Abstract
The δ-proteobacterium Myxococcus xanthus coordinates its motility during aggregation and fruiting body formation. While searching for chemotaxis genes in M. xanthus, we identified a third chemotaxis-like gene cluster, the che3 cluster, encoding homologs to two methyl-accepting chemotaxis proteins (MCPs), a CheW, a hybrid CheA, a CheB, a CheR, but no CheY. Mutations in mcp3A, mcp3B, and cheA3 did not show obvious defects in motility or chemotaxis but did affect the timing of entry into development. Mutations in these genes caused early aggregation of starving cells, even at low cell densities. Furthermore, these mutants showed pronounced overexpression of the developmentally regulated Tn5lac fusions Ω4403, Ω4411, and Ω4521 as well as overexpression of mbhA and tps, markers for peripheral rods and aggregating cells, respectively. Divergently transcribed from the che3 promoter region is another gene, crdA (chemosensory regulator of development), predicted to encode a transcriptional activator of σ54-dependent promoters. To test the hypothesis that CrdA functions as the cognate response regulator for the histidine kinase CheA3, CrdA and CheA3 were assayed and found to interact strongly in the yeast two-hybrid system. Mutant analysis showed that crdA cells were delayed in development (12–24 h) and delayed in MbhA production relative to the wild type. An mcp3BcrdA double mutant displayed the crdA phenotype, indicating that crdA is epistatic to mcp3B. We conclude that the Che3 chemotaxis-like system functions to control developmental gene expression by regulating a σ54 transcriptional activator, CrdA.
Keywords: chemotaxis|development|MCP|CheA|CrdA
Myxococcus xanthus is a model organism for prokaryotic development and gliding motility (1). These bacteria feed collectively on other microorganisms or on nutrients in their environment. When starved, cells aggregate to form macroscopic structures called fruiting bodies containing 105 to 106 cells; the aggregated cells later sporulate. Some cells remain outside of the aggregates as peripheral rods; these cells have a different pattern of gene expression than aggregated or vegetative cells and normally do not sporulate (2). M. xanthus utilizes complex intercellular signaling to regulate different aspects of their multicellular life cycle (3). Cells coordinate their motility to build fruiting bodies in a manner analogous to chemotaxis in Escherichia coli. However, in contrast to E. coli, M. xanthus cells move by gliding motility on a solid surface and direct their motility by controlling cell reversals. In M. xanthus, gliding motility involves extension and retraction of type IV pili (S-motility; refs. 4–6) and a hypothetical slime-extrusion mechanism (A-motility; refs. 7 and 8).
Chemotaxis in bacteria is controlled by specialized two-component signal transduction systems composed of receptors methyl-accepting chemotaxis proteins (MCPs) coupled by CheW to a CheA histidine kinase and a CheY response regulator (for reviews see refs. 9–13). Ligands interact with the chemoreceptor to affect the flow of phosphoryl groups from CheA to CheY. Phosphorylated CheY interacts with components of the flagellar motor, affecting the rotation of the motor. After stimulation, the chemoreceptors are modified by methylation, which attenuates the ligand-induced signaling event and thus brings about adaptation. The ability of cells to adapt to a particular stimulus over a broad concentration range (as much as five orders of magnitude) allows cells to find an optimal chemical environment. A cell's ability to respond to a large number of chemicals depends on the repertoire and specific affinities of the chemoreceptors.
M. xanthus is thought to respond to many chemical signals, including those generated by neighboring cells. Two chemosensory systems, frz and dif, have been described that control cell movements (14, 15) and have been implicated in responding to specific chemical stimuli (16, 17). We hypothesized that additional chemoreceptors might exist to mediate the complex movements of M. xanthus. In this article we report the identification of a third chemosensory system, the Che3 system, in M. xanthus. The Che3 system was not observed to regulate motility or chemotaxis but rather developmental timing and gene expression. Previous observations suggested that chemotaxis systems affect gene expression during swarmer-cell differentiation in Vibrio parahaemolyticus (18) and E. coli (19), pili expression in Synechocystis PCC6803 (20, 21), fibril biogenesis in M. xanthus (22), and virulence gene expression in Vibrio cholerae (23). However, in these previous studies, a direct link between the CheA histidine kinase and a regulator that affects gene expression was not established. In this study, we show that the Che3 system of M. xanthus is a chemotaxis-like signal transduction system that is directly involved in the regulation of developmental gene expression. We hypothesize that the Mcp3A and Mcp3B chemoreceptors regulate the activity of the CheA3 kinase thereby modulating the activity of a σ54-dependent transcriptional activator, CrdA (chemosensory regulator of development), which controls expression of many developmental genes.
Materials and Methods
Bacterial Strains, Plasmids, and Growth.
