Abstract
Hepatitis C virus (HCV) infection is a major cause of chronic liver disease, which can lead to the development of liver cirrhosis and hepatocellular carcinoma. Current therapy of patients with chronic HCV infection includes treatment with IFNα in combination with ribavirin. Because most treated patients do not resolve the infection, alternative treatment is essential. RNA interference (RNAi) is a recently discovered antiviral mechanism present in plants and animals that induces double-stranded RNA degradation. Using a selectable subgenomic HCV replicon cell culture system, we have shown that RNAi can specifically inhibit HCV RNA replication and protein expression in Huh-7 cells that stably replicate the HCV genome, and that this antiviral effect is independent of IFN. These results suggest that RNAi may represent a new approach for the treatment of persistent HCV infection.
Hepatitis C virus (HCV), a member of the Flaviviridae family of viruses, is a major cause of chronic hepatitis and hepatocellular carcinoma (1, 2). Viral clearance during acute HCV infection is usually associated with a multispecific CD4+ and CD8+ T cell response, which is weak or undetectable in subjects who do not control the infection (3–5). Importantly, most chronically infected patients fail to resolve HCV infection after combination therapy with IFN and ribavirin (6–8).
The HCV genome is a positive-stranded ≈9.6-kb RNA molecule consisting of a single ORF, which is flanked by 5′ and 3′ UTR. The HCV 5′ UTR contains a highly structured internal ribosome entry site (8–13). The HCV ORF encodes a single polyprotein that is 3,008–3,037 aa in length and is posttranslationally modified to produce at least ten different proteins: core, envelope proteins E1 and E2, p7, and nonstructural proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B (1, 13, 14). Despite considerable advances in the understanding of the function of these proteins, the basic mechanism(s) of HCV replication still remain unclear because of the absence of a tissue culture system that can sustain productive virus infection (1). The recent development of subgenomic and full-length HCV replicons that replicate and express HCV proteins in stably transfected human hepatoma cell-derived Huh-7 cells has facilitated the analysis of the role of cellular pathways required in HCV replication and the efficacy of antiviral drugs (15–19). For example, by using HCV replicons, the antiviral effects of IFNα and IFNγ have been clearly demonstrated (20–23). Although IFN treatment can efficiently inhibit HCV replication in cultured Huh-7 cells, >60% of patients treated with IFN do not eliminate the virus (24–28). This suggests that HCV may be able to induce a state of IFN resistance in the infected liver. In keeping with this notion, HCV E2 and NS5A proteins have been demonstrated to interfere with the IFN-induced signaling pathway by interacting with protein kinase R (PKR) and inhibiting its kinase activity (29–32). Thus, alternative approaches to the treatment of chronic HCV infection seem to be warranted.
Double-stranded RNA (dsRNA), an intermediate in the replication of many viruses, induces a multifaceted response in higher eukaryotes, including the production of IFN α/β (33). Another antiviral mechanism, RNA interference (RNAi), originally discovered in plants, Caenorhabditis elegans, and Drosophila, is also induced by dsRNA (34–36). In this process, dsRNA is cleaved into 21–23 nucleotide dsRNA molecules (known as short interfering RNA or siRNA) by an RNase III-like enzyme known as Dicer (33, 37–40). These siRNA molecules associate with a multiprotein complex known as the RNA-induced silencing complex and targets homologous mRNA for degradation (33, 37–40). RNAi, which can function independently of IFN-induced pathways, is also effective in mammalian cells (33, 41–43). This suggests that plants and animals share a conserved antiviral mechanism leading to specific destruction of nonself dsRNA (44, 45). RNAi interferes with the replication of a number of animal viruses including HIV-1, flock house virus (FHV), Rous sarcoma virus, dengue virus, and poliovirus (46–56). As an RNA virus, HCV is a prime candidate for RNAi. Indeed, it has been demonstrated recently that HCV-specific siRNA can inhibit levels of a fusion NS5B–luciferase reporter transcript when the siRNA and the target were cotransfected hydrodynamically into mice (57). Although these results illustrate an important principle, the experimental design did not permit the analysis of the effects of RNAi on HCV replication, as they demonstrated the ability of HCV-specific siRNA to inhibit the accumulation of an HCV-containing fusion transcript, not the down-regulation of a preexisting HCV transcript such as those that are present in infected cells. In the current study, we wanted to determine whether RNAi could inhibit ongoing HCV replication using a stably transfected HCV replicon system as a surrogate for HCV-infected hepatocytes. Our results demonstrate that HCV RNA replication and protein expression are efficiently inhibited in Huh-7 cells that stably replicate the HCV replicon and that this effect is independent of IFN.
Materials and Methods
Tissue Culture and Generation of Stable HCV Subgenomic Replicon Cell Line.
The S1179I HCV subgenomic replicon (genotype 1b) was obtained from Charles Rice (The Rockefeller University, New York) and passaged as described (18, 20). Huh-7 cells were transfected with HCV subgenomic replicon RNA and maintained as described (18, 20). An Huh-7 cell line that stably replicated HCV RNA replication (called S1179I herein) was used in all experiments.
Generation and Transfection of siRNA.
