Abstract
The acid-sensing ion channel-1 (ASIC1) contributes to synaptic plasticity and may influence the response to cerebral ischemia and acidosis. We found that cAMP-dependent protein kinase phosphorylated heterologously expressed ASIC1 and endogenous ASIC1 in brain slices. ASIC1 also showed significant phosphorylation under basal conditions. Previous studies showed that the extreme C-terminal residues of ASIC1 bind the PDZ domain of the protein interacting with C-kinase-1 (PICK1). We found that protein kinase A phosphorylation of Ser-479 in the ASIC1 C terminus interfered with PICK1 binding. In contrast, minimizing phosphorylation or mutating Ser-479 to Ala enhanced PICK1 binding. Phosphorylation-dependent disruption of PICK1 binding reduced the cellular colocalization of ASIC1 and PICK1. Thus, the ASIC1 C terminus contains two sites that influence its binding to PICK1. Regulation of this interaction by phosphorylation provides a mechanism to control the cellular localization of ASIC1.
The degenerin/epithelial Na+ channel (DEG/ENaC) cation channel family includes five H+-gated members: acid-sensing ion channel (ASIC)1a, -1b, -2a, -2b, and -3 (“a” and “b” refer to alternatively spliced isoforms) (1–3). ASIC1a, -2a, and -2b share similar expression patterns in the central nervous system, with transcripts most abundant in the hippocampus, cortex, and Purkinje layer of the cerebellum (4–6). ASIC3 is predominately expressed in the peripheral nervous system (7). The molecular diversity of these channels is further expanded when individual ASIC subunits heteromultimerize to generate cation channels with properties different from homomultimers.
Like other DEG/ENaC subunits, ASICs have two transmembrane sequences, a large extracellular domain containing several conserved cysteines, and intracellular N and C termini (1–3). Previous studies have shown that the C termini of ASIC1, -2, and -3 participate in protein interactions. ASIC1 and -2 C termini share homology with type-II PDZ binding motifs and bind the protein interacting with C-kinase-1 (PICK1) (8, 9). PICK1 and ASIC1 are coexpressed throughout the brain, colocalize at synaptic sites in hippocampal neurons, and copurify from synaptosomes (8, 9). The ASIC3 C terminus binds channel-interacting PDZ domain protein (CIPP) (10). The function of these interactions is not well understood, although it has been suggested that they regulate the transport and distribution of the channels in nerve terminals.
ASIC1 contributes to the normal function of central neurons. Disrupting the mouse ASIC1 gene markedly reduced H+-gated currents in hippocampal neurons, impaired long-term potentiation of synaptic transmission, and produced learning and memory deficits (11). The observation that extracellular acidosis activates ASIC1 has suggested that it may also be activated in pathologic conditions, for example, when extracellular pH falls during cerebral ischemia (12, 13). Because protein kinases regulate the function of many brain ion channels, we asked whether ASIC channels are phosphorylated. We focused specifically on ASIC1, because it contains a conserved consensus site for phosphorylation by protein kinase A (PKA) and because it is the most pH-sensitive subunit in the brain.
Materials and Methods
Materials.
PKA and protein kinase C (PKC) isoforms were purchased from Promega. Purified recombinant calcium/calmodulin-dependent protein kinase II (CaMKII) was kindly provided by J. W. Hell (University of Iowa, Iowa City). [32P]ATP (111 TBq/mmol) was obtained from New England Nuclear/Dupont. Protein A-Sepharose was purchased from Amersham Pharmacia–Pharmacia. Forskolin and 3-isobutyl-1-methylxanthine (IBMX) were purchased from Sigma. Tetrodotoxin, microcystin-LR, the PKA inhibitor KT5720, and protein kinase A inhibitor peptide were purchased from Calbiochem–Novabiochem. PICK1 antibodies were purchased from Santa Cruz Biotechnology. Secondary horseradish peroxidase-conjugated antibodies were received from Bio-Rad. Streptavidin-coated plates and all developing reagents were purchased from Pierce. ASIC1 wild-type peptide (ASIC1/PEP) (biotin–RRGKCQKEAKRSSADKGVALSLDD), ASIC1/PEP-S478A peptide (biotin–RRGKCQKEAKRASADKGVALSLDD), ASIC1/PEP-S479A peptide (biotin–RRGKCQKEAKRSAADKGVALSLDD), and ASIC3/PEP (biotin–FWNRRSSQRRSGNTLLQE) were synthesized at the Howard Hughes Medical Institute/Keck Facility at Yale University. All other reagents were purchased from commercial suppliers and were of standard biochemical quality.
