Abstract
No transgenic cystic fibrosis (CF) mouse model developed to date mimics the major clinical phenotype found in humans with CF, chronic Pseudomonas aeruginosa lung infection. In a transgenic CF transmembrane conductance regulator (cftr) mouse colony, we found WT, heterozygous, and homozygous CF mice housed in the same cage became chronically colonized in the oropharynx with environmental P. aeruginosa when the bacterium was present in drinking water. Elimination of P. aeruginosa from drinking water resulted in clearance in most WT and CF heterozygous, but not homozygous mice. For experimental evaluation, a combination of specific animal husbandry techniques and an oral infection route showed cftr−/− mice but not WT mice can be chronically colonized by P. aeruginosa with subsequent lung translocation, yielding a pathologic picture indicative of chronic lung infection. In some instances, mucoid isolates of P. aeruginosa were recovered from lungs, indicating conditions were present for conversion to mucoidy. Overexpression of human CFTR in the lungs of WT mice markedly accelerated the clearance rate of P. aeruginosa, demonstrating that lung levels of CFTR play an important role in defense against infection. P. aeruginosa mutants unable to express the surface polysaccharide alginate or the global regulator GacA were deficient in their ability to colonize the mice. CF mice made potent immune responses to P. aeruginosa outer membrane antigens. Overall, we found that under the proper conditions, transgenic CF mice are hypersusceptible to P. aeruginosa colonization and infection and can be used for evaluations of lung pathophysiology, bacterial virulence, and development of therapies aimed at treating CF lung disease.
Cystic fibrosis (CF), a common and devastating human genetic disease, is caused by mutations in the CF transmembrane conductance regulator (Cftr) gene. Transgenic mice with different mutations in the mouse cftr homolog (1–5) mostly show abnormalities in ion transport that are similar to the gastrointestinal manifestations of CF in humans. The most serious feature of human CF disease, chronic lung infection by mucoid strains of Pseudomonas aeruginosa, has not yet been described in CF mice (1, 6). Some lines of CF mice show abnormalities in overall lung physiology and in various aspects of their response to challenge with microbial pathogens (2, 7–9). In CF mice challenged with P. aeruginosa, an association between excessive inflammation and the resultant lung pathology has been seen, but only in the context of a short-term P. aeruginosa infection. The failure to observe hypersusceptibility to chronic P. aeruginosa infection in CF mice has limited the utility of CF mice to model human CF disease. Here we describe experimental conditions in which transgenic CF mice do indeed manifest increased susceptibility to chronic P. aeruginosa infection when an oral challenge route is used. We also show that CF mice can acquire chronic P. aeruginosa infections from the environment and that translocation of P. aeruginosa to the lungs and emergence of mucoid phenotypes of P. aeruginosa can occur in CF mice after months of P. aeruginosa colonization, similar to what is observed in human CF patients.
Methods
Animals.
ΔF508 cftr mice were previously described (3). Cftrtm1Unc-TgN(FABPCFTR) [fatty acid-binding protein (FABP)-CFTR] mice have a stop codon in the murine cftr gene (S489X) but also express human CFTR in the gut epithelium due to transgenic introduction of Cftr under the control of the FABP promoter (10). Breeding pairs of these noninbred mice were obtained from The Jackson Laboratory and further bred in our facility. FABP-CFTR mice have also been bred into the FVB genetic background and breeding pairs of these inbred mice were provided by Jeffrey Whitsett (University of Cincinnati, Cincinnati), as were transgenic FVB-strain mice overexpressing human CFTR in the respiratory epithelium under the control of transcriptional elements derived from the human lung-specific surfactant protein C (SP-C-CFTR mice) (11). Wild-type C57BL/6 mice were used as controls for the noninbred FABP-CFTR mice, whereas wild-type FVB mice were used as controls for the inbred FABP-CFTR and SP-C-CFTR mice.
Bacterial Strains.
Wild-type P. aeruginosa strains used were: PA14 (12), a clinical isolate from a human burn infection; PAO1-V, a chloramphenicol-sensitive substrain of PAO1 obtained from Michael Vasil (University of Colorado, Denver); N6 and N13, clinical isolates from two different CF patients <3 years of age provided by Jane Burns (University of Washington, Seattle), and lipopolysaccharide-rough, mucoid clinical isolate FRD-1, provided by Dennis Ohman (Medical College of Virginia, Richmond). Inactivation of the algD and gacA genes in strains PA14 and of algD in FRD-1 has been described (12, 13).
Mouse Oropharyngeal Colonization Model.