Target sequences for the siRNAs were determined by using the Ambion web-based criteria, followed by generation of the HCV- and human GAPDH-specific siRNAs using the Silencer siRNA construction kit (Ambion, Austin, TX). Seven HCV-specific siRNAs, based on the prototype sequence present in the HCV replicon (see Fig. 1A for their respective locations in the HCV replicon), were originally tested by using real-time RT-PCR. We selected two siRNAs, NS3-1948 and NS5B-6133 (named on the basis of their location and nucleotide start site in the HCV subgenome), as they had the greatest specific effect on HCV RNA replication. The primers used to generate the HCV-specific siRNAs were NS3-1948 5′-AAGACAGTCCAACACACGCCACCTGTCTC-3′ (sense) and 5′-AATGGCGTGTGTTGGACTGTCCCTGTCTC-3′ (antisense) and NS5B-6133 5′-AACAGTCTGTCAAAGGTGACCCCTGTCTC-3′ (sense) and 5′-AAGGTCACCTTTGACAGACTGCCTGTCTC-3′ (antisense). The scrambled siRNA (scr) was obtained from Qiagen–Xeregon (Germantown, MD). Briefly, 7–8.5 × 104 S1179I cells were plated in 12-well cluster plates. The following day, 1.25 μg of siRNA was transfected by using Oligofectamine reagent (Invitrogen). Total RNA was harvested at various times posttransfection by using TRIZOL reagent (Life Technologies, Grand Island, NY). For experiments using IFNα (PBL Biomedical Laboratories, Piscataway, NJ), S1179I cells were treated with IFNα at a final concentration of 100 units/ml, and total RNA was harvested 2 days later as discussed above.
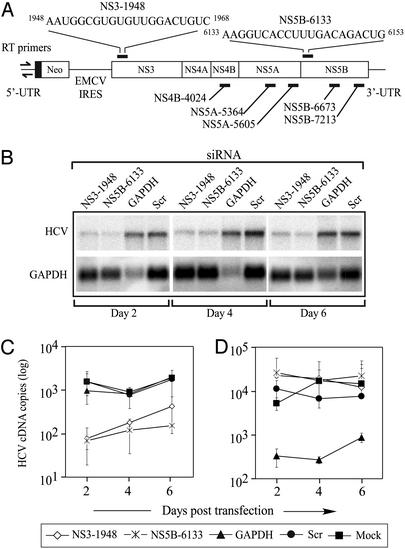
Figure 1.
(A) Schematic representing the HCV plus stranded subgenomic replicon. The ribonucleotide sequences represent the siRNA target sequences for NS3-1948 and NS5B-6133 siRNAs. Other HCV-specific siRNAs tested are also shown. The primers for real-time PCR of HCV RNA are denoted by arrows in the 5′ UTR. (B) Northern blot analysis of the effect of various siRNAs on HCV and GAPDH RNA transcript levels. Total RNA was isolated from S1179I cells that were transfected with siRNA specific for NS3, NS5B, GAPDH, or an irrelevant scrambled (scr) siRNA 2, 4, and 6 days previously. Real-time RT-PCR analysis of HCV (C) and GAPDH (D) RNA content was performed on similar RNA samples and compared with mock-transfected S1179I cells. Representative data (mean ± SEM) from at least three independent experiments are shown.
Reverse Transcription and Real-Time PCR Analysis.
One microgram of total RNA was incubated with DNase 1 by using the DNA-free kit (Ambion). cDNA was generated by using the TaqMan reverse transcription reagents kit (Applied Biosystems) according to manufacturer recommendations. Reactions with no reverse transcriptase enzyme added were performed in parallel with most experiments and yielded no PCR products. Real-time PCR (iCycler, Bio-Rad) was performed as described (58) with the following exceptions. Briefly, reactions were carried out in 25-μl volumes containing either 5 μl (when assessing HCV cDNA copy number) or 2 μl [when assessing human PKR, RNase L, MxA, and 2′,5′-oligoadenylate synthetase (OAS) cDNA copy number] of RT product. To quantitate HCV transcript levels, dilutions of plasmids containing the HCV subgenomic ORF or the human GAPDH gene were always run in parallel with cDNA from the S1179I cells for use as standard curves (dilutions ranged from 108 to 100 copies of each plasmid). The PCR primers for GAPDH are based on the human GAPDH mRNA sequence (GenBank accession no. NMX002046), and spans introns two and three of the GAPDH gene (base pairs 1,457–3,412). The PCR primers for quantitative real-time PCR were HCV 5′-ATGGCGTTAGTATGAGTGTC-3′ (sense) and 5′-GGCATTGAGCGGGTTGATC-3′ (antisense) and GAPDH 5′-GAAGGTGAAGGTCGGAGTC-3′ (sense) and 5′-GAAGATGGTGATGGGATTTC-3′ (antisense). To perform analysis of relative expression of IFN-induced genes using real-time PCR, we used the 2−ΔΔCT method (59), by normalizing to GAPDH expression levels. The PCR primers used were PKR 5′-CAAGAGGTTTGGCATGGATT-3′ (sense) and 5′-GCTCCGCCTTCTCGTTATTA-3′ (antisense); RNase L 5′-AGGCATCTACCTGGGGTCT-3′ (sense) and 5′-GACTGTTCTCTCGGCTGCTT-3′ (antisense); 2,5 OAS 5′-GCACCATCTTGGAATGTCCT-3′ (sense) and 5′-TTTGTTGGGCTGGAGAAA-3′ (antisense); and MxA 5′-ACAGGACCATCGGAATCTTG-3′ (sense) and 5′-CCCTTCTTCAGGTGGAACAC-3′ (antisense). As levels of OAS transcripts from mock- or siRNA-transfected S1179I cells were undetectable by real-time PCR, the PCR products were run on a 1.5% agarose gel to compare relative inductions of OAS levels (see Fig. 4B).
