Abstract
The minimal, active core of human telomerase is postulated to contain two components, the telomerase RNA hTER and the telomerase reverse transcriptase hTERT. The reconstitution of human telomerase activity in vitro has facilitated the identification of sequences within the telomerase RNA and the RT motifs of hTERT that are essential for telomerase activity. However, the precise role of residues outside the RT domain of hTERT is unknown. Here we have delineated several regions within hTERT that are important for telomerase catalysis, primer use, and interaction with the telomerase RNA and the telomerase-associated protein TEP1. In particular, certain deletions of the amino and carboxy terminus of hTERT that retained an interaction with telomerase RNA and TEP1 were nonetheless completely inactive in vitro and in vivo. Furthermore, hTERT truncations lacking the amino terminus that were competent to bind the telomerase RNA were severely compromised for the ability to elongate telomeric and nontelomeric primers. These results suggest that the interaction of telomerase RNA with hTERT can be functionally uncoupled from polymerization, and that there are regions outside the RT domain of hTERT that are critical for telomerase activity and primer use. These results establish that the human telomerase RT possesses unique polymerization determinants that distinguish it from other RTs.
INTRODUCTION
Telomeres are the specialized nucleoprotein complexes at the physical ends of eukaryotic chromosomes (Greider and Blackburn, 1996). Telomerase is a ribonucleoprotein (RNP) enzyme that uses an internal RNA template to specifically direct telomere synthesis (Greider and Blackburn, 1985, 1987). Telomerase plays an essential role in the dynamic process of telomere length regulation in vivo by restoring telomeric sequences that are lost during semiconservative DNA replication (Watson, 1972; Olovnikov, 1973). Telomerase activity has been purified from a number of different organisms, including mammals (Greider, 1996). The telomerase RNA component (TER, telomerase RNA) has been cloned from many different species (Greider, 1996), and the secondary structure of the ciliate and mammalian telomerase RNAs is highly conserved (Romero and Blackburn, 1991; Lingner et al., 1994; McCormick-Graham and Romero, 1995; Chen et al., 2000).
The first mammalian telomerase proteins (TEPs) were identified based on homology with previously cloned telomerase components from ciliates and yeast. TEP1 is a Tetrahymena p80 homolog that binds TER and is associated with telomerase activity in cell extracts (Collins et al., 1995; Harrington et al., 1997a; Nakayama et al., 1997). The mammalian telomerase reverse transcriptase (TERT) shares amino acid sequence similarity with the catalytic telomerase subunit previously identified in ciliates and yeast (Harrington et al., 1997b; Kilian et al., 1997; Lingner et al., 1997; Meyerson et al., 1997; Nakamura et al., 1997). Human TERT is a limiting component for telomerase activity: the hTERT mRNA is often up-regulated in cells containing telomerase activity, and the introduction of hTERT confers telomerase activity to primary human cells (Meyerson et al., 1997; Weinrich et al., 1997; Bodnar et al., 1998; Counter et al., 1998; Nakayama et al., 1998; Vaziri and Benchimol, 1998).
The first telomerase reconstitution assay was accomplished in Tetrahymena, where the functional requirements for the telomerase RNA were delineated by the addition of recombinant telomerase RNA to micrococcal nuclease-treated extracts (Autexier and Greider, 1994; Gilley et al., 1995; Gilley and Blackburn, 1996; Autexier and Greider, 1998; Autexier and Triki, 1999). With a similar assay to study human telomerase, all 5′ nucleotides leading up to the telomerase RNA template, or the last 240 nucleotides of the telomerase RNA are dispensable for telomerase activity (Autexier et al., 1996) (Figure 1B). More recently, a human telomerase reconstitution assay has been developed in rabbit reticulocyte lysates (RRL) that requires only the addition of recombinant hTERT and the telomerase RNA (Weinrich et al., 1997; Beattie et al., 1998). In this assay, the telomerase RNA requirements for catalysis in the presence of recombinant hTERT are similar to the hTER requirements in the nuclease-based reconstitution assay (Autexier et al., 1996; Beattie et al., 1998). For example, nucleotides +10 to +159 of the hTER, which contains the telomeric template, are sufficient to reconstitute weak telomerase activity (Beattie et al., 1998). Two nonoverlapping pieces of the telomerase RNA (constructs +33 to +147 and +164 to +325) are also active when combined with hTERT in a similar rabbit reticulocyte reconstitution assay (Tesmer et al., 1999).
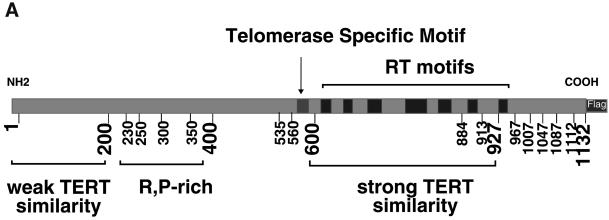
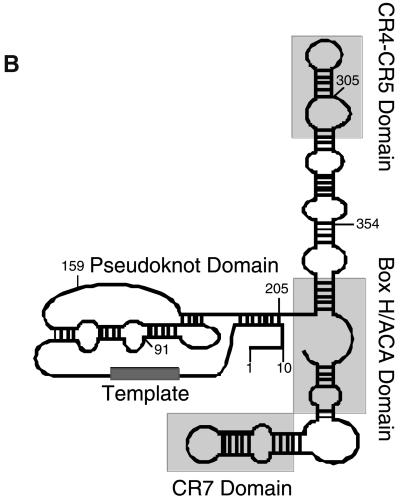
Figure 1.
hTERT and hTER. Schematic of the human telomerase RT and telomerase RNA. (A) The first 200 amino acids contain very weak sequence similarity to the TERT proteins from other species, and therefore we were interested in elucidating the importance of the N terminus. The region between amino acids 200 and 400 contains high arginine and proline content, which is thought to be important in protein–protein interactions. The region between amino acids 536 and 600 has a weak homology to cyclin E. Amino acid 560 begins the region of highest similarity to other TERT family members, and includes the telomerase specific motif. The region of highest similarity to other RTs begins at amino 601. C-terminal truncations of hTERT included a truncation at amino acid 884, which deletes RT motif 6 (GVPEYGCVVNLRKTVV) of hTERT, a truncation at amino acid 913, and a truncation at amino acid 927, which removes motif 7 (hLGxxh), and ends at the highly conserved RT motif. Other C-terminal truncations were made at increments of ∼50 amino acids from 927 to the C terminus of the protein. Each of these specific regions was deleted to ascertain its role in the telomerase complex with regard to activity, hTER binding, and interaction with TEP1. (B) Secondary structure of the human telomerase RNA adapted from Chen et al. (2000). The template region of hTER falls between nucleotides 44 and 52. Highly conserved motifs such as the pseudoknot domain; conserved regions (CR) 4, 5, and 7; and the H/ACA box are indicated in gray. Specific 5′ and 3′ truncations were made at the positions indicated to ascertain the roles of specific RNA sequences and structures in the reconstitution of human telomerase activity.