Five to seven days before an experiment, mice were treated with 0.25 mg of levofloxacin per ml of drinking water as a means of eliminating an indigenous upper respiratory tract Enterobacter spp. that interfered with the colonization and detection of P. aeruginosa. Mice were then taken off the levofloxacin water 24 h before the study and placed on 0.2 mg of gentamicin per ml of drinking water for 1 day, prior to which oropharyngeal cultures were taken to ensure clearance of any indigenous P. aeruginosa and/or Enterobacter spp. from the upper respiratory tract. After this treatment, mice were given drinking water containing P. aeruginosa at a final concentration of 2 × 107 colony-forming units per ml for a period of 5–7 days. Throat cultures were then taken to confirm oropharyngeal colonization, and mice were placed for 1 week on sterile water containing 0.5 mg of nitrofurantoin per ml to eliminate contamination with the Enterobacter spp. For the remainder of the study, mice were placed either on antibiotic maintenance water consisting of 0.1 mg of gentamicin per ml, 0.01 mg of ceftazidime per ml, 0.5 mg of nitrofurantoin per ml, or on sterile water containing 0.2 M sodium acetate, pH 4.0, with 0.2 mg of nitrofurantoin per ml placed into sterile water bottles. Weekly or biweekly throat cultures were taken to screen for colonization with the infecting strain of P. aeruginosa in the oropharyngeal cavity.
Screening for Microbial Colonization of the Oropharynx.
Throat cultures were taken by using an alginate swab inserted into the oropharynx of mice anesthetized by exposure to the inhalant anesthetic isofluorane. The swab was then placed in 1 ml of tryptic soy broth to incubate at 37°C for 5–7 h. Next, 1 ml of 2 mg nitrofurantoin per ml was added to suppress the growth of any contaminating Enterobacter spp. The culture was allowed to incubate overnight at 37°C and was subcultured onto Pseudomonas isolation medium the next day. The growth from the throat cultures that appeared to be P. aeruginosa was confirmed by an oxidase test.
Lung Cultures.
To determine bacterial levels in lungs, mice were killed by inhalation of carbon dioxide, the lungs removed aseptically, and samples taken for histology and quantification of the bacteria after homogenization of a weighed sample.
Serology.
Sera obtained from mice were evaluated by ELISA by using a purified outer membrane preparation from P. aeruginosa strain PAO1, prepared as described (14). Titers were the reciprocal of the serum dilution giving an OD reading >0.2 after 30 min of substrate incubation.
Histology.
Lung tissue was fixed in formalin and embedded in paraffin. Sections were stained with hematoxylin/eosin at the Animal Resources Center of Harvard Medical School. Light microscopic examination was conducted by a trained pathologist unaware of the experimental conditions associated with the samples.
Statistical Analysis.
The marginal probability of oropharyngeal colonization of the mice by P. aeruginosa was estimated on the basis of irregularly timed repeated measures by solving generalized estimating equations (15) using r software for statistical computing (www.r-project.org). The throat culture results obtained over time were used to calculate an overall probability of colonization of a given strain of mouse with a given strain of P. aeruginosa. Two-sided significance tests of the differences in the probability of colonization between mouse strains were obtained by using the ratio of the estimated difference to its robust standard error (15), which follows a standard normal distribution. A one-way ANOVA based on probability of colonization determined by the generalized estimating equation was used to compare colonization among CF, WT, and SP-C-CFTR mice.
Results and Discussion
Natural Occurrence of Chronic Oropharyngeal Infection.
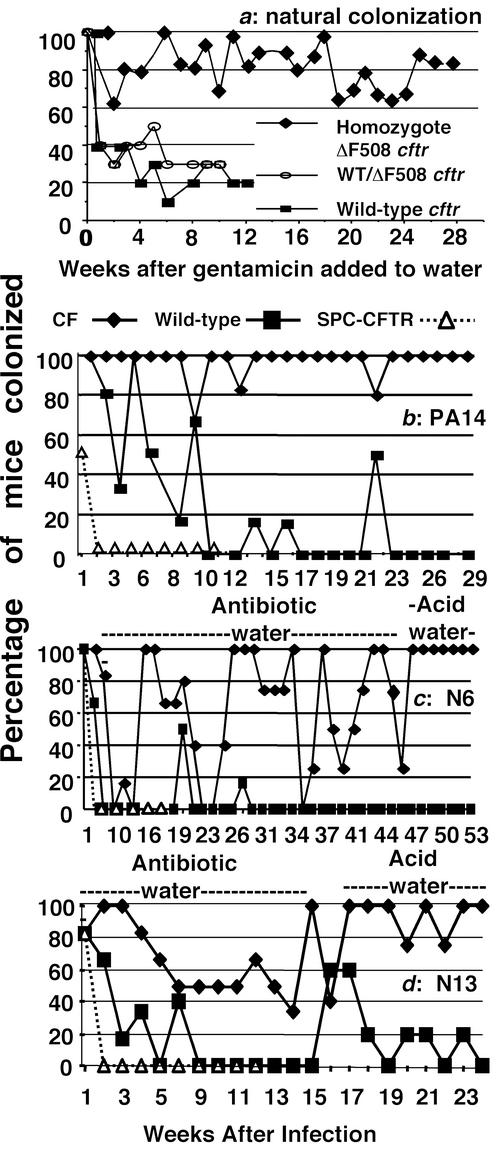
We first noted the presence of P. aeruginosa in cultures of the oropharynx of breeding groups of female WT or heterozygous and male homozygous ΔF508 cftr mice housed in the same cages, tracing the source to a stock solution of an oral osmotic laxative used to fill individual water bottles on animal cages. Heterozygous female ΔF508 cftr mice not housed with homozygous CF mice did not become colonized with P. aeruginosa, likely because they did not receive the oral osmotic laxative. To prevent P. aeruginosa growth in these osmotic laxative solutions, we added the orally nonabsorbable antibiotic gentamicin (0.1 mg/ml) to the solutions and to the animals' liquid diet. This addition resulted in no detectable P. aeruginosa in any subsequent food or laxative/water sample. Once P. aeruginosa environmental contamination was controlled, we noted that <30% of WT and heterozygous female Cftr mice housed with homozygous male CF mice had detectable P. aeruginosa in their subsequent oropharyngeal cultures, whereas >60% to >80% of the homozygous ΔF508 Cftr mice continued to carry P. aeruginosa in their throats (Fig. 1a). Thus, the P. aeruginosa in the oropharynx of the CF mice served to continually contaminate the drinking water bottles, leading to apparent oropharyngeal colonization of WT and heterozygous animals housed in the same cage. Homozygous CF offspring of the colonized mice maintained since birth in cages with water and liquid food containing gentamicin did not acquire oropharyngeal colonization with P. aeruginosa. Therefore, once the contamination of the drinking solutions by P. aeruginosa was controlled, most WT and heterozygous mice could clear their oropharyngeal P. aeruginosa even when still housed in the same cage as a colonized CF sire.