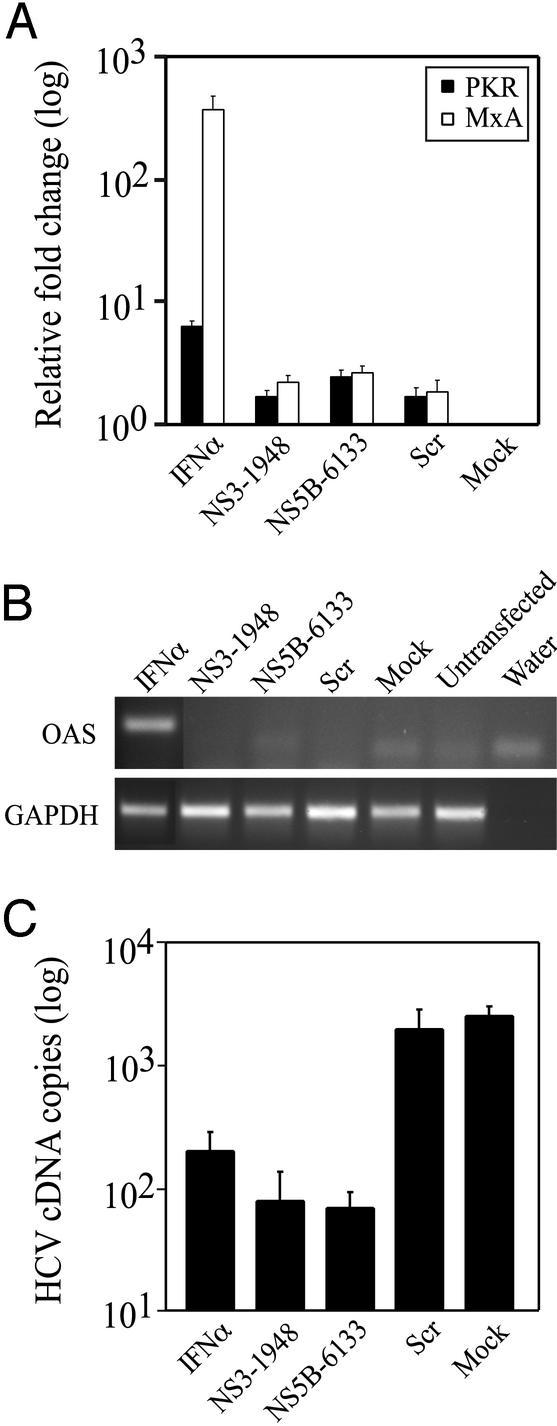
Figure 4.
(A) Effect of RNAi on IFN-induced genes. Real-time RT-PCR was used to compare the content of PKR and MxA transcripts in S1179I cells transfected with siRNA or incubated with IFNα. The relative fold induction of each transcript normalized to that seen in mock-transfected S1179I cells is shown. (B) Determination of expression of OAS and GAPDH mRNA by RT-PCR. (C) Effect of IFN treatment and RNAi on HCV replication was assessed on day 2 after IFN treatment or siRNA transfection in S1179I cells, and compared with mock-transfected S1179I cells. Representative data (mean ± SEM) from two independent experiments are shown.
Northern and Western Blot Analysis.
Total RNA was extracted from transfected cells at days 2, 4, and 6 posttransfection, and 5 μg of total RNA was loaded onto the gel. A minus stranded HCV probe was generated from a BglII fragment (base pairs 6,275–7,320) of the HCV replicon by using the MAXIscript In vitro transcription kit (Ambion). Probing for the GAPDH transcript was performed as described (60). Both probes were purified by using MicroSpin G-50 columns (Amersham Pharmacia). Blots were visualized and quantitated as described (61). Western blot analysis was performed on total lysates from mock- or siRNA-transfected S1179I cells as described (62). The gels were transferred to Hybond-N membranes (Amersham Pharmacia), blocked, and incubated with antibodies to NS3 and NS5B (M5 and M15 rabbit polyclonal antibodies, which were gifts from Darius Moradpour, University of Freiburg, Freiburg, Germany) and GAPDH (Novus Biologicals, Littleton, CO), followed by incubation with a horseradish peroxidase-conjugated goat anti-rabbit antibody (Pierce). The blots were developed with SuperSignal West Pico chemiluminescent substrate (Pierce).
Immunofluorescence Analysis of S1179I Cells.
