3D organization of cells rather than the malignant phenotype per se that correlates with CAR expression.
CAR Expression in Human Breast Cancer.
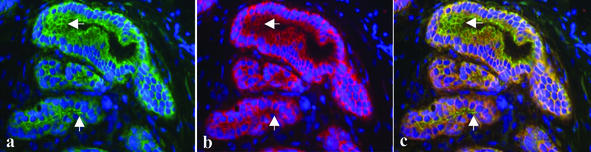
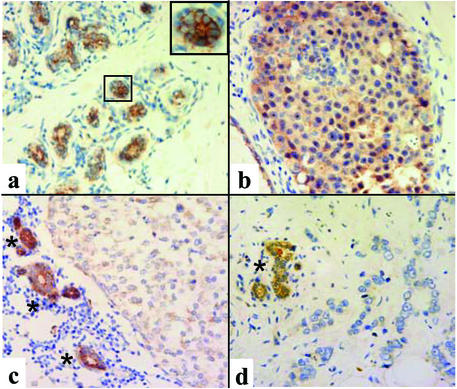
To address the question of whether primary human breast cancer specimens show changes of CAR expression similar to those observed in the 3D tumor progression model, we performed immunofluorescence microscopy of frozen tissue and immunohistochemistry on formalin-fixed, paraffin-embedded sections from normal breast tissue, ductal carcinoma in situ (DCIS), and invasive carcinoma. Normal acini and ducts showed strong CAR expression at cell–cell junctions (Figs. 4 and 5a) where it colocalized with E-cadherin; this colocalization was similar to what was seen in S1 clusters inside basement membrane (shown in Figs. 1 b–d and 4). In addition, CAR expression was seen at the apical portion of the lateral cell–cell contacts where only a weaker E-cadherin signal was detected (Fig. 4). In contrast, DCIS lacked this pattern and CAR was expressed evenly at all cell surfaces and diffusely in the cytoplasm (Fig. 5b). CAR expression in the three infiltrating breast cancer samples included in this study varied substantially. In two of the three cases, low levels of CAR protein were seen in the cytoplasm without preferential accentuation at the cell surface (Fig. 5c). In one case of infiltrating carcinoma, CAR protein was not detectable with these assays (Fig. 5d). These results indicate that up-regulation of CAR is an early event associated with loss of the physiologic tissue organization. As malignant cells become invasive, CAR levels decline dramatically, along with E-cadherin.
Figure 4.
Detection of CAR (green) and E-cadherin (red) in normal human mammary tissue by immunofluorescence microscopy. The arrows indicate CAR expression at the apical cell contacts. 4′,6-Diamidino-2-phenylindole staining (blue) was used to visualize DNA in the cell nuclei. (Magnifications: ×400.)
Figure 5.
Detection of CAR protein in human tissue samples by immunohistochemistry. (a) Normal breast tissue. (Inset) A representative acinus is shown at higher magnification. (b) DCIS. (c and d) Two cases of invasive ductal carcinomas. Asterisks indicate adjacent or entrapped normal ducts. (Magnifications: ×200.)
Discussion
Despite the important role of CAR for the effectiveness of adenovirus-based therapies of cancer, the regulation of this transmembrane protein in normal and cancer cells is poorly understood. Here we demonstrate that malignant breast cells grown in a 3D cell culture model have higher levels of CAR than their clonally related, nonmalignant counterpart and that this difference correlates with loss of tissue integrity and polarity and occurs before the cells become tumorigenic. For our studies, we chose the HMT-3522 human breast cancer progression model because it allows analyzing molecular changes occurring during tumor progression under culture conditions that resemble important aspects of the physiological microenvironment (16). It was recently demonstrated that some forms of adhesions, 3D matrix adhesions, are formed exclusively under 3D culture conditions (17). Previous studies using the HMT-3522 model showed that growth factor-mediated signaling events could be uncoupled in conventional tissue culture models while becoming evident in 3D cultures. A striking example of this phenomenon is the coordinated regulation of β1 integrin and the EGFR (12). The EGF dependence of T4-2 cells, which were originally derived from S1 cells by culturing in EGF-depleted medium (18), allowed us also to study the impact of EGFR signaling on CAR expression. Signals from the EGFR are transduced by the Ras/mitogen-activated protein kinase and phosphatidylinositol 3-kinase pathways, which are known to regulate expression and function of the adherens junction molecule E-cadherin and the tight junction protein ZO-1 (19, 20). Initial reports suggesting that CAR could also function as a cell-adhesion molecule (21) were confirmed recently, as CAR was found to participate in the formation of tight junctions (22). We were therefore particularly interested in analyzing the effect of growth factor signaling on expression of CAR in a 3D tissue context. It is noteworthy that the functional consequences of changes in expression of cell-adhesion molecules is strongly tissue dependent. For example, loss of E-cadherin has been widely associated with tumor progression (23). In contrast, in some tumor types, including ovarian cancer, gain of E-cadherin function might be involved in early tumorigenesis (24). Because of this tissue-specific heterogeneity we focused our analysis here entirely on breast cancer.
