Abstract
The 3,308,274-bp sequence of the chromosome of Lactobacillus plantarum strain WCFS1, a single colony isolate of strain NCIMB8826 that was originally isolated from human saliva, has been determined, and contains 3,052 predicted protein-encoding genes. Putative biological functions could be assigned to 2,120 (70%) of the predicted proteins. Consistent with the classification of L. plantarum as a facultative heterofermentative lactic acid bacterium, the genome encodes all enzymes required for the glycolysis and phosphoketolase pathways, all of which appear to belong to the class of potentially highly expressed genes in this organism, as was evident from the codon-adaptation index of individual genes. Moreover, L. plantarum encodes a large pyruvate-dissipating potential, leading to various end-products of fermentation. L. plantarum is a species that is encountered in many different environmental niches, and this flexible and adaptive behavior is reflected by the relatively large number of regulatory and transport functions, including 25 complete PTS sugar transport systems. Moreover, the chromosome encodes >200 extracellular proteins, many of which are predicted to be bound to the cell envelope. A large proportion of the genes encoding sugar transport and utilization, as well as genes encoding extracellular functions, appear to be clustered in a 600-kb region near the origin of replication. Many of these genes display deviation of nucleotide composition, consistent with a foreign origin. These findings suggest that these genes, which provide an important part of the interaction of L. plantarum with its environment, form a lifestyle adaptation region in the chromosome.
Lactic acid bacteria are used for the preservation of food and feed raw materials such as milk, meat, and vegetables or other plant materials. Research carried out in recent years has led to the conviction that certain strains of lactic acid bacteria, in particular strains from the genera Lactobacillus, may promote health in man and animals (1). The genus Lactobacillus encompasses a considerable number of different species that display a relatively large degree of diversity (2). Among these, Lactobacillus plantarum is a flexible and versatile species that is encountered in a variety of environmental niches, including some dairy, meat, and many vegetable or plant fermentations. Moreover, L. plantarum is frequently encountered as a natural inhabitant of the human gastrointestinal (GI) tract (3), and a selected strain, L. plantarum 299v, is marketed as a probiotic that may confer various health beneficial effects to the consumer (4, 5). The ecological flexibility of L. plantarum is reflected by the observation that this species has one of the largest genomes known among lactic acid bacteria (6). Several strains of L. plantarum are genetically accessible, and genetic tools have been developed for this species, including (controlled) gene expression systems (7, 8) and vectors that can be used for the construction of gene disruption or deletion variants (9, 10). The ability to persist in the human GI tract has stimulated research aimed at the use of L. plantarum as a delivery vehicle for therapeutic compounds, including vaccines (11). Here we present the complete genomic sequence of L. plantarum WCFS1, a single colony isolate from L. plantarum NCIMB8826, which was originally isolated from human saliva (National Collection of Industrial and Marine Bacteria, Aberdeen, U.K.) (12). It has been shown to survive the passage of the stomach in an active form and is able to persist for >6 days in the human GI tract (13).
Methods
Sequencing and Annotation.
The L. plantarum WCFS1 genome sequence was determined by using a whole genome sequencing and assembly approach (14). Protein-encoding ORFs and RNA genes were predicted and functionally annotated (Tables S01 and S02 and linear genome map, www.cmbi.kun.nl/lactobacillus). Functional classification of proteins was performed essentially according to the Riley rules (15). Detailed sequencing and annotation procedures and supplementary material for this paper are available at our web site (www.cmbi.kun.nl/lactobacillus). The L. plantarum genome has been submitted to the EMBL database under accession number AL935263.
Global Analysis.
L. plantarum WCFS1 contains a single, circular chromosome of 3,308,274 bp, which is close to the size predicted on basis of classical L. plantarum genome sizing analysis (6). L. plantarum WCFS1 was found to contain two small, cryptic plasmids (2,365 and 1,917 bp) and a larger plasmid (36,069 bp) encoding genes involved in conjugal plasmid transfer and several other functions. The overall G+C content of the chromosome is 44.5%, whereas the plasmids appeared to have a slightly lower G+C content (genome statistics are summarized in table S03, www.cmbi.kun.nl/lactobacillus).
Replication Origin and Terminus.
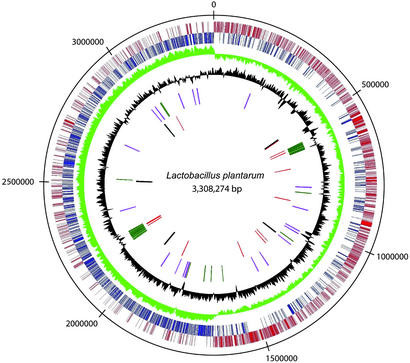
The origin of replication was identified by homology with the chromosomes of Bacillus subtilis (16) and Bacillus halodurans (17), in which the organization of genes around the origin is identical. In L. plantarum WCFS1, 12 of the 14 genes in this region are orthologs of these Bacillus species, organized and oriented in the same manner. Moreover, 11 DnaA binding boxes were found flanking the dnaA gene, providing further evidence for replication initiation (18). Finally, the GC-skew displays a sharp transition in this region (Fig. 1).
Figure 1.