Bacterial strains and plasmids are listed in Table 1. M. xanthus strains were grown vegetatively at 32°C in casitone yeast extract (CYE; 1% casitone/0.5% yeast extract/4 mM MgSO4/10 mM Mops, pH 7.6). Development was assayed at 32°C on clone-fruiting medium [CF; 0.015% casitone/1 mM potassium phosphate/8 mM MgSO4/1.5 mM (NH4)2SO4/6.8 mM sodium citrate/9.1 mM sodium pyruvate/10 mM Mops, pH 7.6] (24).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Ref. |
|---|---|---|
| DZ2 | Wild type | 47 |
| DZ4502 | DZ2 ΔcrdB | This work |
| DZ4504 | DZ2 Δmcp3A | This work |
| DZ4506 | DZ2 Δmcp3B | This work |
| DZ4507 | DZ2 cheA3∷pJK404, KmR | This work |
| DZ4512 | DZ2 ΔcheBR3 | This work |
| DZ4513 | DZ2 crdA∷pJK412, KmR | This work |
| DZ4514 | DZ2 Δmcp3B crdA∷pJK412, KmR | This work |
| DZ4516 | DZ2 Tn5lacΩ4403, TcR | This work |
| DZ4517 | DZ2 Tn5lacΩ4411, TcR | This work |
| DZ4518 | DZ2 Tn5lacΩ4521, TcR | This work |
| DZ4519 | DZ2 Δmcp3A Tn5lacΩ4403, TcR | This work |
| DZ4520 | DZ2 Δmcp3A Tn5lacΩ4411, TcR | This work |
| DZ4521 | DZ2 Δmcp3A Tn5lacΩ4521, TcR | This work |
| DZ4522 | DK1217 mcp3A∷pJK402, KmR | This work |
| DZ4523 | DK1300 mcp3A∷pJK402, KmR | This work |
| DZ4524 | DK1217 mcp3B∷pJK403, KmR | This work |
| DZ4525 | DK1300 mcp3B∷pJK403, KmR | This work |
| DK1217 | aglB1 | 48 |
| DK1300 | sglG1 | 48 |
| DK6620 | DK1622 Tn5lacΩ4521, TcR | 49 |
| DK7826 | DK1622 Tn5lacΩ4411, TcR | 50 |
| DK7827 | DK1622 Tn5lacΩ4403, TcR | 51 |
| pKY481 | Used to create lacZ fusion, KmR | 29 |
| pBJ113 | Used to create insertion/deletions, GalK, KmR | 28 |
| pBJ114 | Used to create insertion/deletions, GalK, KmR | 28 |
| pJK402 | pKY481∷mcp3A (internal fragment), KmR | This work |
| pJK403 | pKY481∷mcp3B (internal fragment), KmR | This work |
| pJK404 | pKY481∷cheA3 (internal fragment), KmR | This work |
| pJK405 | pBJ113 with deletion construct for crdB, KmR | This work |
| pJK406 | pBJ113 with deletion construct for mcp3A, KmR | This work |
| pJK407 | pBJ113 with deletion construct for mcp3B, KmR | This work |
| pJK409 | pBJ113 with deletion construct for cheB3cheR3, KmR | This work |
| pJK412 | pBJ114∷crdA (internal fragment), KmR | This work |
DNA Manipulations and Sequence Analysis.
Failsafe (Epicentre Technologies, Madison, WI) DNA polymerase was used in all PCR reactions. Primers were obtained from Operon Technologies (Alameda, CA). Sequencing reactions were performed at the Division of Biological Sciences facility at the University of California, Davis, and analyzed with editview software. ORFs were identified by blast searches against the nonredundant database at the National Center for Biotechnology Information (NCBI).
Cosmid Library Screen.
A cosmid library containing 35-kb fragments of M. xanthus chromosomal DNA was constructed by Ron Gill (University of Colorado Health Sciences Center, Denver; ref. 25). The library was grown, transferred to Hybond-N membrane (Amersham Biosciences), treated, and probed using standard techniques for Southern analysis (26). A PCR probe was prepared with digoxigenin (DIG)-dNTPs (Roche Diagnostics), generating a fragment encompassing the highly conserved domain of frzCD encoding residues 192 to 378 of FrzCD. The membrane was then treated with α-DIG-horseradish peroxidase diluted 1:5,000 and detected by using Renaissance (NEN) chemiluminescence substrate. Fragments of interest were identified by Southern analysis using the same probe and detection method.
Construction of Mutants.