In 3D cultures, we found a striking reduction in the total CAR protein in comparison to T4-2 cells. Furthermore, whereas immunofluorescence microscopy revealed a strong diffuse staining pattern for CAR protein without accentuation of cell–cell junctions in T4-2 cells, in nonmalignant S1 cells, the signal was restricted to sites of cell–cell contact. A similar staining pattern was observed for E-cadherin, which is in agreement with previously reported results in the same cell system (9), and supports the hypothesis that internalization of E-cadherin is one mechanism of inhibition of E-cadherin-mediated adhesion in breast cancer (25). Although both CAR and E-cadherin show this shift in their subcellular localization in T4-2 cells, an increase in protein levels was observed only for CAR, indicating that regulation of CAR expression levels is independent of the mechanisms regulating E-cadherin expression.
To determine whether the deregulation of CAR in T4-2 cells was correlated with the malignant phenotype or whether loss of tissue organization was sufficient, we examined CAR expression in “premalignant” S2 cells, which show disturbances of their growth pattern in 3D basement membrane cultures but do not form tumors in nude mice. CAR protein expression in these cell lines was similar to the levels observed in T4-2 cells, indicating that a malignant phenotype is not a prerequisite for deregulation of CAR but loss of polarity correlates with increased CAR. This hypothesis was further supported by the finding that CAR levels increased significantly when S1 cells were grown either in a 3D collagen matrix, a nonphysiologic microenvironment for mammary epithelial cells, or on 2D plastic substratum. These findings demonstrate that normal breast epithelial cells also show deregulation of CAR expression on disturbances of their cellular tissue organization. If S1 cells are placed in a functional microenvironment, polarity is reestablished and CAR expression is restricted to sites of cell–cell contact. Malignant cells, however, do not respond to basement membrane; they reorganize tissue structure and CAR expression only after inhibition of growth factor or integrin signal transduction. Lack of response of T4-2 cells is reminiscent of observations in normal human endothelial and HeLa cells. Although CAR levels increased with increasing confluence of the normal cells, malignant HeLa cells showed no changes of receptor levels under the same conditions (26).
To elucidate the mechanisms involved in deregulation of CAR in T4-2 cells, we examined the role of EGFR signaling for regulation of CAR protein expression. T4-2 cells are characterized by a marked increase in EGFR level and a corresponding overactivation of downstream signaling pathways (12). In these cells, inhibition of EGFR signaling with the receptor-blocking antibody mAb225 or the receptor tyrosine kinase inhibitor tyrphostin leads to a significant reduction of cell proliferation, inhibition of downstream signals, and morphologic reversion into a phenotype that resembles S1 cells (12). We found that this phenotypic reversion was accompanied by a reduction of CAR protein levels in reverted cells to levels equal or even lower than those in S1 cells and that CAR protein was relocated to cell–cell junctions. The data on CAR mRNA expression mirrored these findings, suggesting that CAR expression is regulated, at least in part, by pretranslational steps, which is in agreement with previous reports (13). Redistribution of E-cadherin to the cell–cell contact sites also was observed, in agreement with previous experiments (9, 12). Redistribution of E-cadherin and ZO-1 to the cell surface has been reported after inhibition of Ras/mitogen-activated protein kinase signaling in MDCK cells harboring oncogenic Ras (20). Accordingly, the current data and our previous investigations demonstrate that the failure of T4-2 cells to form polarized epithelial structures and the concomitant up-regulation of CAR are correlated with an excess of signaling through the EGFR and its downstream targets.
CAR functions as the primary receptor for adenovirus and is crucial for the success of adenovirus-based therapies of cancer. It is therefore of possible practical therapeutic relevance that a considerable proportion of T4-2 cells grown in 3D cultures were infected at a multiplicity of infection that left S1 and T4-2rev cells essentially untouched. This finding bodes well for adenovirus gene therapy of breast cancer as long as the cancer cells continue to express CAR because normal breast structures appear to be resistant to infection. This finding is in concordance with the differences in CAR protein levels and distributional patterns between S1, T4-2, and T4-2rev cells. Our data are in support of the hypothesis that the CAR protein, when participating in the formation of cell–cell adhesion in normal cells, is only to a limited extend accessible for binding to adenovirus. In contrast, deregulation of CAR in early breast cancer cells, as represented by T4-2 cells, might render these cells more susceptible to infection with adenoviruses by disruption of the integrity of cell–cell adhesion complexes. As this is a common phenomenon in cancer, this mechanism provides a possible additional mode of tumor selectivity for therapeutic adenoviruses.