The determinants within the telomerase RNA that are important for catalysis have been extensively characterized in ciliates and yeast. Tetrahymena TERT and the telomerase RNA are sufficient to reconstitute telomerase activity in RRL; however, there are additional sequences within the telomerase RNA that are required for activity in RRL that are dispensable within the native Tetrahymena telomerase enzyme (Autexier and Greider, 1998; Autexier and Triki, 1999; Licht and Collins, 1999). With an RNP gel shift assay, the highly conserved pseudoknot in the telomerase RNA is important for tTERT binding but is not required for association with the Tetrahymena telomerase-associated proteins p80 or p95 (Autexier and Greider, 1994, 1995; Gilley and Blackburn, 1999; Licht and Collins, 1999). In the yeasts Saccharomyces cerevisiae and Kluyveromyces lactis, certain mutations outside the telomerase RNA template domain are critical for telomerase RNP assembly, polymerization, and telomere length maintenance in vivo (McEachern and Blackburn, 1995; Bhattacharyya and Blackburn, 1997; Prescott and Blackburn, 1997b; Roy et al., 1998). These studies illustrate a critical role for the telomerase RNA distinct from its templating function in telomerase catalysis in humans, ciliates, and yeast.
Much less is known about the role of specific domains within the TERT in telomerase function in vitro and in vivo. Point mutations within the conserved RT domain of Est2p and hTERT abolish telomerase activity, which demonstrates that the RT domain is essential for telomerase function (Counter et al., 1997; Harrington et al., 1997b; Lingner et al., 1997; Weinrich et al., 1997; Beattie et al., 1998; Nakayama et al., 1998). In addition, residues within the N terminus of Est2p are required to bypass senescence of est2Δ cells in S. cerevisiae (Friedman and Cech, 1999). For the majority of the N-terminal Est2p mutations, the loss of telomere length maintenance can be explained by a reduction in telomerase RNA binding (Friedman and Cech, 1999). However, one mutant appears to be defective in recruitment to the telomere because telomere shortening occurs despite robust telomerase activity and telomerase RNA binding (Friedman and Cech, 1999).
The minimal requirements for hTERT that are necessary for telomerase activity and telomerase RNA binding have not yet been examined. Here we demonstrate that certain deletion mutations of hTERT that contain the entire RT domain are completely inactive in vivo and in vitro. These truncated proteins do not appear to be grossly misfolded because they are competent for binding to both telomerase RNA and TEP1. We have shown that the amino terminus of hTERT is also important for the ability to extend telomeric and nontelomeric primers. These findings suggest that there are regions within the telomerase RT that are dispensable for telomerase RNA binding but are essential for primer use and polymerization in vivo.
MATERIALS AND METHODS
Cloning of hTER and hTERT
The full-length human telomerase RNA, spanning nucleotides 1 to 451 (Feng et al., 1995) was cloned as described in Bryan et al. (1997). DNA fragments containing hTER sequences were obtained by amplification of HeLa genomic DNA by using the polymerase chain reaction (PCR). A subsequent PCR amplification was performed with a 5′ primer containing the T7 polymerase promoter sequence. This PCR product was cloned into pUC19 for the template for in vitro transcription (T7-hTER-pUC). The full-length hTERT cDNA plasmid was described previously (Harrington et al., 1997b; Beattie et al., 1998). With this plasmid, N-terminal and C-terminal derivatives of hTERT were generated and replaced by using the appropriate PCR primers, with the sequence coding Flag epitope (DYKDDDDK) at the end of all 3′ primers (for specific hTERT derivatives see figure legends).
In Vitro Transcription
All transcription experiments were carried out at the Ontario Cancer Institute similar to methods previously described (Beattie et al., 1998). hTER plasmid DNA (T7-hTER-pUC) was linearized by digestion with BssHII (1/10–91), XbaI (1/10–159), SmaI (1/10–205), EarI (1/10–305) StuI (1–354), or EcoRI (1–451) to obtain template for either full-length or the respective hTER truncation. T7 transcription reactions (200 μl) contained 10 μg of linearized template DNA, 40 mM Tris-HCl, pH 8.0, 2 mM spermidine, 10 mM dithiothreitol, 1 mM each NTP, 350 U RNA guard, 100 U T7 RNA polymerase, and 6 mM MgCl2. After 2 h of incubation at 37°C, the template was inactivated by adding 200 U DNase I and incubated for 20 min at 37°C. The reactions were extracted with phenol and chloroform: isoamyl alcohol (24:1), ethanol precipitated, and resuspended in water. RNAs were purified from 8 M urea/4% wt/vol polyacrylamide (19:1 acrylamide:bis) by elution in water, filtered through a Supor membrane 0.8 μm/0.2-μm filter (Gelman Sciences, Ann Arbor, MI), precipitated in ethanol, and dissolved in water.
In Vitro Reconstitution
Rabbit reticulocyte T7-coupled transcription/translation reactions were performed as per the manufacturers' instructions (Promega, Madison, WI). Full-length or different hTERT truncations containing a flag-epitope at the C terminus were synthesized in RRL in the presence of either 0.01 μg/μl full-length hTER or 0.1 μg/μl hTER truncations. Each of the hTERT cDNAs was added to the reticulocyte lysates at a concentration of 0.01 μg/μl and incubated at 30°C for 90 min.
Cell Lines and Transfections
hTERT and TEP1 constructs cloned into PCR3 were transfected into human embryonic kidney 293T cells with either Lipofectamine (Life Technologies, Gaithersburg, MD) or Superfect (Qiagen, Chatsworth, CA) as per the manufacturers' instructions.
Immunoprecipitation
Twenty-five microliters of reticulocyte lysates or 200 μg of 293T-transfected cell extract was immunoprecipitated with 15 μl of M2 affinity resin from Sigma (St. Louis, MO) in 0.5% 3-([3-cholamidopropyl]dimethylammino)-2-hydroxy-1-propanesulfonate, 10 mM Tris-HCl, pH 7.5, 1 mM MgCl2, 1 M NaCl, 5 mM β-mercaptoethanol, and 10% glycerol. Two microliters of beads was analyzed for telomerase activity by the telomere repeat amplification protocol (TRAP) assay, and 10 μl of beads was examined by Western blot analysis. For reticulocyte lysate assays used for Northern analysis, 100 μl of lysate was immunoprecipitated with 60 μl of M2 affinity resin (anti-flag beads; Sigma).
Western Analysis
After immunoprecipitation, beads were resuspended in SDS-PAGE–loading dye, the samples were heated for 5 min at 100°C, and electrophoresed on a 4–12% wt/vol Tris glycine SDS page gel. The gel was transferred to polyvinylidene difluoride membrane in 39 mM glycine, 48 mM Tris base, and 20% vol/vol methanol. After the electrotransfer, the membrane was treated with methanol, blocked in 5% wt/vol milk, and probed with 0.2 μg/ml anti-TERT peptide antisera (Harrington et al., 1997b) to detect hTERT, or 0.5 μg/ml anti-myc monoclonal antibody (PharMingen, San Diego, CA) to detect TEP1.
Northern Analysis
Twenty microliters of beads from the anti-flag immunoprecpitations of hTERT–hTER complexes synthesized in RRL was extracted once with phenol, once with phenol:chloroform/isoamyl alcohol (25:24:1), and precipitated with 0.1 volumes 3 M sodium acetate and 2.5 volumes of ethanol in the presence of 1 μl of Gen-elute linear polyacrylamide (Sigma). The RNA pellets were then analyzed by Northern blot as described (Feng et al., 1995).
Telomerase Elongation Assays
The TRAP was performed according to manufacturer's instructions (Intergen, Inc., Purchase, NY) (Kim et al., 1994). The two different primers used for the elongation step were the TS primer 5′-AATCCGTCGAGCAGAGTT-3′, and an oligonucleotide identical to the TS primer, but lacking the terminal (GTT) 3′ nucleotides. Unless otherwise indicated, 0.1 μg of primer and 2 μl of the anti-FLAG immunoprecipitates, or 1 μl of reconstituted reticulocyte lysate was assayed, and amplified for 25 PCR cycles. Extract titrations, as indicated in certain figures and our unpublished data, were used to confirm that the telomerase assays were in the linear range. Five microliters of TRAP reactions was subsequently loaded onto a 12% wt/vol 29:1 acrylamide:bis, nondenaturing gel and electrophoresed for 1 h at 800 V.