Genome-atlas view of the L. plantarum WCFS1 chromosome, with the predicted origin of replication at the top. The outer to inner circles show (i) positive strand ORFs (red); (ii) negative strand ORFs (blue); (iii) GC-skew (green); (iv) G+C content (black); (v) prophage-related functions (green) and IS-like elements (purple); and (vi) rDNA operons (black) and tRNA encoding genes (red). The GC% and GC skew (C-G)/(C+G) were calculated in a window of 4,000 nt, in steps of 75 nt. The G+C percentage was plotted as the number of G+C nucleotides in the plus strand divided by the window size, i.e., (G+C)/4,000; lowest and highest values are 30.8% and 51.8%. The upper and lower values of the GC skew were 0.22 and −0.27.
The genes encoded by the L. plantarum genome are predominantly transcribed in the direction of replication, which is a feature observed in many genomes of low G+C Gram-positive bacteria (Fig. 1). The replication terminus appeared to be positioned diametrically opposite to the origin of replication and is characterized by a sharp transition in the GC-skew. Moreover, a dif-like termination sequence was found starting at base pair 1,669,020. The dif-like sequence in combination with the activities of the site-specific XerC- and XerD-like recombinases are most likely involved in chromosomal resolution during replication (19).
Sequence Repeats.
The L. plantarum genome contains five rRNA operons that are distributed evenly around the chromosome (Fig. 1) and display only a very limited number of sequence polymorphisms. A total of 62 tRNA encoding genes was identified, most of which appear to be genetically linked to some of the rRNA clusters (table S02 and genome maps, www.cmbi.kun.nl/lactobacillus). Several other repeated sequence elements were found, including two classes of transposase-encoding regions that are likely to represent mobile genetic elements. These repeated sequences were designated ISP1 (eight complete copies and two interrupted copies) and ISP2 (four complete copies and one interrupted copy) (Fig. 1). ISP1 represents a classical IS-element, containing a transposase-encoding gene flanked by terminal inverted repeats, and shares homology with previously described IS1165 of Leuconostoc mesenteroides (20). ISP2 appears to lack the terminal inverted repeat sequences, but could code for a protein with homology to the transposase in the so-called SCCmec family of mobile genetic elements of Staphylococcus aureus (21).
Prediction of ORFs.
We identified 3,052 protein-encoding genes, of which only 39 appeared to be pseudogenes. Comparison of the predicted proteins with those of other completely sequenced genomes showed that L. plantarum proteins are most similar to predicted proteins from other low-G+C Gram-positive bacteria, with most hits to Listeria, Streptococcus, and Lactococcus, followed by Bacillus, Clostridium, and Staphylococcus (figure S01, www.cmbi.kun.nl/lactobacillus).
Comparative analysis of the Listeria spp, B. subtilis, and S. aureus genomes has revealed a conserved, colinear organization of their genes, indicating a certain stability of the genomes of this group of Gram-positive bacteria (22). L. plantarum appears to follow this trend, although the synteny is less than between Listeria and Bacillus, and only at a local rather than global level. We have found 16 clusters with a conserved, colinear organization of more than eight genes between L. plantarum, Listeria monocytogenes, and B. subtilis, whereas the synteny between the genomes of L. plantarum and Lactococcus lactis IL1403 was much less.
Putative biological functions were assigned to 2,120 of the predicted proteins, and another 588 predicted proteins in L. plantarum are homologous to conserved proteins of unknown function in other organisms (table S01B, www.cmbi.kun.nl/lactobacillus). The remaining 344 hypothetical proteins had no database match; 57 of these proteins are putative membrane proteins, and another 111 are <100 aa. At least 440 multigene (paralog) families were identified, containing 1,443 predicted proteins.
Prediction of Highly Expressed Genes.
The codon adaptation index (CAI) and equivalent indices are useful indicators for the likelihood that a certain gene is highly expressed in an organism. This correlation is based on the fact that genes with high expression levels have strongly biased usage of synonymous codons (23). CAI values were determined for each gene of L. plantarum by using the ribosomal protein genes as a reference set. In addition to the expected housekeeping genes (23), the set of potentially highly expressed (PHX) genes from L. plantarum (table S04, www.cmbi.kun.nl/lactobacillus) contains genes of the complete Embden–Meyerhoff–Parnas (EMP) pathway and a number of enzymes involved in the degradation of pentoses and hexoses. The focus of L. plantarum on sugar catabolism is also reflected in the observation that a number of phosphotransferase systems (PTSs), and the general PTS enzymes HPr (ptsH) and Enzyme-I (ptsI) are PHX. In particular, all components of the mannose and fructose PTS systems are a member of this set. A further interesting case is the route of N-acetylglucosamine catabolism, which is also entirely PHX. The bias of synonymous codon usage of PHX genes is expressed in the extremely low frequency of the codons ATA (Ile), AGA and AGG (Arg), and the frequent use of CGG and CGT (Arg).
Sugar Import and Central Carbon Metabolism
Sugar Transport.