Our examination of CAR expression in normal human breast tissue and samples of breast cancer revealed some similarities with the model system. We found that in normal breast epithelium, CAR was expressed along the circumference of the cells, including an accentuated staining at the apical portion of the lateral cell surface, a localization that is known to be the site of tight junction formation (27). Cases of DCIS showed CAR expression at the cell surface combined with high cytoplasmatic protein levels. In contrast, our preliminary analysis of infiltrating adenocarcinomas demonstrated a diffuse, low-level cytoplasmic staining or even complete loss of CAR expression. We conclude that the loss of physiological tissue organization as it occurs in early stages of breast cancer development (DCIS) is similar to the situation in T4-2 cells and is associated with an up-regulation of CAR. At more advanced stages during tumor progression, however, CAR expression appears to be reduced. Although this is likely to impede the susceptibility of such tumors to adenovirus infection, the localization and organization of CAR that is expressed is not similar to the normal tissues and lack of polarity may still allow selective infection. Undoubtedly, regulation of CAR in different tissues would be tissue specific. Nevertheless, some of these observations have relevance to other tumor types. In a separate investigation in liver tumors, we found a correlation between loss of CAR expression and histological tumor grade (W.M.K. and K.A.R., unpublished work). Similar findings have been reported for prostate cancer where a statistically significant decrease in CAR surface expression in prostate and in bladder cancer was found (10, 28). These observations mirror expression changes of E-cadherin in tumors, which is frequently lost in high-grade cancers while preservation of its expression is correlated with a favorable prognosis (29).
In conclusion, our data provide insight into the mechanisms regulating expression of CAR in breast epithelial cells, linking this protein to processes involved in maintenance of tissue integrity. We further demonstrate that disruption of this integrity, e.g., by aberrant EGF signaling, may lead to deregulation of CAR expression. This observation has implications for our understanding of the tumor biology of breast cancer and suggests the possibility that disruption of CAR-mediated cell–cell adhesion could promote an invasive phenotype, which would be consistent with the hypothesis that, in prostate and bladder cancer, CAR may be a tumor suppressor (30, 31). Our results indicate that the localization and organization of CAR is an important determinant of the susceptibility of tissues to infection. Our study indicates in addition that the development of adenovirus-based therapies for breast cancer has to take into account variability and localization of CAR expression, which might render subtypes of tumors particularly susceptible to infection whereas others may become resistant. Further investigations are necessary to define this potential window of therapeutic opportunity. In addition, possible effects of signaling inhibitors on receptor expression might interfere with susceptibility of cancer cells to infection with adenovirus and should be considered in the design of combination treatments.
Acknowledgments
We thank J. M. Bergelson, F. McCormick, and F. M. Waldman for helpful comments. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (to M.A.), funds from the U.S. Department of Energy, Office of Biological and Environmental Research (DE-AC03 SF0098), the National Cancer Institute (CA64786), and an Innovator Award from the Department of Defense Breast Cancer Research Program (BC012005) (to M.J.B.), grants from the University of California, San Francisco Research Evaluation and Allocation Committee and an American Cancer Society Individual Research Award (to W.M.K.), and National Institutes of Health Grant P50-CA89520 (to K.A.R.).