RESULTS
It was previously demonstrated that in RRL, only two exogenous components, hTERT and hTER, are required for telomerase activity (Weinrich et al., 1997; Beattie et al., 1998). These components are also sufficient to reconstitute human telomerase activity in S. cerevisiae (Bachand and Autexier, 1999). Currently, there are no reports of reconstituted telomerase activity from completely purified components. This could be due to several reasons, including the necessity for other factors for catalysis or protein folding. For example, formation of the duck hepatitis B RT–RNP complex requires the heat shock protein Hsp90 and p23 activity (Hu and Seeger, 1996; Hu et al., 1997). In support of this hypothesis, it has recently been demonstrated that p23 and Hsp90 in the RRL facilitate the folding of hTERT and hTER into an active complex (Holt et al., 1999). In that study, purified telomerase RNA is added after the translation of hTERT. In our studies, purified telomerase RNA is added to the reticulocyte lysate mixture before synthesis of hTERT, which we found to increase the levels of telomerase activity relative to the addition of telomerase RNA after hTERT synthesis (Beattie et al., 1998; Beattie and Harrington, unpublished data).
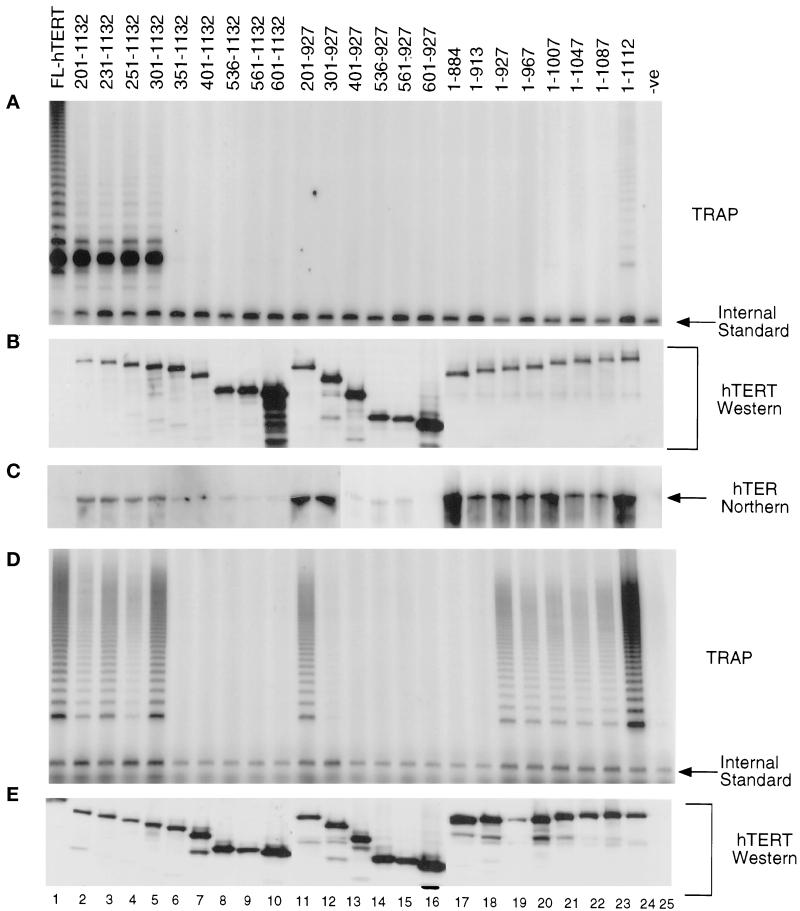
To investigate the regions of hTERT critical for telomerase activity in vitro, we synthesized systematic amino- and carboxy-terminal truncations of hTERT (Figure 1A) and compared each for its ability to reconstitute telomerase activity (Figure 2). After translation of hTERT in the presence of purified full-length telomerase RNA, the lysates were immunoprecipitated with an anti-flag antibody and assayed for telomerase activity by the TRAP (Figure 2A). A deletion of the first 300 amino acids of hTERT retained telomerase activity, however, this truncated protein catalyzed predominately short elongation products, even after PCR amplification of the telomerase extension products (Figure 2A, lanes 2–5). Deletion of the 20 C-terminal amino acids of TERT significantly reduced telomerase activity, and deletion of the 45 C-terminal residues abolished activity (Figure 2A, lanes 24 and 23). With the exception of full-length hTERT, Western analysis of the immunoprecipitates indicated that comparable levels of each hTERT truncation were present (Figure 2B). These results suggest that there are critical regions outside the conserved RT domain (which roughly corresponds to amino acids 601–927, lane 16) that are important for the reconstitution of catalytic activity in the RRL assay. Curiously, full-length hTERT, when combined with the telomerase RNA, appears to be extremely labile and difficult to detect by Western analysis, even after a long exposure of the hTERT immunopreciptations (Figure 2B; our unpublished results). For this reason, and because we cannot determine the percentage of hTERT in the immunoprecipitated complex that is active, we have not attempted to make quantitative comparisons between the apparent specific activity of full-length hTERT relative to the truncation derivatives of hTERT.
Figure 2.
Regions outside the conserved RT domain of hTERT are important for telomerase activity. Numbering refers to the positions of the amino acids of hTERT. (A) Results of telomerase assays from hTERT truncations synthesized in RRL. Different amino- and carboxy-terminal truncations of hTERT (as indicated) containing a flag epitope tag were synthesized in RRL in the presence of purified hTER, and were subjected to an anti-flag immunoprecipitation and analyzed for telomerase activity. (B) Analysis of protein production in the reticulocyte lysates by Western analysis with an α-hTERT peptide polyclonal antibody (Harrington et al., 1997b). Full-length hTERT is not visible due to low levels of the protein below the limit of detection. However, equal amounts of hTERT cDNA were added to the reticulocyte lysates and the presence of telomerase activity indicated that hTERT is present. (C) Northern blot analysis of the anti-flag immunoprecipitations of the hTERT truncations probed with an end-labeled oligonucleotide complementary to the RNA template region (see MATERIALS AND METHODS). hTER is not detected with full-length hTERT because of reduced levels of hTERT (B). The hTERT truncations were transfected into human 293T cells, immunoprecipitated with anti-flag antibody, and analyzed for telomerase activity by the TRAP assay (D), and hTERT protein levels by Western analysis (E).
To determine whether there is a correlation between loss of hTER binding and loss of catalytic activity of the hTERT truncations, the anti-flag immunoprecipitates containing hTERT were analyzed for association with hTER (Figure 2C). Many of the inactive C-terminal hTERT truncations were competent to bind the telomerase RNA (Figure 2C, lanes 11, 12, 17–23). The shortest C-terminal hTERT truncations that retained telomerase RNA binding spanned amino acids 1–884 and 301–927 (lanes 12 and 17). Therefore, the loss of activity associated with certain hTERT truncations in vitro is not due to the inability of the protein to bind the telomerase RNA, or a gross disruption in the overall conformation of the truncated protein. These results demonstrate that the catalytic activity of hTERT and the RNA-binding activity of hTERT are functionally distinct.