Approximately 1.8 × 104 S1179I cells were seeded in Lab-Tek eight-chamber coverglasses (Nalge). The siRNA was labeled with Cy3 by using the Silencer siRNA labeling kit (Ambion). The next day, 1.25 μg of Cy3-labeled siRNA was transfected by using the procedure described above. At specific days posttransfection, the cells were washed three times with PBS and fixed with 4% paraformaldehyde at room temperature for 10 min. Cells were washed and incubated with PBS containing 3% BSA, 0.3% Triton X, and 10% FCS for at least 30 min. Cells were then incubated with a monoclonal antibody to HCV NS5B protein (clone 12B7, obtained from Darius Moradpour) at a final dilution of 1:100, washed three times with PBS, and incubated with an anti-mouse antibody conjugated to Cy5 at a dilution of 1:500. The cells were then incubated with bisbenzimide (0.5 μg/ml) to stain the nuclei blue and then analyzed on a DeltaVision deconvolution microscope using SOFTWORX 2.5 software. Specific deconvolved images were volume-viewed to observe more detailed relationships between various labeled components.
Statistical Analyses.
All statistical analyses were performed by using Microsoft EXCEL software. All graphs represent the mean ± SEM. P values for real-time PCR data were determined by using paired t test.
Results
RNAi Inhibits HCV Replication in Cultured Huh-7 Cells.
To determine whether siRNA specific to the HCV subgenomic replicon sequence could inhibit HCV replication, we screened the activity of seven HCV-specific siRNA molecules by real-time RT-PCR of HCV RNA, and three of the seven siRNAs inhibited HCV replication at least 10-fold. We used two of them, NS3-1948 and NS5B-6133 (named on the basis of their nucleotide location in the subgenomic replicon, Fig. 1A), as they had the greatest specific inhibition of HCV RNA replication (data not shown). We performed Northern blot analysis to determine the kinetics of RNAi using a single strand RNA probe to detect positive strand HCV transcripts (Fig. 1B). Huh-7 cells stably replicating the HCV subgenomic replicon were transfected with scrambled (scr), GAPDH-specific, or HCV-specific (NS3-1948 and NS5B-6133) siRNAs. NS3-1948 and NS5B-6133 significantly inhibited HCV transcript levels as compared with either mock- or scr siRNA-transfected S1179I cells (Fig. 1B). No HCV RNA cleavage intermediates were detected in RNA samples from NS3-1948 and NS5B-6133-treated cells (data not shown). Maximal inhibition of HCV transcript levels was detected on day 2 posttransfection (5.7- and 8.3-fold for NS3-1948 and NS5B-6133, respectively; Fig. 1B). No significant inhibition of HCV transcript levels was detected in cells transfected with the negative control scr siRNA (P = 0.4927, Fig. 1B). In addition, the GAPDH siRNA specifically down-regulated the GAPDH transcript without affecting HCV RNA levels 2 and 4 days posttransfection. These data demonstrate that RNAi can efficiently and specifically down-regulate HCV RNA levels in S1179I cells.
To more accurately quantitate the levels of HCV RNA, we performed real-time RT-PCR by using primers specific to HCV (Fig. 1A) and GAPDH. Quantitative analysis revealed that HCV transcript levels were decreased ≈21-fold (P = 0.0012) and ≈23-fold (P = 0.00044) in cells transfected with NS3-1948 and NS5B-6133, respectively, on day 2 posttransfection (Fig. 1C). HCV transcript levels were not significantly changed in S1179I cells transfected with GAPDH-specific (P = 0.1766) or scr (P = 0.4927) siRNAs (Fig. 1C). In addition, the GAPDH-specific siRNA down-regulated the GAPDH transcript ≈16- and ≈63-fold on days 2 and 4, respectively, without affecting HCV RNA levels (P = 0.032, Fig. 1D). These results demonstrate that (i) RNAi can effectively inhibit HCV replication in cultured Huh-7 cells, (ii) the down-regulation in HCV levels is sequence-specific, and (iii) the RNAi effect is effective at least 6 days posttransfection in cultured cells.
RNAi Inhibits HCV Protein Expression in Cultured Huh-7 Cells.
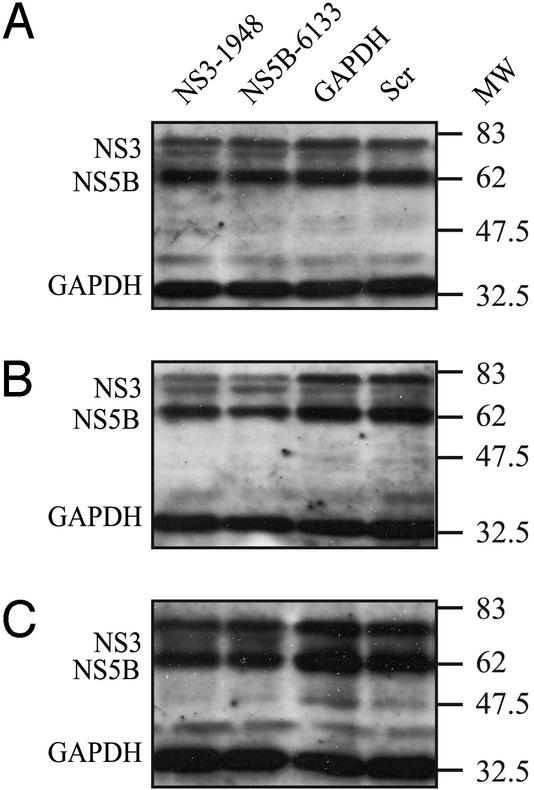
We then examined the levels of HCV protein expression in siRNA-transfected S1179I cells by Western blot analysis. As shown in Fig. 2, levels of NS3 and NS5B proteins were unchanged at 2 days posttransfection (Fig. 2A), but they decreased on day 4 (Fig. 2B) and day 6 (Fig. 2C) posttransfection in S1179I cells transfected with NS3-1948 or NS5B-6133 compared with GAPDH-specific and scr siRNAs. At all time points analyzed, HCV protein expression in mock-transfected S1179I cells was not significantly different compared with S1179I cells transfected with the scr siRNA (data not shown).