Figure 1.
Natural and experimental oropharyngeal colonization of WT or transgenic CF mice by P. aeruginosa. (a) Percentage of mice with the indicated genotype having positive throat cultures of P. aeruginosa initially acquired from contaminated drinking water after the addition of 0.1 mg of gentamicin per ml (arrow) to control bacterial growth in drinking water. (b) Oropharyngeal colonization of P. aeruginosa PA14 in transgenic CF and WT mice, and transgenic SP-C-CFTR mice. (c and d) Colonization of WT, FABP-CFTR, or SP-C-CFTR transgenic mice with P. aeruginosa clinical isolates, N6 and N13, switched from antibiotic water to acid water as indicated. Six to eight mice per group were used initially, although over time some animals died, usually with P. aeruginosa cultured from the lung.
Interestingly, environmental acquisition of chronic P. aeruginosa oropharyngeal colonization was also found in the FABP-CFTR strain of CF mouse that does not need special diets or oral laxatives. Drinking water bottles from these mice yielded high levels of P. aeruginosa morphologically identical to that isolated from the oropharyngeal cultures. Once gentamicin was added to drinking water of the FABP-CFTR mice, there was no further newly acquired oropharyngeal colonization due to P. aeruginosa in this colony. Negative oropharyngeal cultures for P. aeruginosa were found in >80 mice with WT cftr in a variety of genetic backgrounds that were given the same water as the FABP-CFTR mice and housed in the same room. Thus, the water served as the initial source of P. aeruginosa and the oropharynx of the CF mice served as a reservoir to continually transmit the bacterium to drinking water. To test this idea, we individually housed heterozygous or homozygous ΔF508 cftr mice that had at least three positive oropharyngeal cultures for P. aeruginosa followed by negative cultures over several weeks in a cage with sterile water lacking antibiotics. Each homozygous CF mouse had subsequent oropharyngeal cultures positive for P. aeruginosa that was also found contaminating their water, whereas positive cultures were never obtained from the heterozygous ΔF508 cftr mice (not shown).
Experimental Colonization with P. aeruginosa.
Deliberate colonization with specific strains of P. aeruginosa was next attempted via limited oral exposure in the drinking water. The results from the natural colonization showed P. aeruginosa spread from chronically colonized CF mice to heterozygotes in the same cage that were, in reality, only transiently colonized due to drinking contaminated water. This study also indicated that adding an orally nonabsorbable antibiotic, gentamicin, to water maintained its sterility, prevented spread of colonization from colonized to uncolonized mice housed in the same cage, and did not interfere with detection of P. aeruginosa in the oropharynx of chronically colonized CF mice.
Experimental comparisons were first made between WT-C57BL/6 mice and FABP-CFTR mice lacking CFTR in the lung in a mixed background or between WT-FVB mice and the FABP-CFTR mice bred into the FVB background and also with CFTR-overexpressing SP-C-CFTR mice. Critical factors for establishing reproducible colonization results were controlling environmental exposure to P. aeruginosa before infection, preventing cross-colonization by P. aeruginosa in water bottles, and eliminating oropharyngeal colonization with an endogenous Enterobacter sp. that interfered with both establishment and detection of P. aeruginosa colonization. Problematically, the Enterobacter was able to grow on selective P. aeruginosa isolation medium. Placing P. aeruginosa strain PA14 into drinking water for 7 days followed by removal and replacement with drinking water containing 1 mg of gentamicin per ml showed that noninbred FABP-CFTR mice were unable to clear the oropharyngeal colonization over a 29-week period, whereas colonization in the oropharynx of WT C57 Bl/6 mice was reduced to essentially zero by 10 weeks (Fig. 1b). The positive cultures in WT mice obtained after 10 weeks were attributed to animal care personnel not properly handling water bottle changes, a problem further avoided by having animal maintenance performed by study personnel. Importantly, the SP-C-CFTR mice overexpressing CFTR in the lung rapidly cleared the P. aeruginosa, exhibiting positive throat cultures only on the day the contaminated drinking water was removed from the cages (Fig. 1b).