It is possible that the requirement for catalytic activity of the telomerase complex differs between the reconstituted system and the native human telomerase complex. To compare the catalytic activity of several hTERT truncations in human cells, various hTERT deletion constructs containing a flag epitope were transfected into human 293T cells, immunoprecipitated by using an anti-flag antibody, and assayed for telomerase activity (Figure 2, D and E). As a control, we showed previously that wild-type, flag-tagged full-length hTERT was fully functional when transfected into human cells (Harrington et al., 1997b; Figure 2D, lane 1). Similar to the reconstitution of telomerase activity in reticulocyte lysates, the first 300 amino acids could be deleted and still retained telomerase activity (Figure 2D, lanes 2–5). Interestingly, unlike the activity associated with these hTERT truncations in RRL, amino-terminal truncations of hTERT when expressed in 293T cells did not show predominantly shorter elongation products (Figures 2D, lanes 2–5, and 5). Several of the C-terminal truncations of hTERT were active when transfected into 293T cells, whereas they were completely inactive in the reticulocyte reconstitution assay (Figure 2, cf. A and D, lanes 11 and 19–23). For example, carboxy-terminal truncations of hTERT that lacked the last 205 amino acids remained active when transfected into 293T cells (Figure 2D, lane 19). Moreover, an hTERT truncation spanning amino acids 201–927 was still active when transfected into 293T cells, although it was completely inactive in reticulocyte lysates (Figure 2, cf. A and D, lane 11). Deletion of hTERT at amino acid 913 completely abrogated the catalytic activity of this truncation in 293T cells (Figure 2B, lane 17). Thus, N-terminal hTERT deletions were competent for catalysis in vitro and in vivo; however, the catalytic properties of C-terminal hTERT deletions differ considerably between reticulocyte lysates and human cells.
The inability of certain C-terminal hTERT truncations to support catalytic activity in reticulocyte lysates did not reflect an inability to bind the telomerase RNA. All of the C-terminal hTERT truncations that were active in 293T cells could still bind the telomerase RNA when they were synthesized in reticulocyte lysates (Figure 2C). Thus, the differences in telomerase activity between reticulocyte lysates and cells could not be attributed to the inability of the truncations to bind the telomerase RNA. Interestingly, three of the hTERT truncations that were inactive in both 293T cells and in reticulocyte lysates (301–927, 1–884, and 1–913), still retained the ability to bind hTER (Figure 2C, lanes 12, 17, and 18). These results suggest that the loss of catalytic activity of many of the hTERT truncations, either in human cells or in reticulocyte lysates, is more complex than loss of association with the telomerase RNA, and additional determinants are important for the observed activity in cells. Furthermore, there are regions of hTERT outside the RT region that are critical for telomerase activity both in RRL and in human immortalized cells.
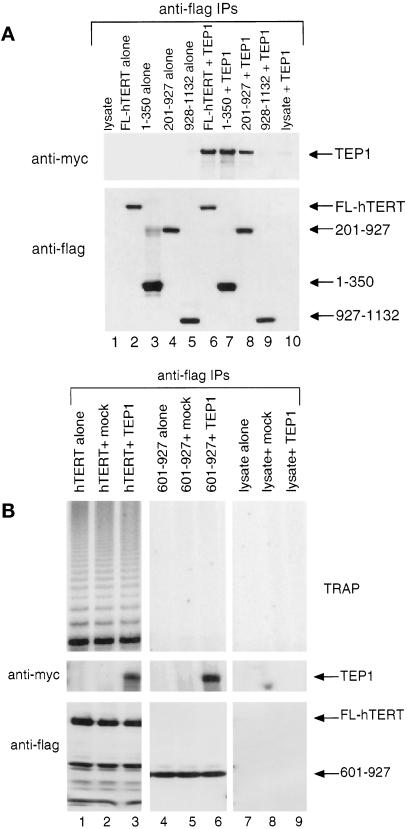
A previous study demonstrates that hTERT and the RNA-binding protein TEP1 could interact in human 293T or HeLa cell extracts (Harrington et al., 1997b). However, the addition of TEP1 is not required to reconstitute telomerase activity in vitro (Weinrich et al., 1997; Beattie et al., 1998; Bachand and Autexier, 1999). To further verify that the hTERT truncations were correctly folded, and to delineate the regions of hTERT that were necessary for the interaction with TEP1, we tested the hTERT truncations for their ability to interact with TEP1. Human 293T cells were cotransfected with each of the flag-epitope–tagged hTERT truncations and with full-length myc-epitope–tagged TEP1. Cell extracts were then immunoprecipitated with anti-flag antibody and analyzed by Western blot with anti-flag and anti-myc antibodies. Each of the hTERT truncations (all of which maintain an intact RT domain, except for constructs 1–350 and 927-1132) were able to bind TEP1 (Figure 3A; our unpublished results). However, an hTERT truncation that spanned amino acids 927-1132 did not interact with TEP1 (Figure 3A, lane 9). These results suggest that each of the hTERT truncations that contained the conserved RT motifs maintained a conformation that allowed an interaction with TEP1. It is noteworthy that two nonoverlapping regions of hTERT (spanning amino acids 1–350 and 350-1132) were each able to specifically interact with TEP1. Because it is not known whether the interaction of hTERT and TEP1 is direct, it is possible that there are multiple regions of hTERT that are capable of binding to TEP1, either directly or indirectly.
Figure 3.
TEP1 binding of hTERT truncations. (A) hTERT truncations and full-length TEP1 (containing a myc-epitope tag) were cotransfected into human 293T cells. Cell extracts were immunoprecipitated by using an anti-flag antibody (to immunoprecipitate hTERT, bottom) and then analyzed by Western analysis by using anti-myc antibody to examine coimmunoprecipitation of TEP 1 (top). (B) Full-length hTERT was synthesized in RRL in the presence of 0.002 μg/μl purified hTER. The lysate alone (no DNA), containing full-length hTERT (TERT) or containing the RT domain (601–927) was incubated on ice either alone (alone, lanes 1 and 4), with mock 293T-transfected cell extracts (+mock, lanes 2 and 5), or TEP1 (with a myc epitope tag) transfected 293T cell extracts (+TEP1, lanes 3 and 6). The mixtures were then immunoprecipitated with anti-flag antibody and assayed for telomerase activity (top) and blotted for the presence of hTERT by using an anti-hTERT antibody (Western blot, middle) and TEP1 by using an anti-myc antibody (Western blot, bottom).
We also tested whether hTERT truncations synthesized in reticulocyte lysates were competent to interact with TEP1. It was necessary to use TEP1 from human cell extracts because it was not possible to synthesize full-length TEP1 in RRL (our unpublished results). Full-length hTERT and the RT domain of hTERT (601–927) each interacted with full-length Myc-TEP1 (Figure 3B). Similarly, other hTERT truncations that contained an intact RT domain were competent to bind TEP1 (our unpublished results). As a control, TEP1 could not be coimmunoprecipitated onto anti-flag resin in the absence of hTERT (Figure 3B, lane 9). Thus, the hTERT truncations both in RRL and in human immortalized cells are in a conformation that allows their interaction with TEP1. We also observed no change in telomerase activity upon coimmunoprecipitation of TEP1 with hTERT either in RRL or in transfected 293T cells (Figure 3B; our unpublished results). These results further suggest that TEP1 is not required to reconstitute activity in RRL (Weinrich et al., 1997; Beattie et al., 1998).