Figure 2.
The effect of RNAi on HCV protein expression in S1179I cells. Western blot analysis was performed on total cell lysates harvested from mock- or siRNA-transfected S1179I cells at various days posttransfection by using NS3- and NS5B-specific antibodies as described in Materials and Methods. Molecular mass markers (in kilodaltons) are shown to the right of each gel. Shown are representative data of three independent experiments.
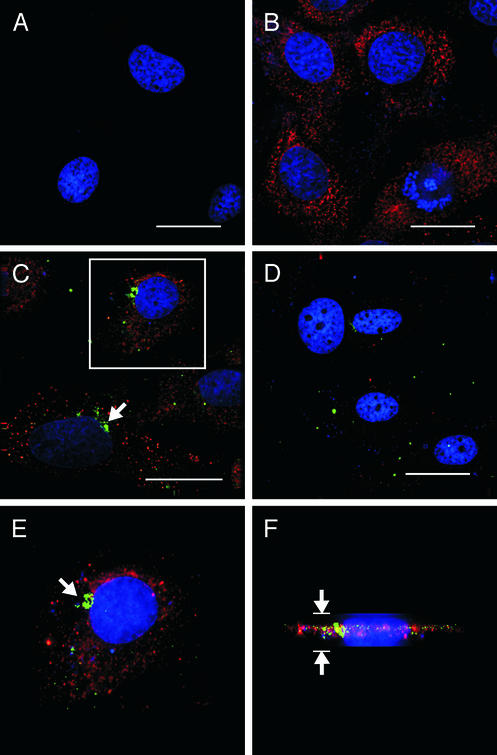
To further characterize the system at a single cell level and determine the presence of siRNA inside the cell, we transfected S1179I cells with Cy3-labeled NS3-1948 or NS5B-6133 and analyzed these cells for NS3 (data not shown) and NS5B (Fig. 3) protein expression by immunofluorescence analysis. Granular cytoplasmic NS5B staining was detected in untransfected (Fig. 3B) and scr-transfected S1179I cells (data not shown) but not in parental Huh-7 cells (Fig. 3A). On day 2, labeled NS5B-6133 siRNA could be detected in most S1179I cells as punctate cytoplasmic foci in the cytoplasm adjacent to the nucleus (Fig. 3 C and E). Using the deconvolution microscope software to rotate the image of the cell, we could demonstrate that the Cy3-labeled siRNA was present inside the cell (Fig. 3F). Interestingly, despite the presence of the siRNA, there was no detectable decrease in the level of NS5B protein expression in cells containing NS5B-6133 2 days after siRNA transfection (Fig. 3C), in keeping with the Western blot analysis (Fig. 2A). On day 4 posttransfection, however, NS5B protein expression decreased strongly in cells transfected with NS3-1948 and NS5B-6133 (Fig. 3D) as compared with mock- and scr-transfected cells (data not shown), even though the labeled NS3-1948 and NS5B-6133 siRNA foci were barely detectable (Fig. 3D).
Figure 3.
HCV NS5B and siRNA localization and expression in S1179I cells. (A) Parental Huh-7 cells were stained with an antibody to NS5B (red) and bisbenzimide, a nuclear stain (blue). Note the absence of cytoplasmic NS5B staining in these cells. (B) S1179I cells were stained with the anti-NS5B antibody and bisbenzimide. Note the granular cytoplasmic NS5B-specific staining. (C) S1179I cells transfected with NS5B-6133 siRNA were stained 2 days posttransfection with the anti-NS5B antibody. Note the perinuclear localization of the labeled siRNA (green foci) and normal NS5B protein expression (red). (D) S1179I cells transfected with Cy3-labeled NS5B-6133 were stained for NS5B protein expression 4 days posttransfection. Note the significant decrease in NS5B protein expression and the lack of perinuclear staining of the siRNA compared with that seen on day 2 posttransfection. (E) Magnification of the cell in the boxed region from C. (F) Volume view of the cell shown in E to determine a more detailed relationship between siRNA and localization in the cell. The arrows in C and E represent localization of Cy3-labeled NS5B-6133. The arrows in F represent the height of the cell. (Scale bars, 30-μm distances.)
siRNA Transfection Does Not Activate Known IFN-Inducible Pathways or Alter Huh-7 Cell Cycle Progression.