A method based on Wu and Kaiser (27) was used to create in-frame deletions in crdB, mcp3A, and mcp3B. Deletion constructs were generated by overlap extension PCR of two primary PCR products that correspond to 5′ and 3′ flanking regions of the target gene, respectively. The 5′ flanking region was ≈750 bp up to and including the seventh codon of the ORF. The 3′ flanking region was ≈750 bp downstream from and including the last 13 codons of the ORF. The constructs were subcloned into pBJ113; deletions were generated by counterselection on galactose (28). Plasmid constructs were verified by sequencing. Chromosomal insertions and deletions were verified by PCR and Southern analysis. The resulting construct encodes a deletion protein with the N-terminal 7 residues fused with the C-terminal 12 residues. The mutation in cheA3 was created by insertion of pJK404 carrying a 750-bp fragment internal to cheA subcloned into pKY481 (29). The mutations in mcp3A and mcp3B in the aglB1 and sglG1 strains were created by insertion of plasmids (pJK402 and pJK403, respectively) with a 750-bp fragment internal to the mcp subcloned into pKY481. The ΩlacZ reporter constructs were generated by electroporating chromosomal DNA from the donor strains and selecting for oxytetracycline resistance. Chromosomal DNA was isolated by using DNeasy Mini Kits (Qiagen, Valencia, CA). Electroporation was carried out on a Bio-Rad Gene Pulser at 0.65 kV, 400 Ω, and 25 μF in 0.2-cm cuvettes. Cells were plated in 0.7% agar on CYE plates containing kanamycin (100 μg/ml) or oxytetracycline (10 μg/ml).
Assay for Development, Sporulation, lacZ Expression, and Motility.
For the time course and density-dependent assays, development was induced by spotting the number of cells indicated in a volume of 10 μl onto CF agar plates and incubated for the times indicated at 32°C. Fruiting bodies were observed under a Zeiss stereoscope, photographed with an MTI (Michigan City, IN) charge-coupled device (CCD)-72 camera, and processed with nih image software and photoshop (Adobe Systems, San Jose, CA). For lacZ expression and sporulation assays, development was induced by plating 200 μl of cells at 1 × 109 cells per ml on CF agar plates and harvesting the entire plate at the times indicated. The culture was resuspended in water, disrupted by mild sonication, and assayed for β-galactosidase levels as described (30). For spore viability, the culture was resuspended in water, incubated at 55°C for 2 h, sonicated, and plated in 0.7% agar on CYE plates. A-motility and S-motility were assessed by analyzing swarm diameter on hard (1.5%) and soft (0.3%) agar surfaces, respectively (31). For reversal frequency and velocity, cells were grown in CYE, harvested during exponential phase, and plated at low density (≈107 cells per ml) on CF 1.5% agar surface (32). Individual cells were observed with a Nikon Labophot-2 microscope (×40 objective) and photographed every 10 sec for 30 min with an MTI CCD-72 camera. nih image software was used to quantify velocities, and reversals were counted over the 30-min period. At least 30 cells for each mutant were assayed.
Dot Blot for RNA.
The dot blot for mRNA was performed by using standard techniques (26). Total RNA was isolated with RNeasy Mini Kits (Qiagen). One microgram of total RNA was spotted for each sample in the assay shown. The vegetative samples were obtained from cells harvested during exponential growth (100 Klett units, red filter) in liquid CYE. The 24 h developmental sample was obtained by harvesting cells from CF plates 24 h after spotting. A DIG-labeled probe equivalent to the tps gene was generated by using PCR with DIG-dNTPs. The blot was incubated with α-DIG-horseradish peroxidase antibody diluted 1:5,000 and detected by chemiluminescence.
Immunoblot Analysis.
All immunoblot assays were performed with a Bio-Rad semidry transfer apparatus. Two micrograms of total protein was loaded for each sample analyzed. After electrophoresis, the gel was blotted onto poly(vinylidene difluoride) and incubated with α-Protein S or α-MbhA antisera diluted 1:5,000. Secondary horseradish-peroxidase-coupled antibody (Pierce) was diluted 1:10,000 and detected by chemiluminescence.
Yeast Two-Hybrid Assay.
Yeast strain PJ69-4A was obtained from Philip James (University of Wisconsin, Madison) and used to test interactions as previously described (33). Transformations were performed by using LiOAc. Both cheA3 and crdA were subcloned into the activation domain pGAD (LEU2 selectable marker) and the binding domain pGBDU (URA3 selectable marker) plasmids. Because the promoters contain only the minimal consensus sequence for interaction with the binding domain, false positives were minimized and positive interactions were discernable by growth on media lacking both adenine and histidine (33). The positive interactions were confirmed with liquid β-galactosidase assays. A positive control was tested with pGAD∷sipL and pGBD∷snf1 (data not shown), and negative interactions are shown in Fig. 4.
Figure 4.
Yeast-two-hybrid assay for CheA3 and CrdA interaction. The assay was performed as described in Materials and Methods. The full-length cheA3 gene and a truncated version (cheA3s) were subcloned into both the activation domain pGAD and the binding domain pGBDU plasmids. Yeast cells (PJ69-4A) were transformed with the plasmids indicated and assayed for growth in the absence of histidine and/or adenine. Only those transformants that contained both cheA3 and crdA were capable of growing (++) in the absence of both adenine and histidine. nd, not determined.
Results
To identify new chemoreceptor genes, we probed a cosmid library containing M. xanthus chromosomal DNA (25) with probes containing the highly conserved domains from the receptor genes frzCD and difA. By using these probes, we identified two previously uncharacterized genes encoding chemoreceptor homologs, designated Mcp3A and Mcp3B. We subcloned and sequenced a 20-kb fragment of DNA and discovered that these genes are located within a cluster of chemotaxis-like genes encoding homologs to CheW, a hybrid CheA, CheB, and CheR (Fig. 1). No gene encoding a CheY homolog was identified within the 35-kb cosmid clone containing these genes.