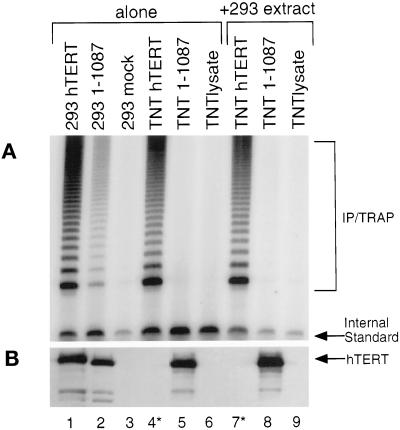
In an effort to determine why the hTERT C-terminal truncations were inactive in vitro but active in human cell extracts, we tested whether a diffusible factor present in 293T cell extracts could restore activity to the longest inactive hTERT C-terminal truncation (a truncation spanning amino acids 1–1087). We added 293T cell extracts to hTERT proteins synthesized in RRL, and immunoprecipitated the complex by using an anti-flag antibody and assayed for telomerase activity. Addition of the human cell extract could not confer activity to the hTERT truncation spanning amino acids 1–1087 (Figure 4, cf. lanes 2 and 8). Moreover, activity could not be restored to the hTERT truncation when 293T cell extracts were added to the reticulocyte lysate during the translation of hTERT, or when added to a shorter, inactive C-terminal truncation spanning amino acids 1–927 (our unpublished results). Therefore, the ability of the hTERT truncation spanning residues 1–1087 to support activity in human 293T cells is not simply due to diffusible factors in the cell extract.
Figure 4.
Cell extracts cannot restore activity to an inactive C-terminal hTERT truncation synthesized in reticulocyte lysates. To examine differences observed with the hTERT truncations made in RRL and human 293T cells, cell extracts were incubated in ice with RRL containing either full-length hTERT, an hTERT truncation containing amino acids 1–1087 (1–1087), or a RRL lysate only control (TNT lysate). The individual cell extracts (lanes 1–3) and the RRL reactions (lanes 4–6), as well as the mixtures (lanes 7–9) were immunoprecipitated with an α-flag antibody and assayed for telomerase activity (A) and hTERT by Western blot analysis by using an anti-hTERT antibody (B). As mentioned previously, those lanes marked with an ∗ possessed levels of FL-hTERT that, although active, were below the limit of detection on a Western blot.
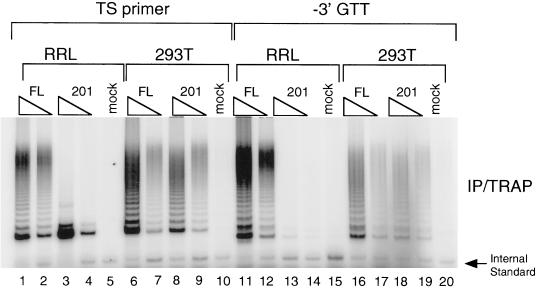
The activity conferred by full-length hTERT in the RRL shares many properties with native human telomerase (Weinrich et al., 1997; Beattie et al., 1998). However, we did find one notable difference: in RRL; hTERT truncations lacking the first 200–300 amino acids catalyzed the production of considerably shorter elongation products compared with the same truncation produced in 293T cells (Figure 2, cf. A and D, lanes 2–5). To further investigate the polymerization defect of the 201-1132 hTERT truncation, we compared the ability of full-length hTERT and the truncation spanning amino acids 201-1132 to elongate different primers (Figure 5). The first primer tested was an oligonucleotide of random sequence ending in GTT (see MATERIALS AND METHODS), and the second was identical to the previous oligonucleotide except that the 3′ GTT nucleotides were omitted. Full-length hTERT expressed in 293T cell extracts or in RRL was able to elongate both primers (Figure 5). However, an hTERT truncation spanning amino acids 201-1132 was able to elongate the shorter primer only when present in 293T cell extracts (Figure 5, lanes 18 and 19). When synthesized in RRL, the truncation derivative of hTERT was unable to elongate the shorter primer (Figure 5, lanes 13 and 14). This result suggests a decrease in the ability of the hTERT truncation to bind or elongate nontelomeric oligonucleotide substrates. Despite the fact that deletion of the first 200 amino acids of hTERT did not affect telomerase RNA or TEP1 binding, this deletion derivative was unable to use nontelomeric primers in vitro. Thus, the first 200 nucleotides of hTERT may be necessary to facilitate an interaction between hTERT and certain primer sequences. Alternatively, the absence of the first 200 amino acids of hTERT may subtly affect the conformation of the RNP that is only revealed upon challenge with a nontelomeric oligonucleotide substrate.
Figure 5.
Primer use by full-length and 201-1132 hTERT. Anti-flag immunoprecipitations containing full-length hTERT (FL), an hTERT truncation spanning amino acids 201-1132 (201), or lysate only (mock), from either RRL (lanes 1–5 and 11–15) or transfected human 293T cells (293T; lanes 6–10 and 16–20) were analyzed for telomerase activity with 0.1 μg of either the TS primer (lanes 1–10) or a TS primer that lacked the 3′ GTT nucleotides (−3′ GTT, lanes 11–20). The triangles represent 2-fold dilutions of each of the immunoprecipitates to demonstrate that the telomerase reactions were in the linear range.
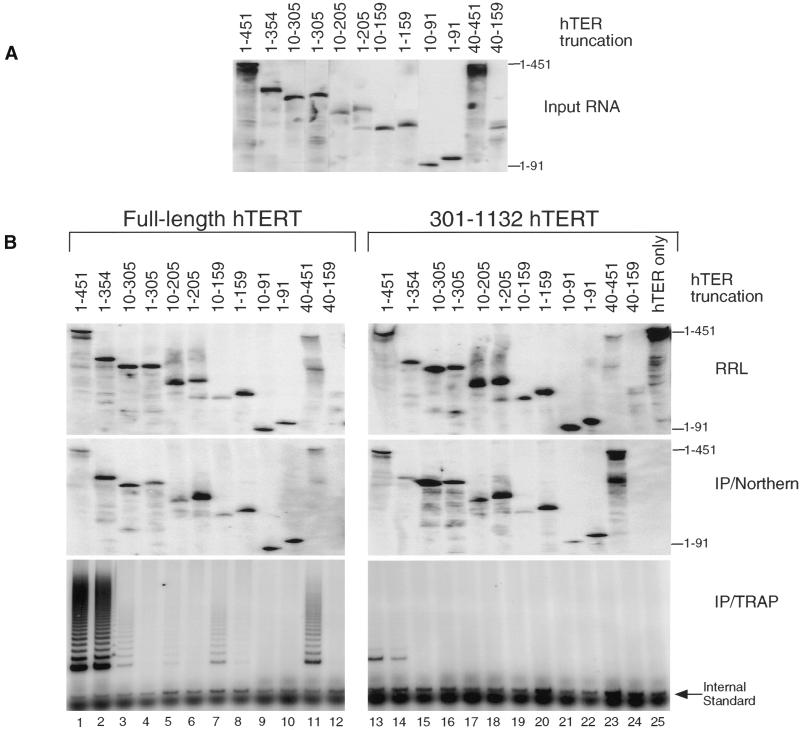
If the different catalytic properties of full-length and certain hTERT truncations were the result of subtle conformational differences, we reasoned that full-length and hTERT truncations may exhibit different abilities to bind and use shorter telomerase RNAs. We previously showed that a telomerase RNA truncation spanning nucleotides 10–159 was active in the presence of full-length hTERT in vitro (Beattie et al., 1998). In that study, the shortest telomerase RNA deletion that was sufficient for catalytic activity in reticulocyte lysates spanned nucleotides +10 to +159. Interestingly, upon repeating our analysis with telomerase RNAs beginning at nucleotide +1, we found that some of these telomerase RNA truncations were not active (Figure 6B, bottom). For example, the telomerase RNA spanning +10 to +159 is active, as demonstrated previously, whereas an RNA spanning nucleotides +1 to +159 is not active in RRL (Figure 6B lanes 7 and 8). Some of the longer telomerase RNAs beginning at residue 1 were active, albeit weakly compared with full-length telomerase RNA (Figure 6B, bottom, lanes 4 and 6). These results suggest that, in the context of some shorter telomerase RNAs, the first 10 nucleotides are inhibitory to the reconstitution of telomerase activity. These results are qualitatively similar to those of Tesmer et al. (1999) who observed that some telomerase RNA deletion derivatives are more active when initiated at nucleotide +33 than when initiated at nucleotide +1. Although we observed weak telomerase activity with a telomerase RNA spanning nucleotides 1–205, Tesmer et al. (1999) concluded that the minimal RNA necessary for efficient reconstitution of telomerase activity spans nucleotides 1–325. Because TRAP is not a quantitative assay, we have not quantified the levels of telomerase activity between the different telomerase RNA deletions. However, our results are clearly consistent with the idea that telomerase RNAs <325 nucleotides are much less active than full-length telomerase RNA (Beattie et al., 1998; Tesmer et al., 1999).