To determine whether the RNAi-mediated inhibition of HCV RNA replication in Huh-7 cells could be explained by dsRNA-induced activation of the IFN pathway, we compared levels of three well known IFN-induced genes in IFN-treated S1179I cells and siRNA-transfected S1179I cells 2 days after administration and transfection, respectively. IFNα treatment of cells induced an ≈367- and ≈6.4-fold increase in levels of MxA (P = 0.038) and PKR (P = 0.0404), respectively, compared with mock-transfected S1179I cells (Fig. 4A). In contrast, MxA and PKR mRNA expression was induced <2.7-fold (P = 0.156 and P = 0.187 for NS3-1948 and NS5B-6133, respectively) relative to mock-transfected S1179I cells (Fig. 4A). Because the scr-transfected cells displayed the same low level of induction of PKR and MxA (Fig. 4A) in the face of normal levels of HCV RNA (Fig. 1 B and C), we conclude that IFN induction does not explain the decrease in HCV replication induced by NS3-1948 and NS5B-6133 siRNAs. This was confirmed by RT-PCR analysis of IFNα-treated and siRNA-transfected cells for OAS mRNA content (Fig. 4B). We then compared the extent to which HCV RNA replication was inhibited by RNAi and IFN treatment. As shown in Fig. 4C, HCV RNA replication was inhibited ≈12.5-fold (P = 0.0052) 2 days after the addition of IFNα, whereas it was inhibited ≈32-fold (P = 0.0046) and ≈36-fold (0.0047) 2 days after transfection with NS3-1948 and NS5B-6133 siRNA (Fig. 4C). Because replication of the HCV replicon is known to be cell cycle-dependent (17), we stained S1179I cells with propidium iodide and performed flow cytometry analysis to determine whether the siRNA had any effect on the cell cycle. Cell cycle profiles of mock- or siRNA-transfected cells were identical, demonstrating that the inhibition of HCV replication was not due to an effect on cell cycle progression (data not shown).
Discussion
We have demonstrated that RNAi can block HCV replication in stably transfected Huh-7 cells in vitro. From the seven HCV-specific siRNAs tested, the greatest inhibition was detected by using the NS3-1948 and NS5B-6133 siRNAs, which inhibited HCV replication in an IFN- and cell cycle-independent manner. The HCV- and GAPDH-specific siRNAs were specific for their respective RNA transcripts, whereas the scr siRNA did not affect either HCV or GAPDH mRNA levels. By demonstrating that constitutively expressed viral and cellular transcripts can be efficiently suppressed by RNAi in Huh-7 cells, these results suggest that RNAi can be used as a tool in future experiments to study the ability of host genes to regulate HCV replication in this system.
HCV RNA replication was inhibited within 2 days of siRNA transfection, and the effect lasted at least 6 days. In contrast, the inhibition of NS3 and NS5B was delayed to day 4 posttransfection, suggesting that these proteins have relatively long half lives. The global decrease in HCV protein expression in all of the transfected S1179I cells that we detected by immunofluorescence analysis suggests that either all of the cells were transfected with the siRNA or that RNAi spreading may have occurred, as demonstrated in plants and C. elegans (63, 64). Further experiments are required to address this interesting possibility.
Labeled HCV-specific siRNA could be detected in the cytoplasm of S1179I cells on day 2, consistent with previous data demonstrating the perinuclear localization of siRNA (65). The significance of this pattern of perinuclear localization and the disappearance of these perinuclear foci by day 4 is still unknown, but it may reflect the association of the siRNA with RNA-induced silencing complexes and the relative instability of the input labeled material. Additional experiments are needed to understand the significance of these results.
Recently, many viruses have been shown to be susceptible to inhibition by RNAi, suggesting that RNAi may play an adaptive antiviral role in controlling these viral infections (46–48, 50, 51, 54, 55). In addition, some viruses have evolved mechanisms to evade RNAi. For example, the FHV induces RNAi in Drosophila cells, yet the FHV B2 protein suppresses this antiviral effect (49), presumably reflecting an attempt to escape from this endogenous antiviral host response. The mechanism for suppression of RNAi by the FHV B2 protein is currently unknown. Further studies analyzing the ability of RNAi to inhibit replication of a full-length HCV replicon are needed to determine whether HCV proteins that are not expressed by the subgenomic replicon can inhibit the antiviral effects of RNAi as described in this report.
Based on the results presented herein, it is theoretically possible that RNAi may play a role in viral clearance during natural HCV infection. These data also suggest that therapeutic induction of RNAi either alone or in combination with IFN treatment might represent an alternative approach for the treatment of chronic HCV infection.
Acknowledgments
We thank Dr. Darius Moradpour for supplying the monoclonal antibodies to NS3 and NS5B proteins and Dr. Charles Rice for providing the HCV subgenomic replicon. We also thank Dr. Stefan Wieland for helpful discussions and critiques of the manuscript and Dr. Malcolm Wood for help with the deconvolution microscope. This study was supported by National Institutes of Health Grants CA76403 and CA40489. In addition, S.B.K. was supported by a postdoctoral fellowship grant from The Skaggs Institute for Chemical Biology. This is manuscript number 15460-MEM from The Scripps Research Institute.