Statistical analysis comparing the probability of P. aeruginosa strain PA14 colonization among the various mouse strains showed that the CF mice had a 97% probability of colonization, the WT mice a 22% probability of colonization, and the SP-C-CFTR mice a 5% probability of colonization (P < 0.001 for all pair-wise comparisons). In a similar experiment in FABP-CFTR, WT C57BL/6, and SP-C-CFTR mice using P. aeruginosa PAO1-V, the probabilities of colonization were 79%, 38%, and 13%, respectively (data not shown, P < 0.001 for FABP-CFTR vs. WT mice and P = 0.003 for SP-C-CFTR mice vs. both FABP-CFTR and WT mice). The somewhat higher P value for the comparison of FABP-CFTR with SP-C-CFTR mice was attributed to a shorter experimental period of observation with the later group, resulting in fewer data points.
Two clinical isolates of P. aeruginosa obtained from CF patients early in the course of infection showed a more variable pattern of colonization of noninbred FABP-CFTR mice when antibiotics were present in the drinking water. Replacing this antibiotic-containing drinking water with acidified water (pH 4.0, 0.2 M Na acetate), which also prevented P. aeruginosa survival in the water, showed that colonization in the FABP-CFTR mice became nearly 100% (Fig. 1 c–d), indicating interference with detection of these strains by oral antibiotics. Even taking into account the results obtained when antibiotics were present in the drinking water, the probability of colonization of the CF mice (67% for strain N6, 74% for strain N13) was still significantly (P < 0.001) higher than for WT (5% for strain N6, 21% for strain N13) or SP-C-CFTR mice (11% for strain N6, 8% for strain N13).
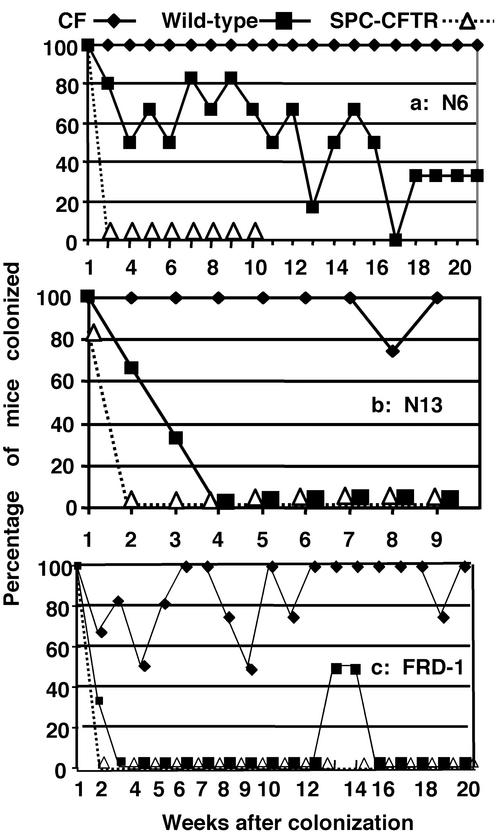
Repeat studies with P. aeruginosa strains N6 and N13 using acid water only to maintain sterility showed both P. aeruginosa strains could establish chronic oropharyngeal colonization in the inbred FABP-CFTR mice (Fig. 2 a and b) with probabilities of colonization of these mice of 100% for strain N6 and 98% for strain N13. WT FVB mice had significantly fewer positive throat cultures (probabilities of colonization 54% for strain N6 and 28% for strain N13, P < 0.001 for all comparisons), although strain N6 was somewhat more resistant to clearance in the WT mice in the absence of antibiotics (see Fig. 1c) The SP-C-CFTR mice rapidly cleared both strains N6 and N13 (Fig. 2 a and b, probabilities of colonization of 10% and 9%, respectively, P < 0.001 for comparison with respective group of FABP-CFTR mouse; for strain N6, P < 0.001 for comparison of SP-C-CFTR with WT mice; for strain N13, P = 0.003 for comparison of SP-C-CFTR with WT mice).
Figure 2.
Colonization of WT, FABP-CFTR, or SP-C-CFTR transgenic mice by different strains of P. aeruginosa: N6 (a), N13 (b), and FRD-1 (c). Colonization experiments were done with acidified water to prevent growth of P. aeruginosa.
Finally, a mucoid clinical isolate of P. aeruginosa, overexpressing alginate and having a rough lipopolysaccharide, was also able to colonize the FABP-CFTR mice with a probability of colonization of 86% (Fig. 2c), whereas both WT and SP-C-CFTR mice rapidly cleared this strain (probabilities of colonization 13% and 6%, respectively, P < 0.001 for both comparisons with FABP-CFTR mice and P < 0.001 for comparison of WT and SP-C-CFTR mice). The positive cultures for strain FRD-1 obtained at weeks 13 and 14 in 50% of the WT mice were attributed to technical errors.