Abbreviations
- CAR
coxsackievirus and adenovirus receptor
- EGF
epidermal growth factor
- EGFR
EGF receptor
- DCIS
ductal carcinoma in situ
References
- 1.Bischoff J R, Kirn D H, Williams A, Heise C, Horn S, Muna M, Ng L, Nye J A, Sampson-Johannes A, Fattaey A, McCormick F. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 2.Lebedeva S, Bagdasarova S, Tyler T, Mu X, Wilson D R, Gjerset R A. Hum Gene Ther. 2001;12:763–772. doi: 10.1089/104303401750148685. [DOI] [PubMed] [Google Scholar]
- 3.Ogawa T, Kusumoto M, Mizumoto K, Sato N, Tanaka M. Breast Cancer. 1999;6:301–304. doi: 10.1007/BF02966444. [DOI] [PubMed] [Google Scholar]
- 4.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 5.Hemmi S, Geertsen R, Mezzacasa A, Peter I, Dummer R. Hum Gene Ther. 1998;9:2363–2373. doi: 10.1089/hum.1998.9.16-2363. [DOI] [PubMed] [Google Scholar]
- 6.Wickham T J, Mathias P, Cheresh D A, Nemerow G R. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 7.Petersen O W, Ronnov-Jessen L, Howlett A R, Bissell M J. Proc Natl Acad Sci USA. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briand P, Petersen O W, Van Deurs B. In Vitro Cell Dev Biol. 1987;23:181–188. doi: 10.1007/BF02623578. [DOI] [PubMed] [Google Scholar]
- 9.Weaver V M, Petersen O W, Wang F, Larabell C A, Briand P, Damsky C, Bissell M J. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rauen K A, Sudilovsky D, Le J L, Chew K L, Hann B, Weinberg V, Schmitt L D, McCormick F. Cancer Res. 2002;62:3812–3818. [PubMed] [Google Scholar]
- 11.Godfrey T E, Kim S H, Chavira M, Ruff D W, Warren R S, Gray J W, Jensen R H. J Mol Diagn. 2000;2:84–91. doi: 10.1016/S1525-1578(10)60621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F, Weaver V M, Petersen O W, Larabell C A, Dedhar S, Briand P, Lupu R, Bissell M J. Proc Natl Acad Sci USA. 1998;95:14821–14826. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Pong R C, Bergelson J M, Hall M C, Sagalowsky A I, Tseng C P, Wang Z, Hsieh J T. Cancer Res. 1999;59:325–330. [PubMed] [Google Scholar]
- 14.Howlett A R, Bailey N, Damsky C, Petersen O W, Bissell M J. J Cell Sci. 1995;108:1945–1957. doi: 10.1242/jcs.108.5.1945. [DOI] [PubMed] [Google Scholar]
- 15.Gudjonsson T, Ronnov-Jessen L, Villadsen R, Rank F, Bissell M J, Petersen O W. J Cell Sci. 2002;115:39–50. doi: 10.1242/jcs.115.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walpita D, Hay E. Nat Rev Mol Cell Biol. 2002;3:137–141. doi: 10.1038/nrm727. [DOI] [PubMed] [Google Scholar]
- 17.Cukierman E, Pankov R, Stevens D R, Yamada K M. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 18.Briand P, Nielsen K V, Madsen M W, Petersen O W. Cancer Res. 1996;56:2039–2044. [PubMed] [Google Scholar]
- 19.Potempa S, Ridley A J. Mol Biol Cell. 1998;9:2185–2200. doi: 10.1091/mbc.9.8.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Lu Q, Schneeberger E E, Goodenough D A. Mol Biol Cell. 2000;11:849–862. doi: 10.1091/mbc.11.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honda T, Saitoh H, Masuko M, Katagiri-Abe T, Tominaga K, Kozakai I, Kobayashi K, Kumanishi T, Watanabe Y G, Odani S, Kuwano R. Brain Res Mol Brain Res. 2000;77:19–28. doi: 10.1016/s0169-328x(00)00036-x. [DOI] [PubMed] [Google Scholar]
- 22.Cohen C J, Shieh J T, Pickles R J, Okegawa T, Hsieh J T, Bergelson J M. Proc Natl Acad Sci USA. 2001;98:15191–15196. doi: 10.1073/pnas.261452898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perl A K, Wilgenbus P, Dahl U, Semb H, Christofori G. Nature. 1998;392:190–193. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- 24.Auersperg N, Pan J, Grove B D, Peterson T, Fisher J, Maines-Bandiera S, Somasiri A, Roskelley C D. Proc Natl Acad Sci USA. 1999;96:6249–6254. doi: 10.1073/pnas.96.11.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boterberg T, Vennekens K M, Thienpont M, Mareel M M, Bracke M E. Cell Adhes Commun. 2000;7:299–310. doi: 10.3109/15419060009015001. [DOI] [PubMed] [Google Scholar]
- 26.Carson S D, Hobbs J T, Tracy S M, Chapman N M. J Virol. 1999;73:7077–7079. doi: 10.1128/jvi.73.8.7077-7079.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoover K B, Liao S Y, Bryant P J. Am J Pathol. 1998;153:1767–1773. doi: 10.1016/S0002-9440(10)65691-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sachs M D, Rauen K A, Ramamurthy M, Dodson J L, De Marzo A M, Putzi M J, Schoenberg M P, Rodriguez R. Urology. 2002;60:531–536. doi: 10.1016/s0090-4295(02)01748-x. [DOI] [PubMed] [Google Scholar]
- 29.Heimann R, Lan F, McBride R, Hellman S. Cancer Res. 2000;60:298–304. [PubMed] [Google Scholar]
- 30.Okegawa T, Li Y, Pong R C, Bergelson J M, Zhou J, Hsieh J T. Cancer Res. 2000;60:5031–5036. [PubMed] [Google Scholar]
- 31.Okegawa T, Pong R C, Li Y, Bergelson J M, Sagalowsky A I, Hsieh J T. Cancer Res. 2001;61:6592–6600. [PubMed] [Google Scholar]