Figure 1.
Genetic organization of the che3 cluster. crdA (yellow) encodes a σ54-dependent transcriptional activator. crdB (red) encodes a hybrid lipoprotein receptor/peptidoglycan binding protein. Green arrows represent ORFs encoding proteins with predicted transmembrane domains (crdC, mcp3A, and mcp3B). ORFs encoding cytoplasmic chemotaxis protein homologs (cheW3, cheA3, cheB3, and cheR3) are blue. The white arrows encode a nifS homolog, an ORF with no similarity to any sequence in the database, and a hypothetical protein (left to right, respectively). The transcription start site (black arrow) was identified by using primer extension, and a putative terminator was identified by using GCG StemLoop. RT-PCR confirmed that crdB-cheA3 comprise a single transcript. crdC, mcp3A, mcp3B, and cheA3 appear to be translationally coupled.
To analyze the potential roles of these genes, we constructed an insertion mutation in cheA3, and in-frame deletion mutations in crdB, mcp3A, and mcp3B. Surprisingly, none of these mutations produced any observable defects in motility. The mcp3A and mcp3B mutations also were tested in the A+S− and A−S+ mutant backgrounds to identify any motility defects not discernable in the wild-type background and none were observed. Furthermore, the crdB, mcp3A, mcp3B, and cheA3 mutants each displayed normal gliding velocities (4 μm/min on 1.5% agar) and reversal frequencies (every 8 min) (34, 35). None of these mutants showed any observable growth defects. These results suggest that CrdB, Mcp3A, Mcp3B, and CheA3 do not directly control motility under vegetative conditions.
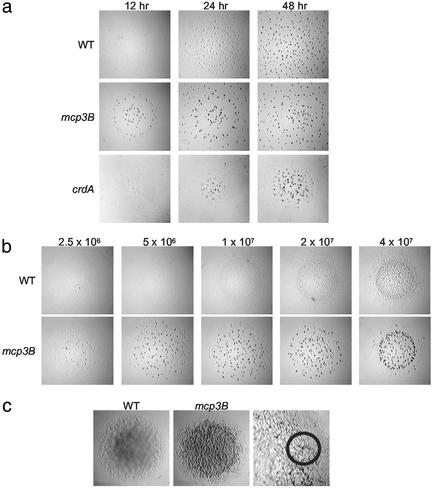
Analysis of these mutants under developmental conditions, however, showed that these strains formed fruiting bodies much earlier and at lower cell densities than the wild type (Fig. 2). Wild-type cells formed fruiting bodies after 48 h of starvation and required a high cell density (about 2 × 109 cells per ml). The crdB, mcp3A, mcp3B, and cheA3 mutants formed fruiting bodies as early as 12 h (Fig. 2a) even at low initial cell densities (Fig. 2b), as low as 2.5 × 107 cells per ml (mcp3B is shown in Fig. 2). However, early fruiting resulted in defective sporulation, as the number of viable spores formed by these mutants was <0.1% that of the wild type. Surprisingly, these mutants showed some characteristics of development on rich media. Cells growing vegetatively on standard nutrient-rich agar plates displayed the rippling phenomenon associated with development (refs. 36 and 37; Fig. 2c) and formed fruiting-body-like aggregates containing some spores (about 200 spores from the area indicated; Fig. 2c). Together, these results indicate that the Che3 system is involved in regulation of entry into development. We therefore hypothesize that this chemosensory pathway controls developmental gene expression.
Figure 2.
Effect of the che3 mutations on development. The assays were performed as described in Materials and Methods. (a) Effect of the mcp3B and crdA mutations on the timing of aggregation. Cells were plated at an initial concentration of 2 × 109 cells per ml. Each black dot is an aggregate or fruiting body containing ≈105 to 106 cells. Note that aggregates remain translucent before sporulation [wild type (WT) at 24 h and mcp3B at 12 h]. (b) Effect of the mcp3B mutation on density-dependent aggregation. The plates were photographed at 24 h. Wild-type aggregates remain translucent (Upper Right), indicating that refractile spores have not yet formed. (c) Effect of the mcp3B mutation on cells growing on rich medium. The colonies were ≈2 cm in diameter when photographed. Right shows higher magnification (×5) of the mcp3B mutant. Small semitranslucent mounds (circle) were observed after 3 days in the outer edge of the growing mcp3B colony; none were observed in the wild-type colony. The corresponding area for both colonies was harvested and analyzed for heat- and sonication-resistant spores.