Abbreviations
- HCV
hepatitis C virus
- RNAi
RNA interference
- PKR
protein kinase R
- dsRNA
double-stranded RNA
- siRNA
short interfering RNA
- FHV
flock house virus
- OAS
2′,5′-oligoadenylate synthetase
References
- 1.Bradley D W. Curr Top Microbiol Immunol. 2000;242:1–23. doi: 10.1007/978-3-642-59605-6_1. [DOI] [PubMed] [Google Scholar]
- 2.Alter M J, Margolis H S, Krawczynski K, Judson F N, Mares A, Alexander W J, Hu P Y, Miller J K, Gerber M A, Sampliner R E, et al. N Engl J Med. 1992;327:1899–1905. doi: 10.1056/NEJM199212313272702. [DOI] [PubMed] [Google Scholar]
- 3.Diepolder H M, Jung M C, Keller E, Schraut W, Gerlach J T, Gruner N, Zachoval R, Hoffmann R M, Schirren C A, Scholz S, Pape G R. Clin Exp Immunol. 1998;113:244–251. doi: 10.1046/j.1365-2249.1998.00665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gruner N H, Gerlach T J, Jung M C, Diepolder H M, Schirren C A, Schraut W W, Hoffmann R, Zachoval R, Santantonio T, Cucchiarini M, et al. J Infect Dis. 2000;181:1528–1536. doi: 10.1086/315450. [DOI] [PubMed] [Google Scholar]
- 5.Chisari F V. J Clin Invest. 1997;99:1472–1477. doi: 10.1172/JCI119308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lanford R E, Bigger C. Virology. 2002;293:1–9. doi: 10.1006/viro.2001.1316. [DOI] [PubMed] [Google Scholar]
- 7.Christie J M, Chapman R W. Hosp Med. 1999;60:357–361. doi: 10.12968/hosp.1999.60.5.1116. [DOI] [PubMed] [Google Scholar]
- 8.McCaffrey A P, Ohashi K, Meuse L, Shen S, Lancaster A M, Lukavsky P J, Sarnow P, Kay M A. Mol Ther. 2002;5:676–684. doi: 10.1006/mthe.2002.0600. [DOI] [PubMed] [Google Scholar]
- 9.Trowbridge R, Gowans E J. J Viral Hepat. 1998;5:95–98. doi: 10.1046/j.1365-2893.1998.00090.x. [DOI] [PubMed] [Google Scholar]
- 10.Wang C, Sarnow P, Siddiqui A. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blight K J, Rice C M. J Virol. 1997;71:7345–7352. doi: 10.1128/jvi.71.10.7345-7352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reed K E, Rice C M. Curr Top Microbiol Immunol. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- 14.Bartenschlager R, Lohmann V. J Gen Virol. 2000;81:1631–1648. doi: 10.1099/0022-1317-81-7-1631. [DOI] [PubMed] [Google Scholar]
- 15.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda M, Yi M, Li K, Lemon S M. J Virol. 2002;76:2997–3006. doi: 10.1128/JVI.76.6.2997-3006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pietschmann T, Lohmann V, Rutter G, Kurpanek K, Bartenschlager R. J Virol. 2001;75:1252–1264. doi: 10.1128/JVI.75.3.1252-1264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blight K J, Kolykhalov A A, Rice C M. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 19.Kishine H, Sugiyama K, Hijikata M, Kato N, Takahashi H, Noshi T, Nio Y, Hosaka M, Miyanari Y, Shimotohno K. Biochem Biophys Res Commun. 2002;293:993–999. doi: 10.1016/S0006-291X(02)00342-X. [DOI] [PubMed] [Google Scholar]
- 20.Guo J T, Bichko V V, Seeger C. J Virol. 2001;75:8516–8523. doi: 10.1128/JVI.75.18.8516-8523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frese M, Pietschmann T, Moradpour D, Haller O, Bartenschlager R. J Gen Virol. 2001;82:723–733. doi: 10.1099/0022-1317-82-4-723. [DOI] [PubMed] [Google Scholar]
- 22.Frese M, Schwarzle V, Barth K, Krieger N, Lohmann V, Mihm S, Haller O, Bartenschlager R. Hepatology. 2002;35:694–703. doi: 10.1053/jhep.2002.31770. [DOI] [PubMed] [Google Scholar]
- 23.Cheney I W, Lai V C, Zhong W, Brodhag T, Dempsey S, Lim C, Hong Z, Lau J Y, Tam R C. J Virol. 2002;76:11148–11154. doi: 10.1128/JVI.76.21.11148-11154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis G L, Esteban-Mur R, Rustgi V, Hoefs J, Gordon S C, Trepo C, Shiffman M L, Zeuzem S, Craxi A, Ling M H, Albrecht J. N Engl J Med. 1998;339:1493–1499. doi: 10.1056/NEJM199811193392102. [DOI] [PubMed] [Google Scholar]
- 25.McHutchison J G, Gordon S C, Schiff E R, Shiffman M L, Lee W M, Rustgi V K, Goodman Z D, Ling M H, Cort S, Albrecht J K. N Engl J Med. 1998;339:1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- 26.Lindsay K L, Trepo C, Heintges T, Shiffman M L, Gordon S C, Hoefs J C, Schiff E R, Goodman Z D, Laughlin M, Yao R, Albrecht J K. Hepatology. 2001;34:395–403. doi: 10.1053/jhep.2001.26371. [DOI] [PubMed] [Google Scholar]
- 27.Reddy K R, Wright T L, Pockros P J, Shiffman M, Everson G, Reindollar R, Fried M W, Purdum P P, III, Jensen D, Smith C, et al. Hepatology. 2001;33:433–438. doi: 10.1053/jhep.2001.21747. [DOI] [PubMed] [Google Scholar]
- 28.Glue P, Rouzier-Panis R, Raffanel C, Sabo R, Gupta S K, Salfi M, Jacobs S, Clement R P. Hepatology. 2000;32:647–653. doi: 10.1053/jhep.2000.16661. [DOI] [PubMed] [Google Scholar]