Figure 6.
RNA binding and catalytic activities of hTERT and hTER truncation combinations. (A) Northern analysis (as in Figure 2C) of each in vitro transcribed hTER truncation (5 ng) before the addition of RRL. Numbers of the hTER truncations refer to nucleotide positions. (B) Full-length hTERT or 301-1132 hTERT was synthesized in RRL in the presence of different hTER truncations (0.01 μg/μl the specified hTER construct). Each hTER truncation (5 ng) was analyzed by Northern analysis after incubation in the RRL (top). The RRL were subjected to an anti-flag immunoprecipitation and assayed for RNA binding by Northern analysis (IP/Northern, middle) and telomerase activity (IP/TRAP, bottom). (A) Assayed for telomerase activity. (B) RNA binding by Northern analysis (as in Figure 2). Lanes 1–12, full-length hTERT synthesized in the presence of the indicated hTER truncation; lanes 13–24, 301-1132 hTERT synthesized in the presence of the indicated hTER truncation, lane 25, full-length hTER with no hTERT protein.
We examined each of these telomerase RNA truncations for the ability to reconstitute activity with hTER and an hTERT truncation spanning amino acids 301-1132. The hTERT truncation retained telomerase activity with full-length hTER; however, similar to the hTERT truncation spanning amino acids 201-1132, the truncation spanning amino acids 301-1132 also exhibited shorter telomerase extension products. In contrast to full-length hTERT, all of the hTER RNAs that were deleted past nucleotide 354 showed significantly reduced telomerase activity when mixed with the hTERT truncation spanning amino acids 301-1132 (Figure 6B, bottom, lanes 15–24).
In a reconstitution assay with micrococcal nuclease-treated cell extracts followed by the addition of exogenous hTER, the first 40 nucleotides of hTER were not required for activity with native telomerase (Autexier et al., 1996). We also found that the first 40 nucleotides of hTER were not required for activity with full-length hTERT in the RRL reconstitution assay, although the activity observed was reduced relative to the full-length telomerase RNA. (Figure 6A, cf. lanes 1 and 11). However, the first 40 nucleotides were necessary for reconstitution with the hTERT truncation spanning amino acids 301-1132. A telomerase RNA spanning nucleotides +40 to +159 was not active with either full-length hTERT or the 301-1132 hTERT truncation (Figure 6, lane 12 and 24).
To determine whether loss of activity with the truncated telomerase RNAs was due to loss of RNA binding, we examined the association of each of the RNAs with the full-length hTERT and an hTERT truncation (301–1132) by Northern analysis of the anti-flag immunoprecipitates (Figure 6B, middle). Each of the hTERT proteins was able coimmunoprecipitate all of the truncated RNAs, except an RNA spanning nucleotides 40–159. The amount of each telomerase RNA added before the immunoprecipitation was similar, although some degradation of the RNAs did occur after incubation in the RRL (Figure 6, A and B, top). The telomerase RNA-binding profiles for both full-length hTERT and the hTERT deletion spanning amino acids 301-1132 were similar, whereas the telomerase activity profiles were clearly different. These results reveal that there are different RNA requirements for hTERT binding and catalysis, and further support our observation that the catalytic activity and the RNA-binding activity of hTERT are functionally distinct. These results also suggest that any conformational difference between full-length and the 301-1132 hTERT truncation was not different enough to alter the interaction with shorter telomerase RNAs.
DISCUSSION
We have examined the regions of hTERT required for catalytic activity in vitro and in transfected 293T cells. We found that the first 300 amino acids of hTERT were not absolutely essential for telomerase activity either in RRL or transfected 293T cells. However, in RRL, but not in transfected human 293T cells, deletion of the N terminus of hTERT severely compromised the extension of telomeric and nontelomeric primers, suggesting a defect in either enzyme translocation, processivity, or primer binding. In reticulocyte lysates, hTERT C-terminal deletions of 45 amino acids and beyond completely abolished telomerase activity. These results differ from the mutation analysis of HIV-RT, where up one-third of the C terminus (right up to the RT motif) could be disrupted with little effect on polymerase activity (Prasad and Goff, 1989). Unlike HIV-RT, these results indicate that there are protein requirements for catalysis that are outside of the conserved RT domain of hTERT. The differences observed in vitro and in cells also demonstrate that although the reconstitution assay is important for delineating specific RNA and protein requirements, the recombinant enzyme does not completely recapitulate all of the properties of native human telomerase.
The differences in activities of hTERT truncations between cells and reticulocyte lysates are not due to the inability of the hTERT truncations to bind the telomerase RNA hTER in vitro. Each of the truncations that were active in human cells was competent to bind hTER in vitro. Therefore, the inactivity of the hTERT truncations was not likely the result of a grossly misfolded protein that disrupted telomerase RNA binding. These results demonstrate that the catalytic activity of hTERT and the RNA-binding activity of hTERT are separable. Studies with S. cerevisiae Est2p demonstrated the importance of the N terminus in the bypass of senescence and in telomerase RNA binding (Friedman and Cech, 1999). However, in this study, no catalytically inactive mutations were identified that retained the ability to bind the telomerase RNA. However, point mutations within the Est2p and hTERT RT region, although catalytically inactive, still retain the ability to bind the telomerase RNA (Lingner et al., 1997; Zhang et al., 1999; our unpublished data). Our analysis demonstrates that although inactive in vitro and in 293 cells, certain hTERT truncations also retain the ability to interact with the telomerase RNA.
We also found that TEP1 could interact with hTERT truncations that contain the RT domain. Because TEP1 was able to interact with an hTERT truncation (RT domain) that could not bind the telomerase RNA, we suggest that the interaction may occur independent of the telomerase RNA. This result is consistent with our observation that the interaction between TEP1 and TERT is not sensitive to treatment with RNase in cell extracts or in RRL (Harrington et al., 1997b; our unpublished results). These results demonstrate that determinants involved in the hTERT–TEP1 interaction remain intact in RRL, and that the interaction with the RT domain is most likely not dependent on the telomerase RNA. We cannot rule out that other critical structures that are important for catalytic activity have been disrupted. Because we were unable to synthesize full-length TEP1 in RRL, our source of TEP1 was from 293T transfected cells. Therefore, this assay does not allow us to address whether the interaction between hTERT and TEP1 is direct. The addition of TEP1 to telomerase reconstituted in RRL does not affect catalysis in vitro. Although TEP1 is associated with telomerase activity and can specifically bind the telomerase RNA (Harrington et al., 1997; Nakayama et al., 1997), these results suggest that the addition of TEP1 to the telomerase complex does not directly influence catalytic activity, at least as measured in this in vitro assay.