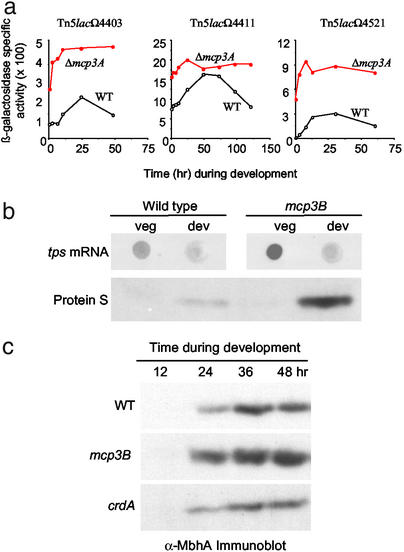
To test this hypothesis, we analyzed the expression patterns of several previously characterized reporter genes, Tn5lacΩ4403, Tn5lacΩ4411, and Tn5lacΩ4521, expressed at 12, 5, and 2.5 h during development, respectively (30). These reporter fusions were analyzed in a strain containing the mcp3A deletion and in the wild-type strain (Fig. 3a). In each case, the timing and level of the reporter gene expression occurred much earlier and at higher levels in the Δmcp3A mutant relative to that in the wild type. In fact, all three reporters were expressed vegetatively in the Δmcp3A mutant background as indicated by the level of expression at 0 h in development.
Figure 3.
Effect of mcp mutations on developmentally regulated genes. Assays were performed as described in Materials and Methods. (a) Effect of the mcp3A mutation on lacZ expression from Tn5lac developmental reporters Ω4403, Ω4411, and Ω4521. Shown are the expression patterns in the DZ2 (wild type) background for Ω4403, Ω4411, and Ω4521 strains (open circles) and the Δmcp3AΩ4403, Δmcp3AΩ4411, and Δmcp3AΩ4521 strains (filled circles). (b) Effect of the mcp3B mutation on the expression of Protein S. Shown is a dot-blot assay for tps mRNA (Upper) and immunoblot assay for Protein S production (Lower). (c) Effect of the mcp3B and crdA mutations on the expression of myxobacterial hemagglutinin. Shown is an immunoblot assay performed with anti-MbhA antibody diluted 1:5,000.
We assayed the expression level of other developmentally controlled genes and found them to be altered as well. For example, Protein S (encoded by tps) is a major spore coat protein produced primarily in aggregating cells (38, 39). Dot-blot analysis showed that tps mRNA was overexpressed in the mcp3B mutant relative to that of the wild type, and immunoblot analysis showed that the corresponding protein levels also were greatly enhanced in the mcp3B mutant (Fig. 3b). Another developmentally regulated gene is mbhA, which encodes myxobacterial hemagglutinin (MbhA). Expression of mbhA is normally localized to the peripheral rods (40), and peak expression occurs 24 h after starvation (41). Again, the mcp3B mutant displayed a dramatic increase in production of MbhA relative to that seen in the wild type (Fig. 3c). The timing of MbhA production in the mcp3B mutant was similar to that of the wild type, even though the level of production was greatly enhanced. Elevated MbhA production also was observed for the crdB, mcp3A, and cheA3 mutants (data not shown). Thus, gene expression is significantly altered in the che3 mutants.
Direct control of gene expression by the Che3 system would require a factor capable of modulating RNA polymerase activity. We identified such a gene encoding a response regulator designated crdA immediately upstream and divergently transcribed from the promoter for the che3 genes (Fig. 1). Sequence analysis shows that CrdA belongs to the NtrC family of transcriptional activators that control expression from σ54-dependent promoters (42, 43). CrdA showed the greatest similarity over its full length to AtoC from E. coli (E = 8e-96). It possesses a AAA ATPase domain and a helix–turn–helix DNA binding domain at its C terminus, hallmark features of this class of transcriptional activators.
To test the possibility that CheA3 might be the cognate kinase for the CrdA response regulator, CheA3 and CrdA were assayed in the yeast two-hybrid system for their ability to interact (Fig. 4). Because CheA3 is a hybrid kinase possessing a receiver domain at its C terminus, both the full-length CheA3 and a truncated version lacking the receiver domain, CheA3s, were subcloned into the binding domain and activation domain plasmids for the yeast two-hybrid analysis. Growth in the absence of histidine and adenine allowed for clear identification of positive interactions, because the strain used (PJ69-4A) has different promoters driving expression from the histidine and adenine biosynthesis genes (33). When CrdA or CheA3 were present alone, no growth occurred. However, when both CrdA and CheA3 (or CheA3s) were present together, growth occurred at apparent wild-type rates, indicating that CheA3 and CrdA strongly interact. The ability for these proteins to interact suggests that CheA3 is the cognate kinase for CrdA.
To assess the role of CrdA in development, an insertion mutation disrupting the N-terminal receiver domain of crdA was prepared and analyzed for its ability to form fruiting bodies after starvation (Fig. 2a). The crdA mutant was delayed by 12 to 24 h in development; in contrast the crdB, mcp3A, mcp3B, and cheA3 mutants were 12 to 24 h early relative to the wild type. In addition, the crdA mutant showed a lag and overall lower level of production of MbhA relative to the wild type (Fig. 3c). Thus, the phenotype of the crdA mutant suggests that CrdA acts as a transcriptional activator. It is worth noting that by 72 h the crdA mutant produced as many fruiting bodies and viable spores as the wild type.