- 29.Abid K, Quadri R, Negro F. Science. 2000;287:1555. doi: 10.1126/science.287.5458.1555a. [DOI] [PubMed] [Google Scholar]
- 30.Taylor D R, Shi S T, Romano P R, Barber G N, Lai M M. Science. 1999;285:107–110. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- 31.Gale M, Jr, Blakely C M, Kwieciszewski B, Tan S L, Dossett M, Tang N M, Korth M J, Polyak S J, Gretch D R, Katze M G. Mol Cell Biol. 1998;18:5208–5218. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gale M J, Jr, Korth M J, Tang N M, Tan S L, Hopkins D A, Dever T E, Polyak S J, Gretch D R, Katze M G. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 33.Cullen B R. Nat Immunol. 2002;3:597–599. doi: 10.1038/ni0702-597. [DOI] [PubMed] [Google Scholar]
- 34.Elbashir S M, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 35.Elbashir S M, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elbashir S M, Lendeckel W, Tuschl T. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hannon G J. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 38.Tuschl T. Chembiochem. 2001;2:239–245. doi: 10.1002/1439-7633(20010401)2:4<239::AID-CBIC239>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 39.Sharp P A. Genes Dev. 2001;15:485–490. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- 40.Moss E G. Curr Biol. 2001;11:R772–R775. doi: 10.1016/s0960-9822(01)00467-5. [DOI] [PubMed] [Google Scholar]
- 41.Yu J Y, DeRuiter S L, Turner D L. Proc Natl Acad Sci USA. 2002;99:6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brummelkamp T R, Bernards R, Agami R. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 43.Sui G, Soohoo C, Affarel B, Gay F, Shi Y, Forrester W C. Proc Natl Acad Sci USA. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cogoni C, Macino G. Curr Opin Microbiol. 1999;2:657–662. doi: 10.1016/s1369-5274(99)00041-7. [DOI] [PubMed] [Google Scholar]
- 45.Sijen T, Kooter J M. BioEssays. 2000;22:520–531. doi: 10.1002/(SICI)1521-1878(200006)22:6<520::AID-BIES5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 46.Lindenbach B D, Rice C M. Mol Cell. 2002;9:925–927. doi: 10.1016/s1097-2765(02)00539-7. [DOI] [PubMed] [Google Scholar]
- 47.Novina C D, Murray M F, Dykxhoorn D M, Beresford P J, Riess J, Lee S K, Collman R G, Lieberman J, Shankar P, Sharp P A. Nat Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- 48.Jacque J M, Triques K, Stevenson M. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Li W X, Ding S W. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 50.Capodici J, Kariko K, Weissman D. J Immunol. 2002;169:5196–5201. doi: 10.4049/jimmunol.169.9.5196. [DOI] [PubMed] [Google Scholar]
- 51.Hu W, Myers C, Kilzer J, Pfaff S, Bushman F. Curr Biol. 2002;12:1301. doi: 10.1016/s0960-9822(02)00975-2. [DOI] [PubMed] [Google Scholar]
- 52.Gitlin L, Karelsky S, Andino R. Nature. 2002;418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- 53.Carmichael G G. Nature. 2002;418:379–380. doi: 10.1038/418379a. [DOI] [PubMed] [Google Scholar]
- 54.Adelman Z N, Sanchez-Vargas I, Travanty E A, Carlson J O, Beaty B J, Blair C D, Olson K E. J Virol. 2002;76:12925–12933. doi: 10.1128/JVI.76.24.12925-12933.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park W S, Miyano-Kurosaki N, Hayafune M, Nakajima E, Matsuzaki T, Shimada F, Takaku H. Nucleic Acids Res. 2002;30:4830–4835. doi: 10.1093/nar/gkf627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Surabhi R M, Gaynor R B. J Virol. 2002;76:12963–12973. doi: 10.1128/JVI.76.24.12963-12973.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCaffrey A P, Meuse L, Pham T T, Conklin D S, Hannon G J, Kay M A. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 58.van Berkel V, Levine B, Kapadia S B, Goldman J E, Speck S H, Virgin H W T. J Clin Invest. 2002;109:905–914. doi: 10.1172/JCI14358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Livak K J, Schmittgen T D. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 60.Guidotti L G, Guilhot S, Chisari F V. J Virol. 1994;68:1265–1270. doi: 10.1128/jvi.68.3.1265-1270.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang P L, Althage A, Chung J, Chisari F V. Proc Natl Acad Sci USA. 2002;99:13825–13830. doi: 10.1073/pnas.202398599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kapadia S B, Molina H, van Berkel V, Speck S H, Virgin H W T. J Virol. 1999;73:7658–7670. doi: 10.1128/jvi.73.9.7658-7670.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 64.Palauqui J C, Elmayan T, Pollien J M, Vaucheret H. EMBO J. 1997;16:4738–4745. doi: 10.1093/emboj/16.15.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bryom M, Pallota V, Brown D, Ford L. Ambion TechNotes. 2002;9:3–5. [Google Scholar]