L. plantarum is a versatile and flexible organism and is able to grow on a wide variety of sugar sources. This phenotypic trait is reflected by the high number of genes encoding putative sugar transporters, which even exceeds that found in Streptococcus mutans (ref. 24; table S01B, www.cmbi.kun.nl/lactobacillus). The majority of these transporters are predicted PEP-dependent sugar PTSs. L. plantarum WCFS1 encodes 25 complete PTS enzyme II complexes, and several incomplete complexes. This high number of PTS is far more than those found in other microbial genomes, and similar only to Listeria monocytogenes (22). The substrate specificities of L. plantarum PTSs have been predicted based on homology to annotated PTS genes and from their genomic context, because in many cases the PTS enzyme II genes are found to be clustered with enzyme and regulatory genes involved in sugar metabolism (figure S02, www.cmbi.kun.nl/lactobacillus). In addition to PTSs, the L. plantarum genome encodes 30 transporter systems that were predicted to be involved in the transport of carbon sources. However, the substrate specificity could not be predicted for some PTS and other carbon-uptake systems, and various sugar transport systems are known to import more than one substrate, thereby expanding the carbon transport capacity of this species even further (table S05, www.cmbi.kun.nl/lactobacillus).
Sugar Metabolism.
Once internalized, sugars are used as carbon source for growth and for the generation of energy through fermentation. Classically, L. plantarum is grouped among the facultative heterofermentative lactobacilli, indicating that sugars can be fermented via the EMP pathway or the phosphoketolase pathway, leading to homolactic and heterolactic fermentation profiles, respectively (25). In agreement with this classification, the genes for an intact phophoketolase pathway were found on the L. plantarum chromosome. The genes encoding enzymes involved in the EMP pathway were found to be organized in two operons. This genetic linkage facilitates efficient, concerted regulation of expression of these enzymes in response to both the level and type of sugar source available in the environment. As expected, the L. plantarum chromosome does not encode an intact citrate acid cycle. However, similar to what has been found in Lactococcus lactis (26), several of the enzymes from this pathway appear to be present, including six copies of fumarate reductase (of which two are truncated). This high degree of fumarate reductase redundancy suggests that L. plantarum harbors a rudimentary electron transport chain. Moreover, the finding of a molybdopterin-dependent nitrate reduction system in L. plantarum (see www.cmbi.kun.nl/lactobacillus) could indicate the utilization of nitrate as the ultimate electron acceptor.
Pyruvate Metabolism.
L. plantarum displays an almost homolactic fermentation pattern during growth on glucose that is degraded via the EMP pathway leading to pyruvate, which is subsequently converted to approximately equimolar amounts of d- and l-lactate by the activities of stereospecific lactate dehydrogenase enzymes (27). In addition to these ldhL and ldhD genes, the chromosome encodes two other putative genes for lactate dehydrogenase and a relatively large number of other pyruvate-dissipating enzymes that are predicted to catalyze the production of other metabolites, including formate, acetate, ethanol, acetoin, and 2,3-butanediol. A remarkable degree of redundancy in the genes encoding these functions is observed. In comparison to Lactococcus lactis IL1403 (26), the pyruvate-dissipating potential in L. plantarum is clearly much larger. Nevertheless, Lactococcus lactis also displays some redundancy in especially its lactate dehydrogenase-encoding genes. These observations support the relative importance of pyruvate-dissipating capacity in these fermentative microbes.
Lifestyle Adaptations.
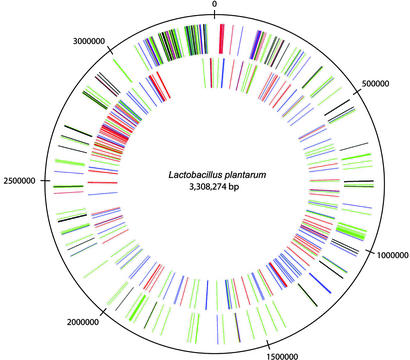
From the large set of genes involved in sugar uptake and utilization, combined with the observation that many of these genes belong to the PHX group of genes, it can be concluded that L. plantarum is programmed for efficient utilization of many different carbon sources. This finding agrees with the observation that L. plantarum is a versatile and flexible microbe that can sustain its growth in a variety of environmental niches. Remarkably, many of the genes for sugar transport and metabolism are clustered near the origin of replication (Fig. 2). In particular, the 213-kb region from 3,072,500 to 3,285,500 encodes almost exclusively proteins for sugar transport, metabolism, and regulation. Moreover, this entire region has a lower GC content (41.5%) than the rest of the genome (Fig. 1), suggesting that many genes may have been acquired by horizontal gene transfer. This would be in agreement with the hypothesis that this part of the L. plantarum chromosome represents a lifestyle-adaptation region that is used to effectively adapt to the changes in conditions encountered in the numerous environmental niches in which this microbe is found.
Figure 2.
Nonrandom distribution of genes belonging to specific functional categories in the L. plantarum chromosome. The outer circle contains all genes encoding proteins involved in sugar transport (PTS are colored black, other transporters are colored blue), sugar metabolism (green), and biosynthesis and/or degradation of polysaccharides (red). The inner circle contains all genes predicted to encode secreted proteins; see also Table 1. Red, signal peptides; green, N-terminal lipoprotein anchor; blue, N-terminal signal anchor sequence.
Biosynthesis and Degradation
Proteolytic System and Amino Acid Biosynthesis.