A statistical analysis of the probabilities of colonization from all seven of the experiments described above (Figs. 1 b–d and 2 and PAO I–V) showed FABP-CFTR mice had a mean ± SE probability of colonization of 85.9 ± 4.9%, WT mice had a mean ± SE probability of colonization of 25.9 ± 6.2%, and SP-C-CFTR mice had a mean ± SE probability of colonization of 8.9 ± 1.1% (overall ANOVA P < 0.001; P < 0.001 for FABP-CFTR vs. the other two groups; P = 0.02 for SP-C-CFTR vs. WT mice). In essentially all of the experiments except for the one shown in Fig. 1c, in which antibiotics were present in the drinking water, the SP-C-CFTR mice overexpressing human CFTR in the lung exhibited accelerated clearance of the P. aeruginosa strains from the oropharynx. Chroneos et al. (16) showed that the SP-C-CFTR mice had an ≈7-fold higher mRNA level for human CFTR in the lung compared with murine Cftr expressed by WT mice. Our findings that CFTR overexpression in the lung also promotes oropharyngeal clearance suggest that upper airway colonization is maintained by reservoirs of bacteria in the lung. Nonetheless, this result affirms that the level of CFTR expression affects resistance to P. aeruginosa colonization and provides support for the use of oropharyngeal colonization of CF mice for evaluating pharmacologic or genetic therapies designed to increase expression of CFTR and to determine whether there is likely to be an effect relevant to susceptibility or resistance to P. aeruginosa infection. Finding that increased CFTR expression enhances clearance of P. aeruginosa supports the hypothesis that P. aeruginosa directly binds to CFTR (17), promoting clearance by activation of innate immunity via rapid nuclear factor κB nuclear translocation in airway epithelial cells (18) and by bacterial internalization into these cells followed by desquamation (19).
Findings in Autopsied Mice.
About 6 months postcolonization, some environmentally and experimentally colonized CF mice started to die, many with lung cultures positive for P. aeruginosa. At least five different isolates obtained at autopsy from mice initially infected with nonmucoid strains clearly demonstrated the mucoid morphology typical of chronic P. aeruginosa infection in CF patients. Quantitation of P. aeruginosa in lungs of dead mice at autopsy was not considered meaningful due to potential artifact caused by postmortem overgrowth. Deliberate sacrifice of mice colonized for 24–53 weeks with strains N6, N13, or FRD-1 followed by cultures of the autopsied tongue, esophagus, trachea, and lungs showed that 60–90% of the CF mice had P. aeruginosa present on the tongue, esophagus, and/or trachea, and about one-third of these mice had low levels of P. aeruginosa in the lungs (Table 1).
Table 1.
Detection of P. aeruginosa in cultures of homogenates of the indicated organ from CF or WT mice at time of death
| Percentage of mice with positive cultures for P. aeruginosa
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P. aeruginosa strain, weeks infected | No. of mice tested
|
Lung
|
Esophagus
|
Trachea
|
Tongue
|
|||||
| CF | WT | CF | WT | CF | WT | CF | WT | CF | WT | |
| N6 (53) | 3 | 6 | 33 | 0 | 33 | 16 | 66 | 0 | 100 | 0 |
| N13 (24) | 11 | 5 | 36 | 0 | 73 | 0 | 45 | 0 | 91 | 0 |
| FRD1 (25) | 5 | 6 | 20 | 0 | 80 | 0 | 80 | 0 | 80 | 16 |
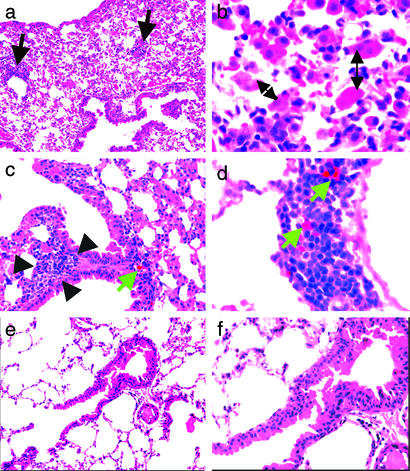
Histopathologic analysis of lung sections from infected mice showed reactive lymphoid hyperplasia, characterized mainly by expanded bronchus-associated lymphoid tissue and occasionally by patchy lymphoid aggregates elsewhere in the interstitium, usually located along lymphatic pathways (Fig. 3). Deposition of hemosiderin was seen in this tissue and in neighboring lymph nodes. Increased numbers of intraalveolar macrophages were also noted. These changes were seen in lungs of CF mice with colonization detectable only in the oropharynx and in mice with P. aeruginosa in both the oropharynx and the lung. End-stage (honeycomb) lung was not observed. Quantitative analysis of bacterial loads in the lung indicated that when P. aeruginosa was present, bacterial levels were low (<100 colony-forming units per g of lung tissue). The mice with only oropharyngeal colonization tended to show less prominent lymphoid expansion. Uninfected age-matched CF mice never exposed to P. aeruginosa had essentially normal-appearing lungs (Fig. 3).
Figure 3.