Interestingly, we found that an hTERT truncation lacking the first 200 amino acids could only synthesize short telomeric products in reticulocyte lysates, and could not elongate partly nontelomeric primers. Due to the inability to detect activity of the hTERT truncations by using a non-PCR–based telomerase elongation assay, we cannot yet precisely define the nature of the polymerization defect (our unpublished results). In cells, however, an hTERT truncation spanning amino acids 201-1132 showed similar catalytic properties and primer use to native human telomerase in vivo. This result implies that there are factors within the cell that allow efficient polymerization of certain hTERT truncations in vivo that are not present in RRL.
Recently, it has been reported that by using a similar in vitro reconstitution assay that the minimal telomerase RNA required for efficient telomerase catalysis spanned nucleotides +33 to +325 of hTER (Tesmer et al., 1999). Our results showed that efficient telomerase activity could also be obtained with hTER truncations of 1–354 or longer. However, under the telomerase reconstitution conditions we have developed, we also observe catalytic activity with RNAs as short as nucleotides +10 to +159. Because certain telomerase RNAs (1–159, 1–91, 10–91, and 40–159) are completely inactive in our assay, it is clear that the 1/10–305, 1/10–205, and 10–159 telomerase RNA truncations are active, albeit weakly. We suggest that the prior addition of the telomerase RNA during the translation of hTERT, as carried out in our experiments, allows the detection of weak telomerase activity (Beattie et al., 1998).
Human telomerase RNA truncations deleted past nucleotide 159 are inactive in the RRL reconstitution assay. These shorter deletions are predicted to significantly disrupt a stem-loop structure involved in a pseudoknot proposed by Chen et al. (2000). This stem is critical for telomerase activity in each of the separate assays developed to analyze human telomerase RNA requirements (Autexier et al., 1996; Beattie et al., 1998; Tesmer et al., 1999). In a truncated RNA spanning nucleotides 10–159, the 3′ stem of the pseudoknot is not present, however, this RNA is still weakly active. When the 3′ stem of the pseudoknot is disrupted in the context of the full-length RNA, catalytic activity it abolished (Autexier et al., 1996). It therefore appears that deletion of this stem is less detrimental than its disruption. Also, although a portion of the pseudoknot structure is disrupted, the 10–159 RNA can still bind hTERT. This result differs slightly from what is observed with Tetrahymena telomerase, in that disruption of the pseudoknot in the telomerase RNA abolishes tTERT binding (Gilley and Blackburn, 1999). There is little structure associated with the first 90 nucleotides of human telomerase RNA as suggested by the secondary structure model proposed by Chen et al. (2000). However, a telomerase RNA spanning nucleotides 1–91 and 10–91 can still bind hTERT in RRL. It therefore remains to be determined what sequences or tertiary structures are critical for hTERT binding in RRL and whether other factors within the RRL may contribute to the ability of hTERT to bind hTER. Although the template region of the telomerase RNA may be important for binding, this region is not sufficient for an interaction with hTERT because a telomerase RNA spanning nucleotides 40–159, which contains the RNA template, cannot interact with hTERT. Based on the secondary structure model for human telomerase RNA (Chen et al., 2000), the partially inhibitory role of the first 10 nucleotides with hTER truncations remains unknown. We cannot rule out, however, that there are subtle conformational differences with each of the hTER truncations that allow for binding to hTERT proteins, but not catalytic activity.
Our results suggest that there are conditions present in human cells, but not present in RRL, that affect the assembly and catalytic activity of human telomerase. For example, posttranslational modification of hTERT necessary for assembly or catalysis could occur in cells but not in reticulocyte lysates. It remains possible that misassembled hTERT truncations in reticulocyte lysates could remain unaffected by the addition of cell extract. However, the ability of these hTERT truncations to bind several regions of hTER and TEP1 argue against the gross misfolding of these proteins. Finally, a nondiffusible factor may be present in the cell extract that is not available to complex with the recombinant hTERT truncations. These factors are not likely to be p23 or hsp90 because foldosome proteins are present both 293T cell extracts and RRL. As yet, no additional “core” telomerase catalytic components, other than the telomerase RNA and hTERT, have been identified.
One intriguing possibility to explain the differences in telomerase reconstitution between RRL and 293T cells is the ability of “inactive” hTERT to form a multimer with endogenous hTERT when transfected into 293 cells. Such a functional interaction has been previously observed by using different mutations of the yeast telomerase RNA, which led to the hypothesis that telomerase may contain two active sites (Prescott and Blackburn, 1997a). These experiments, however, do not rule out other types of functional interactions that do not require multimerization. Future experiments in cells lacking endogenous TERT will enable us to discern whether such a functional interaction is able to confer catalytic activity to some of the hTERT truncations.
There are many different types of conformational changes associated with the formation of an active RNP complex. RNA–protein interactions can function cooperatively to assemble into an active complex. In ribosome assembly, protein binding induces conformational changes within the rRNA (Weeks, 1997), and in the case of the N-protein from bacteriophage λ, much of the protein remains unstructured until it is bound to the nut site RNA (Mogridge et al., 1998). Conformational changes within RNA–protein complexes occur to facilitate biochemical reactions. This is demonstrated by the several conformational changes in the spliceosome that occur before the catalysis of splicing (Sontheimer and Steitz, 1993; Madhani and Guthrie, 1994; Newman et al., 1995; Kim and Abelson, 1996). Our data suggests that cooperative RNA/protein folding and/or conformational changes probably occur within the telomerase RNP. Although many of the hTERT truncations are competent to bind the telomerase RNA it remains unclear whether this RNA–protein complex is competent for telomerase activity. Perhaps subtle conformational changes aside from hTER binding are needed for optimal catalysis of certain hTERT protein truncations. We have identified a number of RNA–protein and protein–protein interactions within the human telomerase complex that are important for the formation of catalytically active telomerase.
This study has demonstrated that the ability of hTER and hTERT to reconstitute telomerase activity in vitro is dependent on both regions of the hTERT protein outside the conserved RT domain and regions of telomerase RNA outside the telomeric template. We observed differences between the reconstituted enzyme and the native enzyme, suggesting that critical factors or conditions are not present in the reticulocyte lysate. Further studies are aimed at identification of the factors critical to reconstitution of telomerase with properties similar to the native enzyme, and understanding the intricacies of the telomerase polymerization mechanism in a highly purified, reconstituted telomerase RNP.
ACKNOWLEDGMENTS
We thank R. Oulton for technical assistance, and R. Collins, B. Blencowe, C. Greider, J-L Chen, and members of the Cech laboratory for critical comments on the manuscript. T.L.B. is a Research Fellow of the National Cancer Institute of Canada with funds provided by the Terry Fox Run. This work was supported in part by a grant to L.H. from the Medical Research Council of Canada.