Because the developmental phenotype of the crdA mutant is the opposite of the che3 mutants in many respects, we hypothesize that CheA3 inhibits CrdA activation. If this model is correct, then a mcp3BcrdA double mutant should display a crdA phenotype. The mcp3BcrdA double mutant was constructed and found to be delayed in fruiting by 12 to 24 h (data not shown), indicating that crdA is epistatic to mcp3B. This result supports the hypothesis that the Che3 system directly regulates CrdA, which in turn regulates developmental gene expression.
Mcp3A and Mcp3B are chemoreceptor homologs and their ability to be methylated should affect signaling. To test this, we made an in-frame deletion in the methylation system genes, cheB3 and cheR3, resulting in a ΔcheBR3 double mutant. This mutant had the same phenotype as the Δmcp3A and Δmcp3B mutants (data not shown). These results show the importance of the methylation system for signaling through Mcp3A and Mcp3B and indicate that the Che3 system functions similarly to other known chemotaxis systems.
Discussion
In this study, we show that the Che3 chemosensory system of M. xanthus seems to be dedicated to the regulation of gene expression. In this respect, the Che3 system is most similar to the vast majority of known two-component systems, which regulate transcription of target genes in response to environmental stimuli. However, it is very different from most previously described chemotaxis systems, which are two-component systems that regulate motility. None of the mutations within the che3 gene cluster showed any defect in motility. Mutations in mcp3A, mcp3B, and cheA3, as well as a ΔcheBR3 deletion, resulted in density-independent, premature entry into development, and showed obvious defects in regulation of developmentally controlled genes including mbhA, tps, and the Tn5lac fusions Ω4403, Ω4411, and Ω4521. These were surprising findings because Mcp3A, Mcp3B, CheA3, CheB, and CheR show the greatest similarity to homologs known to control chemotaxis, but not gene expression, in other organisms.
The identification of CrdA, a member of the σ54-dependent family of DNA binding proteins, and its ability to interact with CheA3, directly links the central chemotaxis components to gene expression. Based on the data, we propose a model where all components of the Che3 chemosensory system regulate the activity of CrdA (Fig. 5). Because mutants in the Che3 system showed increased and premature expression of developmentally regulated genes, the Che3 signal transduction system must be inhibiting the expression of these genes during vegetative growth. Additionally, CheA3 was found to interact strongly with CrdA, a putative NtrC-like transcriptional regulator. Because the crdA mutant is delayed in development and crdA is epistatic to mcp3B, CrdA is most likely the primary response regulator for CheA3. In addition, mutations in the che3 system affect a large array of developmentally regulated genes, suggesting that CrdA controls the expression of a global regulator. Alternatively, the Che3 system may feed into a more elaborate phosphorelay system such as that which controls osmoregulation in Saccharomyces cerevisiae (44) or sporulation in Bacillus subtilis (45). The receiver domain of the hybrid kinase CheA3 may play a role in such a relay.
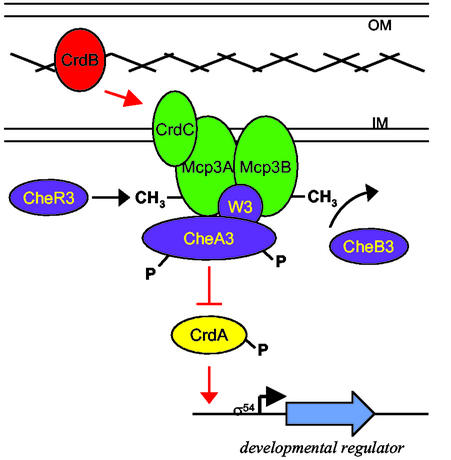
Figure 5.
Model for chemosensory regulation of gene expression. The model is described in Discussion. Proteins predicted to be integral membrane proteins are green, cytoplasmic proteins are blue, and the DNA binding response regulator (CrdA) is yellow. The cross hatch represents the peptidoglycan layer. The light blue arrow indicates a putative target gene whose expression is controlled by the Che3 system. CrdB (red) is depicted as bridging the peptidoglycan layer because the C terminus shows strong similarity to a domain within OprF known to interact with peptidoglycan (46); the N terminus of CrdB shows great similarity to a lipoprotein receptor. CrdC is depicted as an inner membrane protein because it has a putative transmembrane domain in its C terminus; CrdC is shown as part of the complex with the receptors because the genes for crdC, mcp3A, mcp3B, and cheA3 are translationally coupled. A signal, possibly a lipoprotein, may induce a conformational change in CrdB, which is transmitted to the CrdC–chemoreceptor complex. Subsequently, the receptor complex would alter CheA3 autophosphorylation levels in a manner similar to that observed during chemotaxis signal transduction (13). The overall level of CrdA–P would regulate expression of target genes involved in development in M. xanthus.