Histopathologic analysis of lung sections from CF mice with oropharyngeal and/or lung infection with P. aeruginosa. All sections were stained with hematoxylin and eosin. (a) Lung section from an animal with oropharyngeal but not lung infection, showing abundant intraalveolar macrophages and patchy lymphoid aggregates (black arrows) located along lymphatic pathways (original magnification, ×40). (b) Lung section from an animal with oropharyngeal but not lung infection showed increased intraalveolar macrophages (double-headed arrows, original magnification ×100). (c and d) Lung sections from two animals with both oropharyngeal and lung infection with P. aeruginosa. (c) Hyperplastic bronchus-associated lymphoid tissue (BALT) (black arrows); hemosiderin within the BALT stands out as a golden-brown pigment (green arrow, original magnification ×40). (d) Hemosiderin (green arrows) in perilymphatic lymphoid aggregates within the lung (original magnification ×100). (e and f) Lung section from an uninfected CF mouse. No specific pathologic features were noted (original magnifications ×40 and ×100, respectively).
Histologic analyses thus indicate recurrent or chronic lung infection in most of the colonized animals and likely reflect an early stage of CF lung disease, as might be expected in CF infants and children within a few months of the onset of P. aeruginosa infection and before the onset of significant fibrotic changes. Increased numbers of intraalveolar macrophages are a nonspecific finding seen in association with all types of airway obstruction. Hemosiderin is a breakdown product of hemoglobin and indicates prior bleeding. Its presence in the lymph nodes and bronchus-associated lymphoid tissue suggests that the pathology observed resulted from active tissue invasion by P. aeruginosa. It has also been established that increased bronchial-associated lymphoid tissue is a feature of chronic P. aeruginosa lung infection in both CF humans and experimentally infected rats (20–22). We did not see an infiltrate of neutrophils, as is typical of acute bronchitis/bronchiolitis and bronchopneumonia, likely due to the absence of high levels of P. aeruginosa more characteristic of the late stage of CF lung infection from which the most information regarding the pathologic picture of pulmonary disease has been determined for these patients.
Evaluation of P. aeruginosa Mutants for Virulence.
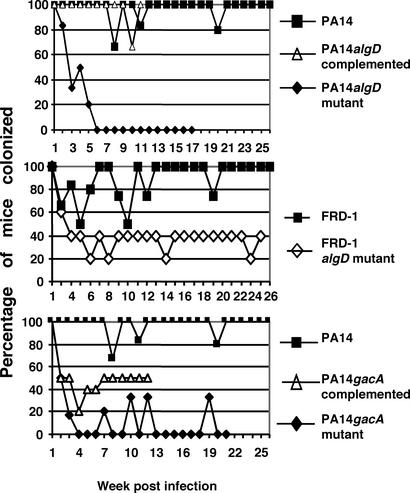
We next tested the ability of the infection model to evaluate changes in the ability to establish oropharyngeal colonization in CF mice using P. aeruginosa strains with mutations in genes encoding virulence factors. Using antibiotics in drinking water, we compared WT PA14 with strains with mutations in the global regulatory gene, gacA, and algD, the gene encoding GDP mannose dehydrogenase needed to produce alginate. The gacA mutant was likely to have an attenuated phenotype, because GacA is needed for biofilm formation, a component of P. aeruginosa disease in CF (23). Using acidified water, we evaluated an algD mutant of mucoid strain FRD-1. Although alginate is a known virulence factor for chronic infection, it was not clear whether alginate production was needed for establishing infection by nonmucoid lipopolysaccharide-smooth strains that produce low levels of alginate (24–26). All of the strains were present in drinking water at ≈107 colony-forming units/ml during exposure of the CF mice. Both of the algD mutant strains were less able to maintain chronic oropharyngeal colonization in CF mice, with the probability of colonization by the PA14 algD mutant being 20% compared with 97% for both the WT (Fig. 1b, reproduced in Fig. 4a) and complemented PA14 strains (Fig. 4a, P < 0.001 for the mutant vs. the WT and complemented strains). The probability of colonization of CF mice with the FRD-1 algD mutant was 44% (Fig. 4b) compared with a probability of 86% with the WT strain (Fig. 2c, reproduced in Fig. 4b, P = 0.014). A complemented strain of FRD-1 algD was not available for study. The PA14 gacA mutant had a probability of colonization of 17% (Fig. 4c) compared with 97% for the WT strain (Fig. 1b, reproduced in Fig. 4c, P < 0.001). Occasional positive cultures with the gacA mutant suggested residual colonization in some mice. Although the complemented gacA strain did not recover full virulence, indicating perhaps other mutations were present in this strain, the probability of colonization was nonetheless 52%, significantly higher than that of the mutant strain (P = 0.016). Overall, these results establish that the model can be used to evaluate the contribution of individual P. aeruginosa virulence factors to chronic infection in CF.
Figure 4.
Colonization of FABP-CFTR mice by the strain of P. aeruginosa indicated next to each graph. Colonization experiments were done with antibiotics for strain PA14 and acid water for strain FRD-1. Six to eight mice per group were used initially.
Immune Response to Infection.