Lactic acid bacteria generally inhabit protein-rich environments (including milk), and are equipped with a protein-degradation machinery to create a selective advantage for growth under these conditions. The L. plantarum genome did not appear to encode the primary enzyme required for large polypeptide utilization, namely the extracellular protease Prt that is involved in primary breakdown of proteins. However, L. plantarum has uptake systems (Opp and Dtp) for peptides, which are the primary protein-degradation products. Once internalized, these peptides are degraded by a variety of peptidases, which have been extensively studied in both lactococci and lactobacilli (28, 29). L. plantarum has 19 genes encoding intracellular peptidases of different specificity (table S06, www.cmbi.kun.nl/lactobacillus). The most obvious difference between L. plantarum and Lactococcus lactis IL1403 is the number of peptidases capable of cleaving N-terminal proline residues, because L. plantarum has three such genes (pepI, pepR), whereas Lactococcus lactis has none. Despite this elaborate protein degradation machinery, the L. plantarum genome encodes complete pathways for biosynthesis of most amino acids, and their genes are generally organized in large clusters or operons. A noticeable exception is the absence of the pathways leading to the branched-chain amino acids valine, leucine, and isoleucine.
Nonribosomal Peptide Synthesis.
A nonribosomal peptide synthesis (NRPS) gene cluster of 25 kb was found in the L. plantarum genome (lp_0578 to lp_0581; linear genome map, www.cmbi.kun.nl/lactobacillus), which is the first example of such a biosynthesis machinery in lactic acid bacteria. The peptide-like products produced by similar systems are highly variable in structure and composition, and include important pharmaceutical compounds. Moreover, several of these compounds have been shown to play a key role in microbial pathogenicity (30). The NRPS machinery characteristically is a multimodular protein and the L. plantarum cluster encodes two NRPSs, consisting of five and one module(s), respectively. The cluster also encodes an essential phosphopantetheinyl transferase, as well as proteins putatively required for regulation, secretion/transport, and enzymes for precursor supply.
Transport.
By far the largest class of proteins in L. plantarum is represented by transport proteins (411 genes). Of these transporters, 57 ATP-binding cassette (ABC) transporters (168 proteins) were identified, of which 27 were importers and 30 were exporters. Many of these importers transport amino acids and peptides, whereas the substrate specificity of most of the exporters is unknown. The L. plantarum chromosome encodes several transporters for uptake of branched-chain amino acids, including an ABC transporter encoded by the livABCDE genes, which is in agreement with the absence of the genes encoding enzymes involved in the biosynthesis of these amino acids in L. plantarum. It is noteworthy that the glutamine-specific ABC-transporters display considerable redundancy, because four complete systems are found in L. plantarum. Interestingly, in B. subtilis the glutamine synthetase (L. plantarum homologue lp_1581) is affected by the presence of glutamine and plays an important role in modulation of global regulation of nitrogen metabolism. Moreover, L. plantarum encodes a homologue of the global repressor glnR (lp_1580) of B. subtilis, of which the activity is modulated by the glutamine synthetase, but lacks the other nitrogen metabolism regulators described for B. subtilis (TnrA and CodY). These findings suggest that glutamine transport could be of critical importance in the regulation of nitrogen metabolism in L. plantarum through its potential effect on the signaling role fulfilled by the glutamine synthetase.
Regulation and Signaling.
Another large class is that of the regulatory functions, containing at least 262 genes (8.5% of total proteins). This class includes three σ factor encoding genes (rpoD, rpoN, and sigH) and at least 13 sensor-regulator pairs that belong to the two-component regulator family (see www.cmbi.kun.nl/lactobacillus). The relatively high proportion of regulatory genes found in L. plantarum is similar to that only of Pseudomonas aeroguinosa (8.4%) and Listeria monocytogenes (7.3%), and could be a reflection of the many different environmental conditions that all these three bacteria face.
Adaptation to Stress.