Why might a regulator of gene expression be coupled to a chemotaxis-like signal transduction pathway? Chemosensory systems may be particularly well suited for the control of developmental gene expression because of their capacity for adaptation. Adaptation is achieved by altering the level of methylation of the chemoreceptors by the CheR methyltransferase and CheB methylesterase (13). Because the M. xanthus ΔcheBR3 double mutant displays premature development, it seems that the ability to adapt is necessary for cells to change their pattern of gene expression in response to the complex set of intercellular signals during development.
Other microorganisms may use chemotaxis pathways to control gene expression. Several chemosensory systems have been implicated in the regulation of gene expression, including swarmer-cell differentiation in V. parahaemolyticus (18) and E. coli (19), pili expression in Synechocystis (20, 21), fibril biogenesis in M. xanthus (22), and virulence gene expression in V. cholerae (23). However, in each of those cases it is not clear whether the effect on transcription is direct or indirect. A recently discovered chemotaxis-like operon from Rhodospirillum centenum suggests a parallel regulatory circuit to that described in this article. Mutations within the R. centenum che3 operon lead to premature cyst formation, and that operon possesses a response regulator with a DNA binding domain (J. Berleman and C. Bauer, personal communication). Together, these observations suggest a more general role for chemotaxis-like pathways in the control of gene expression in bacteria.
Acknowledgments
We thank Hera Vlamakis for her work on the screen of the cosmid library, Philip James for providing the two-hybrid system, Ron Gill for the cosmid library, Dale Kaiser for strains, and Bryan Julien for plasmids, Bruce Birren for providing the sequence of the entire cosmid containing the che3 cluster, and the members of the Zusman lab for valuable discussions. This research was supported by grants from the National Institutes of Health to D.R.Z. (GM20509) and a postdoctoral fellowship to J.R.K. (GM19676).
Abbreviations
- MCP
methyl-accepting chemotaxis protein
- DIG
digoxigenin
- CYE
casitone yeast extract
- CF
clone-fruiting medium
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF448145).
References
- 1.Shimkets L J. Annu Rev Microbiol. 1999;53:525–549. doi: 10.1146/annurev.micro.53.1.525. [DOI] [PubMed] [Google Scholar]
- 2.O'Connor K A, Zusman D R. J Bacteriol. 1991;173:3342–3355. doi: 10.1128/jb.173.11.3342-3355.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim S K, Kaiser D, Kuspa A. Annu Rev Microbiol. 1992;46:117–139. doi: 10.1146/annurev.mi.46.100192.001001. [DOI] [PubMed] [Google Scholar]
- 4.Wall D, Kaiser D. Mol Microbiol. 1999;32:1–10. doi: 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- 5.Sun H, Zusman D R, Shi W. Curr Biol. 2000;10:1143–1146. doi: 10.1016/s0960-9822(00)00705-3. [DOI] [PubMed] [Google Scholar]
- 6.Skerker J M, Berg H C. Proc Natl Acad Sci USA. 2001;98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodgkin J, Kaiser D. Mol Gen Genet. 1979;171:167–176. [Google Scholar]
- 8.Wolgemuth C, Hoiczyk E, Kaiser D, Oster G. Curr Biol. 2002;12:369–377. doi: 10.1016/s0960-9822(02)00716-9. [DOI] [PubMed] [Google Scholar]
- 9.Bourret R B, Borkovich K A, Simon M I. Annu Rev Biochem. 1991;60:401–441. doi: 10.1146/annurev.bi.60.070191.002153. [DOI] [PubMed] [Google Scholar]
- 10.Parkinson J S, Kofoid E C. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 11.Zhulin I B. Adv Microb Physiol. 2001;45:157–198. doi: 10.1016/s0065-2911(01)45004-1. [DOI] [PubMed] [Google Scholar]
- 12.Armitage J P. Adv Microb Physiol. 1999;41:229–289. doi: 10.1016/s0065-2911(08)60168-x. [DOI] [PubMed] [Google Scholar]
- 13.Stock J B, Surette M G. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Ingraham J L, editors. Vol. 2. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1103–1129. [Google Scholar]
- 14.Yang Z, Geng Y, Xu D, Kaplan H B, Shi W. Mol Microbiol. 1998;30:1123–1130. doi: 10.1046/j.1365-2958.1998.01160.x. [DOI] [PubMed] [Google Scholar]
- 15.McBride M J, Weinberg R A, Zusman D R. Proc Natl Acad Sci USA. 1989;86:424–428. doi: 10.1073/pnas.86.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kearns D B, Shimkets L J. Proc Natl Acad Sci USA. 1998;95:11957–11962. doi: 10.1073/pnas.95.20.11957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sogaard-Andersen L, Slack F J, Kimsey H, Kaiser D. Genes Dev. 1996;10:740–754. doi: 10.1101/gad.10.6.740. [DOI] [PubMed] [Google Scholar]