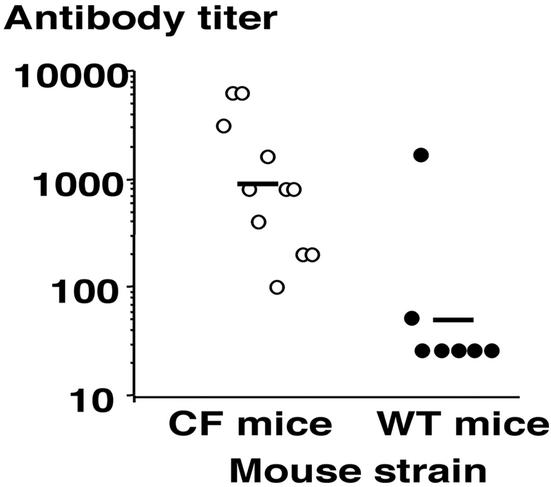
A hallmark of chronic P. aeruginosa infection in CF patients is development of a potent immune response that fails to clear the pathogen (27, 28). It has also been shown that by the age of 3, almost all CF patients have serologic evidence of P. aeruginosa infection as measured by the presence of antibody to outer membrane proteins (29). Analysis of serum antibodies to a P. aeruginosa outer membrane preparation prepared from strain PAO1 (14) showed that chronically colonized CF mice had significantly (P < 0.001, t test) higher levels of antibody to P. aeruginosa compared with low titers in most wild-type mice that rapidly cleared the organisms (Fig. 5). Thus, the level of P. aeruginosa infection in the CF mice was sufficient to provoke a potent antibody response, whereas the exposure of WT mice to P. aeruginosa in the drinking water followed by transient colonization was insufficient to provoke such a response in most animals.
Figure 5.
Antibody titers achieved in sera of CF or WT mice after oropharyngeal colonization for >10 weeks with P. aeruginosa. Each symbol represents the titer for one mouse; bar represents geometric mean.
In this study, we present data in support of the critical role of CFTR in modulating chronic lung infection with P. aeruginosa by showing that transgenic Cftr mice remained colonized with P. aeruginosa for >30 weeks, whereas their wild-type counterparts cleared detectable levels in the oropharyngeal cavity within 4–10 weeks. By infecting transgenic CF mice via their drinking water, we found that these mice developed a disease that mimics the natural history of P. aeruginosa infection in human CF. Likely, the findings most closely resemble what occurs early in CF lung disease due to P. aeruginosa. We also saw that for some mice, translocation of P. aeruginosa to the lower airways of the lung was the natural progression after long-term colonization. Histopathologic analysis found evidence of chronic infection, particularly around the airways, a finding consistent with human CF lung disease. The use of this model also provides evidence of its sensitivity to levels of CFTR expression, as was illustrated through the use of transgenic mice that overexpress CFTR in the respiratory epithelium. We also showed that this transgenic mouse model can measure the expression and importance of various P. aeruginosa phenotypes in a model relevant to CF lung disease. Thus, the model should be useful for evaluating the potential efficacy of various strategies to modulate CFTR expression including gene therapy and chemotherapeutic agents that increase expression of mutant Cftr alleles such as ΔF508.
Acknowledgments
We thank Ervin Meluleni of the Animal Resources Center of Harvard Medical School for sectioning and staining of lung tissues. This work was supported by National Institutes of Health Grants AI48917, HL04277, and HL66678.
Abbreviations
- CF
cystic fibrosis
- CFTR
CF transmembrane conductance regulator
- FABP
fatty acid-binding protein
- FABP-CFTR mice
Cftrtm1Unc-TgN(FABPCFTR) mice
- SP-C mice
FVB mice overexpressing human CFTR in the lung under the control of the surfactant protein C promoter
References
- 1.Grubb B R, Boucher R C. Physiol Rev. 1999;79:S193–S214. doi: 10.1152/physrev.1999.79.1.S193. [DOI] [PubMed] [Google Scholar]
- 2.Davidson D J, Dorin J R, Mclachlan G, Ranaldi V, Lamb D, Doherty C, Govan J, Porteous D J. Nat Genet. 1995;9:351–357. doi: 10.1038/ng0495-351. [DOI] [PubMed] [Google Scholar]
- 3.Colledge W H, Abella B S, Southern K W, Ratcliff R, Jiang C W, Cheng S H, Macvinish L J, Anderson J R, Cuthbert A W, Evans M J. Nat Genet. 1995;10:445–452. doi: 10.1038/ng0895-445. [DOI] [PubMed] [Google Scholar]
- 4.Zeiher B G, Eichwald E, Zabner J, Smith J J, Puga A P, Mccray P B, Capecchi M R, Welsh M J, Thomas K R. J Clin Invest. 1995;96:2051–2064. doi: 10.1172/JCI118253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Doorninck J H, French P J, Verbeek E, Peters R H P C, Morreau H, Bijman J, Scholte B J. EMBO J. 1995;14:4403–4411. doi: 10.1002/j.1460-2075.1995.tb00119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Heeckeren A M, Schluchter M D. Lab Anim. 2002;36:291–312. doi: 10.1258/002367702320162405. [DOI] [PubMed] [Google Scholar]