L. plantarum encodes genes for a number of stress-related proteins, including several proteases involved in stress response such as the energy-dependent intracellular proteases ClpP, HslV, and Lon, which degrade aberrant and nonfunctional proteins. In addition to the groES-groEL chaperonin and the hrcA-grpE-dnaK-dnaJ operons encoding heat shock proteins, L. plantarum also encodes three small heat shock proteins of the HSP20 family (31), and three highly homologous cold-shock proteins (CspL, CspC, CspP) that have previously been characterized (32). In addition to other common stress pathways, lactic acid-producing bacteria must efficiently deal with acidification of their local environment. The F0F1-ATPase presumably serves as a major regulator of intracellular pH. Moreover, 10 encoded sodium-proton antiporters could also be involved in the L. plantarum acid stress response as has been reported for similar genes in Listeria monocytogenes (33). Finally, the L. plantarum chromosome encodes three paralogous alkaline-shock proteins, which are also expected to play a role in pH tolerance in this microbe (34). Previously, the physiological response of L. plantarum to hyperosmotic stress has been studied and it was shown that mainly electrolyte-mediated osmolality up-shifts led to accumulation of compatible solutes (35). The L. plantarum genome encodes at least three systems for the uptake and biosynthesis of the osmoprotectants glycine-betaine/carnitine/choline, including two ABC transporters (opuABCD, choSQ). Furthermore, there are genes encoding various oxidative stress-related proteins such as catalase, thiol peroxidase, glutathione peroxidase, halo peroxidase, four thioredoxins, four glutathione reductases, five NADH-oxidases, and two NADH peroxidases. In agreement with previous observations, the L. plantarum genome does not encode a superoxide dismutase. More than two decades ago it was established that L. plantarum compensates for the lack of this enzyme by high level (20–30 mM) intracellular accumulation of Mn2+ ions, which at these concentrations can act as a scavenger for oxygen radicals (36). The L. plantarum genome encodes a large capacity (55 proteins) for transport of cations, including the recently identified P-type manganese translocating ATPase encoded by mntA (37). The L. plantarum genome encodes at least three additional transport systems that are putatively involved in manganese accumulation, including an ABC-transporter and two highly homologous natural resistance-associated macrophage proteins (NRAMP)-like transporters, which have been shown to be up-regulated under manganese starvation (M.N.N.G., E. Pentcheva, J. C. Verdoes, E. Klaassens, W.M.d.V., J. Delcour, P. Hols, and M.K., unpublished data). The accumulation of manganese observed in L. plantarum is in good agreement with the relative abundance of high-affinity transport systems for this transition metal.
Secretion
Secretion and Processing Machinery.
Components of the secretion machinery found in L. plantarum WCFS1 include the signal-recognition particle proteins Ffh and FtsY, the general chaperone Tf (trigger factor), and the components SecA/SecE/SecG/SecY/YajC (but no SecDF) of the major translocation pathway. Two YidC homologs were found that may also play a role in the secretion pathway, because it has been shown in Escherichia coli that YidC associates with the Sec translocase (38). Furthermore, we found two prsA/prtM-like peptidylprolyl isomerases, three signal peptidases I, a single signal peptidase II for cleavage of lipoprotein signal peptides and coupling to membrane lipids, and a single sortase for cleavage of C-terminal LPxTG-type anchors and coupling to peptidoglycan. No components of a twin-arginine translocation (TAT) pathway were found.
Extracellular Proteins.
There were 217 proteins with N-terminal signal sequences predicted, of which 144 with potential signal peptidase cleavage sites. Most of these proteins are predicted to be anchored to the cell (Table 1) by single N- or C-terminal transmembrane anchors (83 proteins), lipoprotein anchors (47 proteins, including four phage related genes), LPxTG-type anchors (25 proteins), or other cell-wall binding (repeated) domains, such as LysM domains (10 proteins) or choline-binding domains (3 proteins) (detailed in table S07, www.cmbi.kun.nl/lactobacillus). A previously uncharacterized C-terminal domain of ≈120 residues, designated WxL domain because it contains this conserved motif twice, is found in 19 proteins of L. plantarum, but also in some proteins of Lactococcus lactis and Listeria. This domain may also be involved in cell-envelope binding.
Table 1.
Predicted functions of extracellular proteins
| Function | SP | NLP | N-SA | Total |
|---|---|---|---|---|
| Transport | ||||
| ABC transporter, substrate binding | 2 (0) | 22 | 7 | 31 |
| Cell wall | ||||
| Biosynthesis | 2 (0) | 1 | 9 | 12 |
| Degradation | 9 (5) | 1 | 3 | 13 |
| Enzymes (other) | 8 (1) | 5 | 15 | 28 |
| Other (+phage) | 4 (2) | 5 | 7 | 16 |
| Hypothetical | ||||
| Conserved (domain) | 55 (49) | 1 | 18 | 74 |
| Nonconserved | 17 (13) | 12 | 14 | 43 |
| Total | 97 (70) | 47 | 73 | 217 |
SP, signal peptide; NLP, N-terminal lipoprotein anchor; N-SA, N-terminal signal anchor sequence. The number of SP-containing proteins that also have a cell-wall binding domain is indicated in parentheses.
L. plantarum proteins containing LPxTG-type sortase motifs actually have a different and quite distinct consensus sequence LPQTxE (in 22 of the 25 proteins; figure S03, www.cmbi.kun.nl/lactobacillus), which may reflect the specificity of the single sortase encoded by the genome. Such a highly conserved sortase motif has not been described before in other Gram-positive bacteria such as Lactococcus lactis (26), Listeria monocytogenes (22), S. aureus (39), or Streptococcus pneumoniae (40), which have 6, 40, 14, and 13 proteins with LPxTG-type anchors, respectively. Most of the extracellular proteins belong to paralog families, typically containing 3–6 members, and occasionally >10 members. Most of the predicted extracellular enzymes are hydrolases, some of known substrate specificity (signal peptidases, sortase, proteinases), but many of unknown specificity but with hydrolase catalytic residue consensus motifs. However, for the majority of the extracellular proteins, no definite function prediction can be made (Table 1). In general, these proteins have a normal signal peptide and multiple domains, including at least one cell-envelope anchoring domain. It is highly likely that some of these extracellular proteins play a role in adhesion or binding to other cells or proteins, because they contain domains with homology to proteins in databases with predicted functions such as mucus-binding, fibronectin-binding, aggregation-promoting, intercellular adhesion, or cell clumping.