REFERENCES
- Autexier C, Greider CW. Functional reconstitution of wild-type and mutant Tetrahymena telomerase. Genes Dev. 1994;8:563–575. doi: 10.1101/gad.8.5.563. [DOI] [PubMed] [Google Scholar]
- Autexier C, Greider CW. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes Dev. 1995;9:2227–2239. doi: 10.1101/gad.9.18.2227. [DOI] [PubMed] [Google Scholar]
- Autexier C, Greider CW. Mutational analysis of the Tetrahymena telomerase RNA: identification of residues affecting telomerase activity in vitro. Nucleic Acids Res. 1998;26:787–795. doi: 10.1093/nar/26.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autexier C, Pruzan R, Funk WD, Greider CW. Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J. 1996;15:5928–5935. [PMC free article] [PubMed] [Google Scholar]
- Autexier C, Triki I. Tetrahymena telomerase ribonucleoprotein RNA-protein interactions. Nucleic Acids Res. 1999;27:2227–2234. doi: 10.1093/nar/27.10.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachand F, Autexier C. Functional reconstitution of human telomerase expressed in Saccharomyces cerevisiae. J Biol Chem. 1999;274:38027–38031. doi: 10.1074/jbc.274.53.38027. [DOI] [PubMed] [Google Scholar]
- Beattie TL, Zhou W, Robinson MO, Harrington L. Reconstitution of human telomerase activity in vitro. Curr Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A, Blackburn EH. A functional telomerase RNA swap in vivo reveals the importance of nontemplate RNA domains. Proc Natl Acad Sci USA. 1997;94:2823–2827. doi: 10.1073/pnas.94.7.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells [see comments] Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Marusic L, Bacchetti S, Namba M, Reddel RR. The telomere lengthening mechanism in telomerase-negative immortal human cells does not involve the telomerase RNA subunit. Hum Mol Genet. 1997;6:921–926. doi: 10.1093/hmg/6.6.921. [DOI] [PubMed] [Google Scholar]
- Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- Collins K, Kobayashi R, Greider CW. Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- Counter CM, Meyerson M, Eaton EN, Ellisen LW, Caddle SD, Haber DA, Weinberg RA. Telomerase activity is restored in human cells by ectopic expression of hTERT (hEST2), the catalytic subunit of telomerase. Oncogene. 1998;16:1217–1222. doi: 10.1038/sj.onc.1201882. [DOI] [PubMed] [Google Scholar]
- Counter CM, Meyerson M, Eaton EN, Weinberg RA. The catalytic subunit of yeast telomerase. Proc Natl Acad Sci USA. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J, Le S, West MD, Harley CB, Andrews WH, Greider CW, Villeponteau B. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- Friedman KL, Cech TR. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 1999;13:2863–2874. doi: 10.1101/gad.13.21.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley D, Blackburn EH. Specific RNA residue interactions required for enzymatic functions of Tetrahymena telomerase. Mol Cell Biol. 1996;16:66–75. doi: 10.1128/mcb.16.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley D, Blackburn EH. The telomerase RNA pseudoknot is critical for the stable assembly of a catalytically active ribonucleoprotein. Proc Natl Acad Sci USA. 1999;96:6621–6625. doi: 10.1073/pnas.96.12.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley D, Lee MS, Blackburn EH. Altering specific telomerase RNA template residues affects active site function. Genes Dev. 1995;9:2214–2226. doi: 10.1101/gad.9.18.2214. [DOI] [PubMed] [Google Scholar]
- Greider CW. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. Telomeres, telomerase and cancer. Sci Am. 1996;274:92–97. doi: 10.1038/scientificamerican0296-92. [DOI] [PubMed] [Google Scholar]
- Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass MB, Arruda I, Robinson MO. A mammalian telomerase-associated protein. Science. 1997a;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- Harrington L, Zhou W, McPhail T, Oulton R, Yeung DS, Mar V, Bass MB, Robinson MO. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997b;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SE, Aisner DL, Baur J, Tesmer VM, Dy M, Ouellette M, Trager JB, Morin GB, Toft DO, Shay JW, Wright WE, White MA. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 1999;13:817–826. doi: 10.1101/gad.13.7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Seeger C. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc Natl Acad Sci USA. 1996;93:1060–4. doi: 10.1073/pnas.93.3.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Toft DO, Seeger C. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 1997;16:59–68. doi: 10.1093/emboj/16.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian A, Bowtell DD, Abud HE, Hime GR, Venter DJ, Keese PK, Duncan EL, Reddel RR, Jefferson RA. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- Kim CH, Abelson J. Site-specific crosslinks of yeast U6 snRNA to the pre-mRNA near the 5′ splice site. RNA. 1996;2:995–1010. [PMC free article] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Licht JD, Collins K. Telomerase RNA function in recombinant Tetrahymena telomerase. Genes Dev. 1999;13:1116–1125. doi: 10.1101/gad.13.9.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hendrick LL, Cech TR. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C. Randomization-selection analysis of snRNAs in vivo: evidence for a tertiary interaction in the spliceosome. Genes Dev. 1994;8:1071–1086. doi: 10.1101/gad.8.9.1071. [DOI] [PubMed] [Google Scholar]
- McCormick-Graham M, Romero DP. Ciliate telomerase RNA structural features. Nucleic Acids Res. 1995;23:1091–1097. doi: 10.1093/nar/23.7.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Blackburn EH. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, Liu Q, Bacchetti S, Haber DA, Weinberg RA. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- Mogridge J, Legault P, Li J, Van Oene MD, Kay LE, Greenblatt J. Independent ligand-induced folding of the RNA-binding domain and two functionally distinct antitermination regions in the phage lambda N protein. Mol Cell. 1998;1:265–275. doi: 10.1016/s1097-2765(00)80027-1. [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Saito M, Nakamura H, Matsuura A, Ishikawa F. TLP1: a gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell. 1997;88:875–884. doi: 10.1016/s0092-8674(00)81933-9. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Tahara H, Tahara E, Saito M, Ito K, Nakamura H, Nakanishi T, Ide T, Ishikawa F. Telomerase activation by hTRT in human normal fibroblasts and hepatocellular carcinomas. Nat Genet. 1998;18:65–68. doi: 10.1038/ng0198-65. [DOI] [PubMed] [Google Scholar]
- Newman AJ, Teigelkamp S, Beggs JD. snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA. 1995;1:968–980. [PMC free article] [PubMed] [Google Scholar]
- Olovnikov A. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- Prasad VR, Goff SP. Linker insertion mutagenesis of the human immunodeficiency virus reverse transcriptase expressed in bacteria: definition of the minimal polymerase domain. Proc Natl Acad Sci USA. 1989;86:3104–3108. doi: 10.1073/pnas.86.9.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J, Blackburn EH. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 1997a;11:2790–2800. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J, Blackburn EH. Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev. 1997b;11:528–540. doi: 10.1101/gad.11.4.528. [DOI] [PubMed] [Google Scholar]
- Romero DP, Blackburn EH. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- Roy J, Fulton TB, Blackburn EH. Specific telomerase RNA residues distant from the template are essential for telomerase function. Genes Dev. 1998;12:3286–3300. doi: 10.1101/gad.12.20.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer EJ, Steitz JA. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- Tesmer VM, Ford LP, Holt SE, Frank BC, Yi X, Aisner DL, Ouellette M, Shay JW, Wright WE. Two inactive fragments of the integral RNA cooperate to assemble active telomerase with the human protein catalytic subunit (hTERT) in vitro. Mol Cell Biol. 1999;19:6207–6216. doi: 10.1128/mcb.19.9.6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Benchimol S. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr Biol. 1998;8:279–282. doi: 10.1016/s0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- Watson J. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- Weeks KM. Protein-facilitated RNA folding. Curr Opin Struct Biol. 1997;7:336–342. doi: 10.1016/s0959-440x(97)80048-6. [DOI] [PubMed] [Google Scholar]
- Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB, Taylor RD, Carlos R, Andrews WH, Wright WE, Shay JW, Harley CB, Morin GB. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- Zhang X, Mar V, Zhou W, Harrington L, Robinson MO. Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev. 1999;13:2388–2399. doi: 10.1101/gad.13.18.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]