- 18.McCarter L, Hilmen M, Silverman M. Cell. 1988;54:345–351. doi: 10.1016/0092-8674(88)90197-3. [DOI] [PubMed] [Google Scholar]
- 19.Burkart M, Toguchi A, Harshey R M. Proc Natl Acad Sci USA. 1998;95:2568–2573. doi: 10.1073/pnas.95.5.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung Y H, Cho M S, Moon Y J, Choi J S, Yoo Y C, Park Y I, Lee K M, Kang K W, Park Y M. FEBS Lett. 2001;492:33–38. doi: 10.1016/s0014-5793(01)02227-x. [DOI] [PubMed] [Google Scholar]
- 21.Bhaya D, Takahashi A, Grossman A R. Proc Natl Acad Sci USA. 2001;98:7540–7545. doi: 10.1073/pnas.131201098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Z, Ma X, Tong L, Kaplan H B, Shimkets L J, Shi W. J Bacteriol. 2000;182:5793–5798. doi: 10.1128/jb.182.20.5793-5798.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S H, Butler S M, Camilli A. Proc Natl Acad Sci USA. 2001;98:6889–6894. doi: 10.1073/pnas.111581598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bretscher A P, Kaiser D. J Bacteriol. 1978;133:763–768. doi: 10.1128/jb.133.2.763-768.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hager E, Tse H, Gill R E. Mol Microbiol. 2001;39:765–780. doi: 10.1046/j.1365-2958.2001.02266.x. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 27.Wu S S, Kaiser D. J Bacteriol. 1996;178:5817–5821. doi: 10.1128/jb.178.19.5817-5821.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Julien B, Kaiser A D, Garza A. Proc Natl Acad Sci USA. 2000;97:9098–9103. doi: 10.1073/pnas.97.16.9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho K, Zusman D R. Mol Microbiol. 1999;34:268–281. doi: 10.1046/j.1365-2958.1999.01594.x. [DOI] [PubMed] [Google Scholar]
- 30.Kroos L, Kuspa A, Kaiser D. Dev Biol. 1986;117:252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- 31.Shi W, Zusman D R. Proc Natl Acad Sci USA. 1993;90:3378–3382. doi: 10.1073/pnas.90.8.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trudeau K G, Ward M J, Zusman D R. Mol Microbiol. 1996;20:645–655. doi: 10.1046/j.1365-2958.1996.5521075.x. [DOI] [PubMed] [Google Scholar]
- 33.James P, Halladay J, Craig E A. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spormann A M, Kaiser A D. J Bacteriol. 1995;177:5846–5852. doi: 10.1128/jb.177.20.5846-5852.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blackhart B D, Zusman D R. Proc Natl Acad Sci USA. 1985;82:8767–8770. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimkets L J, Kaiser D. J Bacteriol. 1982;152:451–461. doi: 10.1128/jb.152.1.451-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welch R, Kaiser D. Proc Natl Acad Sci USA. 2001;98:14907–14912. doi: 10.1073/pnas.261574598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inouye M, Inouye S, Zusman D R. Proc Natl Acad Sci USA. 1979;76:209–213. doi: 10.1073/pnas.76.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Downard J S, Zusman D R. J Bacteriol. 1985;161:1146–1155. doi: 10.1128/jb.161.3.1146-1155.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Connor K A, Zusman D R. J Bacteriol. 1991;173:3318–3333. doi: 10.1128/jb.173.11.3318-3333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cumsky M, Zusman D R. Proc Natl Acad Sci USA. 1979;76:5505–5509. doi: 10.1073/pnas.76.11.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barrios H, Valderrama B, Morett E. Nucleic Acids Res. 1999;27:4305–4313. doi: 10.1093/nar/27.22.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rombel I, North A, Hwang I, Wyman C, Kustu S. Cold Spring Harbor Symp Quant Biol. 1998;63:157–166. doi: 10.1101/sqb.1998.63.157. [DOI] [PubMed] [Google Scholar]
- 44.Posas F, Wurgler-Murphy S M, Maeda T, Witten E A, Thai T C, Saito H. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 45.Hoch J A. Curr Opin Microbiol. 1998;1:170–174. doi: 10.1016/s1369-5274(98)80007-6. [DOI] [PubMed] [Google Scholar]
- 46.De Mot R, Vanderleyden J. Mol Microbiol. 1994;12:333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 47.Campos J M, Zusman D R. Proc Natl Acad Sci USA. 1975;72:518–522. doi: 10.1073/pnas.72.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hodgkin J, Kaiser A D. Mol Gen Genet. 1979;171:177–191. [Google Scholar]
- 49.Kaplan H B, Kuspa A, Kaiser D. J Bacteriol. 1991;173:1460–1470. doi: 10.1128/jb.173.4.1460-1470.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gorski L, Kaiser D. J Bacteriol. 1998;180:5896–5905. doi: 10.1128/jb.180.22.5896-5905.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gorski L, Gronewold T, Kaiser D. J Bacteriol. 2000;182:2438–2444. doi: 10.1128/jb.182.9.2438-2444.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]