An unusual surface-associated protein in L. plantarum is the 3,360-residue protein, designated Sdr, that contains a nearly perfect SD-repeat (Ser-Asp) of >1,600 residues, in addition to an N-terminal signal peptide, a C-terminal transmembrane anchor, low complexity regions, and a domain of unknown function. Extracellular proteins with a similar domain structure including very long Ser-containing repeats have been found in other Gram-positive bacteria (40–43). It has been suggested (40) that glycosyltransferases, encoded by adjacent genes, could make O-linked glycosylations on the serines, producing structures similar to mucins that may coat the surface of the bacterium or interact with host cell mucins. In L. plantarum there are three tagE-like genes, encoding putative poly(glycerol-phosphate) α-glucosyltransferases, near the sdr gene, which could fulfill such a role.
Overall, this large group of proteins could function in recognition or binding of certain components in the varying environments that L. plantarum occupies. Intriguingly, the genes encoding extracellular proteins are not randomly distributed over the chromosome, because the region from 2,604,000 to 3,063,000 bp has a strong overrepresentation of these genes (Fig. 2). Moreover, many of these genes appear in clusters of three to six genes, and the function of these gene clusters in particular is unknown. This region of the chromosome is adjacent to the region that almost exclusively encodes proteins involved in sugar transport and utilization and the regulation thereof. These findings support the hypothesis that this part of the L. plantarum genome represents a lifestyle adaptation region that overrepresents functions related to flexible interaction with varying environments.
Phages.
The L. plantarum chromosome contains two apparently complete prophage genomes and several prophage remnants. The large prophage regions Lp1 (44 kb) and Lp2a (43 kb) resemble temperate pac-site phages, found in dairy environments, in their genome organization. The closest related phage was L. plantarum phage phig1e (44). Remarkably, prophage Lp2a shares DNA sequence identity with prophage Lp1 over the entire DNA packaging/head/tail gene cluster and the lysis cassette. A detailed analysis of the L. plantarum prophage regions including comparison with phages from lactic acid bacteria is necessary.
Horizontal Gene Transfer.
Horizontal gene transfer between bacteria can occur by means of various mechanisms, including natural competence and bacteriophage infection. Although it has never been reported to be naturally competent, L. plantarum appears to encode components of the machinery required for DNA binding and uptake that have been described in B. subtilis (45).
Genes that were possibly acquired by L. plantarum through horizontal gene transfer were searched by using two methods; the first method is based on sequence homology (see table S08, www.cmbi.kun.nl/lactobacillus), whereas the second method is based on base composition analysis. The gene cluster citR-mae-citCDEF, encoding citric acid cycle proteins, is closely related to Leuconostoc mesenteroides (40–80% amino acid identity), whereas the lactose permease (LacS; 62% identity) and split β-galactosidase subunits (LacL and LacM; 96% identity) are highly related to Leuconostoc lactis (46). Moreover, there are five consecutive genes encoding proteins with nearly 100% identity to the sucrose transport, metabolism, and regulation proteins of Pediococcus pentosaceus (GenBank accession no. Z32771). Another group of genes encodes proteins that display highest homology to Gram-negative bacteria such as Salmonella, Agrobacterium, Rhizobium, Ralstonia, Pseudomonas, and Neisseria. This group includes two small gene clusters (lp_0250 to lp_0252 and lp_0498 to lp_0502) in which six genes have highest homology (E score < e−80) to Salmonella and other Gram-negative microbes, whereas the only homologues found among Gram-positive bacteria are identified in Clostridia. It is noteworthy that one of these genes encodes a putative selenocysteine synthase that catalyses the conversion of seryl-tRNA into selenocysteinyl-tRNA(Sec) that is required for the incorporation of selenocysteine residues into protein. Selenocysteine incorporation appears to be wide-spread and is also found in various Gram-positive bacteria (47, 48). Therefore, it is remarkable that the synthetase found in L. plantarum has no homologue in any of the closely related Gram-positives like B. subtilis, Listeria monocytogenes, or Lactococcus lactis.
Base composition analysis of genes was performed by calculating a χ2 index based on the expected and observed frequency for each nucleotide (40). Very large regions of unusual base composition were found in the L. plantarum genome (see horizontal gene transfer, base composition analysis (table S09) and figure S04, www.cmbi.kun.nl/lactobacillus). Remarkably, a large part of the region enriched in genes involved in sugar uptake and catabolism displayed unusual base composition, consistent with the existence of a region reflecting the flexible and adaptive lifestyle of L. plantarum.
Conclusions
The sequence of the L. plantarum WCFS1 chromosome reveals that this microbe focuses on carbon catabolism, which is illustrated by the capacity to import and use a large variety of carbon sources and is corroborated by the finding that many genes encoding enzymes involved in the central carbon metabolism belong to the group of potentially highly expressed genes. The genome sequence also supports the flexibility and versatility of L. plantarum, which is clearly illustrated by the exceptionally high number of sugar import systems, including many PTSs. Moreover, the discovery of a large collection of surface-anchored proteins also indicates that L. plantarum has the potential to associate with a large variety of surfaces and potential substrates for growth. Finally, the relatively high number of regulatory functions implies that L. plantarum can effectively adapt to many environmental conditions. Environmental flexibility and adaptation by L. plantarum may result from a series of functions concentrated within a defined genomic region, which has been designated the lifestyle adaptation region.