- 7.Kent G, Iles R, Bear C E, Huan L J, Griesenbach U, McKerlie C, Frndova H, Ackerley C, Gosselin D, Radzioch D, et al. J Clin Invest. 1997;100:3060–3069. doi: 10.1172/JCI119861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gosselin D, Stevenson M M, Cowley E A, Griesenbach U, Eidelman D H, Boule M, Tam M F, Kent G, Skamene E, Tsui L C, Radzioch D. Am J Respir Crit Care Med. 1998;157:1253–1262. doi: 10.1164/ajrccm.157.4.9702081. [DOI] [PubMed] [Google Scholar]
- 9.van Heeckeren A, Walenga R, Konstan M W, Bonfield T, Davis P B, Ferkol T. J Clin Invest. 1997;100:2810–2815. doi: 10.1172/JCI119828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou L, Dey C R, Wert S E, Duvall M D, Frizzell R A, Whitsett J A. Science. 1994;266:1705–1708. doi: 10.1126/science.7527588. [DOI] [PubMed] [Google Scholar]
- 11.Whitsett J A, Dey C R, Stripp B R, Wikenheiser K A, Clark J C, Wert S E, Gregory R J, Smith A E, Cohn J A, Wilson J M, et al. Nat Genet. 1992;2:13–20. doi: 10.1038/ng0992-13. [DOI] [PubMed] [Google Scholar]
- 12.Rahme L G, Tan M W, Le L, Wong S M, Tompkins R G, Calderwood S B, Ausubel F M. Proc Natl Acad Sci USA. 1997;94:13245–13250. doi: 10.1073/pnas.94.24.13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chitnis C E, Ohman D E. Mol Microbiol. 1993;8:583–590. doi: 10.1111/j.1365-2958.1993.tb01602.x. [DOI] [PubMed] [Google Scholar]
- 14.Hancock R E W, Mouat E C, Speert D P. J Infect Dis. 1984;149:220–226. doi: 10.1093/infdis/149.2.220. [DOI] [PubMed] [Google Scholar]
- 15.Liang K Y, Zeger S L. Biometrika. 1986;73:13–22. [Google Scholar]
- 16.Chroneos Z C, Wert S E, Livingston J L, Hassett D J, Whitsett J A. J Immunol. 2000;165:3941–3950. doi: 10.4049/jimmunol.165.7.3941. [DOI] [PubMed] [Google Scholar]
- 17.Pier G B. Curr Opin Microbiol. 2002;5:81–86. doi: 10.1016/s1369-5274(02)00290-4. [DOI] [PubMed] [Google Scholar]
- 18.Schroeder T H, Lee M M, Yacono P W, Cannon C L, Gerceker A A, Golan D E, Pier G B. Proc Natl Acad Sci USA. 2002;99:6907–6912. doi: 10.1073/pnas.092160899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schroeder T H, Reiniger N, Meluleni G, Grout M, Coleman F T, Pier G B. J Immunol. 2001;166:7410–7418. doi: 10.4049/jimmunol.166.12.7410. [DOI] [PubMed] [Google Scholar]
- 20.Sato A, Chida K, Iwata M, Hayakawa H. Am Rev Respir Dis. 1992;146:473–478. doi: 10.1164/ajrccm/146.2.473. [DOI] [PubMed] [Google Scholar]
- 21.Kitazawa H, Sato A, Iwata M. Kansenshogaku Zasshi. 1997;71:214–221. doi: 10.11150/kansenshogakuzasshi1970.71.214. [DOI] [PubMed] [Google Scholar]
- 22.Suda T, Chida K, Hayakawa H, Imokawa S, Iwata M, Nakamura H, Sato A. Chest. 1999;115:357–363. doi: 10.1378/chest.115.2.357. [DOI] [PubMed] [Google Scholar]
- 23.Parkins M D, Ceri H, Storey D G. Mol Microbiol. 2001;40:1215–1226. doi: 10.1046/j.1365-2958.2001.02469.x. [DOI] [PubMed] [Google Scholar]
- 24.Pier G B, DesJardins D, Aguilar T, Barnard M, Speert D P. J Clin Microbiol. 1986;24:189–196. doi: 10.1128/jcm.24.2.189-196.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marty N, Dournes J L, Chabanon G, Montrozier H. FEMS Microbiol Lett. 1992;98:35–44. doi: 10.1016/0378-1097(92)90128-b. [DOI] [PubMed] [Google Scholar]
- 26.Tatnell P J, Russell N J, Govan J R, Gacesa P. Biochem Soc Trans. 1996;24:406S. doi: 10.1042/bst024406s. [DOI] [PubMed] [Google Scholar]
- 27.Doring G, Hoiby N. Infect Immun. 1983;42:197–201. doi: 10.1128/iai.42.1.197-201.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fomsgaard A, Hoiby N, Shand G H, Conrad R S, Galanos C. Infect Immun. 1988;56:2270–2278. doi: 10.1128/iai.56.9.2270-2278.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burns J L, Gibson R L, McNamara S, Yim D, Emerson J, Rosenfeld M, Hiatt P, McCoy K, Castile R, Smith A L, Ramsey B W. J Infect Dis. 2001;183:444–452. doi: 10.1086/318075. [DOI] [PubMed] [Google Scholar]