Acknowledgments
This paper is dedicated to the fond memory of our dear friend and colleague Dr. Hans Sandbrink, who passed away during the course of this research. We thank Frank van Enckevort, Jakub Rychter, Maud le Coq, Maarten Arends, Nico Penninkhof, Jornt Bek, Michiel Wels, Ingeborg Boels, Wilbert Sybesma, Patrick van de Boogaard, Elaine Vaughan, Armand Hermans, Bart Pieterse, Mariët van der Werf, Bernadet Renckens, and Mark Sturme for their contribution to annotation. We thank Marleen Abma-Henkens, Marjo van Staveren, and Paul Mooijman for their skillful technical assistance. We thank Harald Brüssow for preliminary analysis of the prophages, and Torsten Stachelhaus for preliminary analysis of the nonribosomal peptide biosynthesis gene cluster. We thank James Brown for identification of the RNase P RNA region.
Abbreviations
- ABC
ATP-binding cassette
- PHX
potentially highly expressed
- EMP
Embden–Meyerhoff–Parnas
- PTS
phosphotransferase system
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the EMBL database (accession no. AL935263).
References
- 1.Kalliomaki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E. Lancet. 2001;357:1076–1079. doi: 10.1016/S0140-6736(00)04259-8. [DOI] [PubMed] [Google Scholar]
- 2.Stiles M E, Holzapfel W H. Int J Food Microbiol. 1997;36:1–29. doi: 10.1016/s0168-1605(96)01233-0. [DOI] [PubMed] [Google Scholar]
- 3.Ahrne S, Nobaek S, Jeppsson B, Adlerberth I, Wold A E, Molin G. J Appl Microbiol. 1998;85:88–94. doi: 10.1046/j.1365-2672.1998.00480.x. [DOI] [PubMed] [Google Scholar]
- 4.Adawi D, Ahrne S, Molin G. Int J Food Microbiol. 2001;70:213–220. doi: 10.1016/s0168-1605(01)00550-5. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham-Rundles S, Ahrne S, Bengmark S, Johann-Liang R, Marshall F, Metakis L, Califano C, Dunn A M, Grassey C, Hinds G, Cervia J. Am J Gastroenterol. 2000;95:22–25. doi: 10.1016/s0002-9270(99)00813-8. [DOI] [PubMed] [Google Scholar]
- 6.Chevallier B, Hubert J C, Kammerer B. FEMS Microbiol Lett. 1994;120:51–56. doi: 10.1111/j.1574-6968.1994.tb07006.x. [DOI] [PubMed] [Google Scholar]
- 7.Pavan S, Hols P, Delcour J, Geoffroy M C, Grangette C, Kleerebezem M, Mercenier A. Appl Environ Microbiol. 2000;66:4427–4432. doi: 10.1128/aem.66.10.4427-4432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bringel F, Frey L, Hubert J C. Plasmid. 1989;22:193–202. doi: 10.1016/0147-619x(89)90002-4. [DOI] [PubMed] [Google Scholar]
- 9.Hols P, Defrenne C, Ferain T, Derzelle S, Delplace B, Delcour J. J Bacteriol. 1997;179:3804–3807. doi: 10.1128/jb.179.11.3804-3807.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferain T, Hobbs J N, Jr, Richardson J, Bernard N, Garmyn D, Hols P, Allen N E, Delcour J. J Bacteriol. 1996;178:5431–5437. doi: 10.1128/jb.178.18.5431-5437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pouwels P H, Leer R J, Shaw M, Heijne den Bak-Glashouwer M J, Tielen F D, Smit E, Martinez B, Jore J, Conway P L. Int J Food Microbiol. 1998;41:155–167. doi: 10.1016/s0168-1605(98)00048-8. [DOI] [PubMed] [Google Scholar]
- 12.Hayward A C, Davis G H G. Br Dent J. 1956;101:43. [Google Scholar]
- 13.Vesa T, Pochart P, Marteau P. Aliment Pharmacol Ther. 2000;14:823–828. doi: 10.1046/j.1365-2036.2000.00763.x. [DOI] [PubMed] [Google Scholar]
- 14.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 15.Karp P D, Riley M, Paley S M, Pellegrini-Toole A, Krummenacker M. Nucleic Acids Res. 1999;27:55–58. doi: 10.1093/nar/27.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, et al. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 17.Takami H, Nakasone K, Takaki Y, Maeno G, Sasaki R, Masui N, Fuji F, Hirama C, Nakamura Y, Ogasawara N, et al. Nucleic Acids Res. 2000;28:4317–4331. doi: 10.1093/nar/28.21.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshikawa H, Ogasawara N. Mol Microbiol. 1991;5:2589–2597. doi: 10.1111/j.1365-2958.1991.tb01967.x. [DOI] [PubMed] [Google Scholar]
- 19.Hill T M. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1602–1615. [Google Scholar]
- 20.Johansen E, Kibenich A. Plasmid. 1992;27:200–206. doi: 10.1016/0147-619x(92)90022-3. [DOI] [PubMed] [Google Scholar]
- 21.Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K. Antimicrob Agents Chemother. 2001;45:1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, et al. Science. 2001;294:849–852. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- 23.Karlin S, Mrazek J, Campbell A, Kaiser D. J Bacteriol. 2001;183:5025–5040. doi: 10.1128/JB.183.17.5025-5040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ajdic D, McShan W M, McLaughlin R E, Savic G, Chang J, Carson M B, Primaeaux C, Tian R, Kenton S, Jia H, et al. Proc Natl Acad Sci USA. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kandler O. Antonie Van Leeuwenhoek. 1983;49:209–224. doi: 10.1007/BF00399499. [DOI] [PubMed] [Google Scholar]
- 26.Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, Ehrlich S D, Sorokin A. Genome Res. 2001;11:731–753. doi: 10.1101/gr.169701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferain T, Schanck A N, Delcour J. J Bacteriol. 1996;178:7311–7315. doi: 10.1128/jb.178.24.7311-7315.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunji E R, Mierau I, Hagting A, Poolman B, Konings W N. Antonie Leeuwenhoek. 1996;70:187–221. doi: 10.1007/BF00395933. [DOI] [PubMed] [Google Scholar]
- 29.Christensen J E, Dudley E G, Pederson J A, Steele J L. Antonie Leeuwenhoek. 1999;76:217–246. [PubMed] [Google Scholar]
- 30.Marahiel M A, Stachelhaus T, Mootz H D. Chem Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 31.Van Montfort R, Slingsby C, Vierling E. Adv Protein Chem. 2001;59:105–156. doi: 10.1016/s0065-3233(01)59004-x. [DOI] [PubMed] [Google Scholar]
- 32.Derzelle S, Hallet B, Francis K P, Ferain T, Delcour J, Hols P. J Bacteriol. 2000;182:5105–5113. doi: 10.1128/jb.182.18.5105-5113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cotter P D, Gahan C G, Hill C. Mol Microbiol. 2001;40:465–475. doi: 10.1046/j.1365-2958.2001.02398.x. [DOI] [PubMed] [Google Scholar]
- 34.Kuroda M, Ohta T, Hayashi H. Biochem Biophys Res Commun. 1995;207:978–984. doi: 10.1006/bbrc.1995.1281. [DOI] [PubMed] [Google Scholar]
- 35.Glaasker E, Tjan F S, Ter Steeg P F, Konings W N, Poolman B. J Bacteriol. 1998;180:4718–4723. doi: 10.1128/jb.180.17.4718-4723.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Archibald F S, Fridovich I. J Bacteriol. 1981;145:442–451. doi: 10.1128/jb.145.1.442-451.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hao Z, Chen S, Wilson D B. Appl Environ Microbiol. 1999;65:4746–4752. doi: 10.1128/aem.65.11.4746-4752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scotti P A, Urbanus M L, Brunner J, de Gier J W, von Heijne G, van der Does C, Driessen A J, Oudega B, Luirink J. EMBO J. 2000;19:542–549. doi: 10.1093/emboj/19.4.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, et al. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 40.Tettelin H, Nelson K E, Paulsen I T, Eisen J A, Read T D, Peterson S, Heidelberg J, DeBoy R T, Haft D H, Dodson R J, et al. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 41.Josefsson E, McCrea K W, Ni Eidhin D, O'Connell D, Cox J, Hook M, Foster T J. Microbiology. 1998;144:3387–3395. doi: 10.1099/00221287-144-12-3387. [DOI] [PubMed] [Google Scholar]
- 42.McCrea K W, Hartford O, Davis S, Eidhin D N, Lina G, Speziale P, Foster T J, Hook M. Microbiology. 2000;146:1535–1546. doi: 10.1099/00221287-146-7-1535. [DOI] [PubMed] [Google Scholar]
- 43.Bensing B A, Sullam P M. Mol Microbiol. 2002;44:1081–1094. doi: 10.1046/j.1365-2958.2002.02949.x. [DOI] [PubMed] [Google Scholar]
- 44.Desiere F, Pridmore R D, Brussow H. Virology. 2000;275:294–305. doi: 10.1006/viro.2000.0498. [DOI] [PubMed] [Google Scholar]
- 45.Dubnau D, Lovett C M., Jr . In: Bacillus subtilis and Its Closest Relatives: From Genes to Cells. Sonenshein A L, Hoch J A, Losick R, editors. Washington, DC: Am. Soc. Microbiol.; 2002. pp. 453–471. [Google Scholar]
- 46.Vaughan E E, David S, de Vos W M. Appl Environ Microbiol. 1996;62:1574–1582. doi: 10.1128/aem.62.5.1574-1582.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bock A, Forchhammer K, Heider J, Baron C. Trends Biochem Sci. 1991;16:463–467. doi: 10.1016/0968-0004(91)90180-4. [DOI] [PubMed] [Google Scholar]
- 48.Gladyshev V N, Kryukov G V. Biofactors. 2001;14:87–92. doi: 10.1002/biof.5520140112. [DOI] [PubMed] [Google Scholar]