Fusion Proteins.
For the production of polyhistidine fusion proteins containing the C-terminal region (ASIC/C-term), oligonucleotides flanking the indicated regions and containing appropriate restriction sites were used to amplify corresponding sequences from cDNA clones of hASIC1a (amino acids 459–528), hASIC2a (amino acids 466–512), and hASIC3 (amino acids 449–513) by PCR. The DNA products were ligated into pET-32a vector (Novagen). Fusion proteins were expressed in Escherichia coli (BL21-codon plus; Stratagene) and purified with Ni2+ chelate resin nickel–nitrolotriacetic acid (Sepharose, Qiagen, Chatsworth, CA) according to the protocols suggested by the manufacturer.
Phosphorylation Assays.
The resins with protein bound were phosphorylated as described (14) with 0.5–1 μg of PKA, PKC, or CaMKII in 50 μl of basic phosphorylation buffer, and 0.2 μM [32P]ATP. The pellets were extracted with SDS sample buffer and used directly for SDS/PAGE. Phosphorylated protein bands were visualized by autoradiography. For quantitative phosphorylation as required for the measurement of the stoichiometry of 32P phosphate incorporation by PKA or PKC, 10 μM unlabeled ATP was added to the phosphorylation reaction. At this ATP concentration, phosphorylation of ASIC subunits by PKA or PKC was saturated. Extending the incubation time to 1 h also did not result in an increase in 32P phosphate incorporation by PKA or PKC. Quantification of 32P incorporation into milligrams of ASIC1/C-term was through scintillation counting.
Immunoprecipitation and Immunoblotting.
COS-7 cells transfected by electroporation were solubilized in 1% Triton X-100/0.5% deoxycholate/0.1% SDS/10 mM Tris⋅Cl, pH 7.4/150 mM NaCl/10 mM EDTA/10 mM EGTA, with a glass/Teflon homogenizer. Pepstatin A (1 μM), leupeptin, and aprotinin (10 μg/ml), Pefabloc (0.2 mM), benzamidine (0.1 mg/ml), p-nitrophenyl phosphate (1 mM), and microcystin-LR (2 μM) were present in all buffers. Unsolubilized material was sedimented by ultracentrifugation (100,000 × g, 30 min; 70.1 Ti rotor, 4°C), and the supernatant was collected and stored at −80°C.
ASIC1 was precipitated by using antibody 6.4 (human ASIC1a), which specifically binds to ASIC1 (11). For control immunoprecipitations, nonspecific rabbit IgG antibodies were obtained from Jackson Laboratories. Then 3–5 mg of protein A Sepharose, preswollen and washed three times with TBS (150 mM NaCl/10 mM Tris⋅Cl, pH 7.4) containing 1% Triton X-100, were added, and the samples were mixed for 2.5 h as before. The immunocomplexes were sedimented by centrifugation and washed three times with 1% Triton X-100 in TBS and once with 10 mM Tris⋅Cl, pH 7.4, before being extracted with 20 μl of SDS sample buffer (2% SDS/20 mM DTT/10% sucrose/125 mM Tris⋅Cl, pH 6.8) for 20 min at 60°C.
After separation by SDS/PAGE, the proteins were transferred onto nitrocellulose, which was blocked by incubation for 2 h with 3% BSA in TBS (TBS-BSA). Blots were incubated with ASIC1 or PICK1 antibody (1:1,000 in TBS-BSA) for 1–2 h, washed five times with TBS-BSA, incubated with horseradish–peroxidase (HRP)-labeled protein A or HRP-labeled donkey anti-goat respectively (diluted 1:1,000 in TBS-BSA), washed for 3–4 h with 0.05% Tween 20 in TBS (8–10 changes), and developed with the enhanced chemiluminescence reagent (Pierce).
Stimulation of Coronal Mouse Brain Slices.
Acute hippocampal slices (0.4 mm) were prepared from 3-wk-old C57-black mice (Harlan Breeders, Indianapolis) by using a vibratome and treated as described (15). Supernatants were used immediately for immunoprecipitation by using hASIC1 or control antibodies.
Association of Fusion Proteins.
Two micrograms of affinity-purified ASIC1/C–term polyhistidine fusion proteins [as estimated by SDS/PAGE and subsequent staining with Coomassie brilliant blue in addition to quantitative bicinchoninic acid- protein assay (Pierce)] were immobilized for binding with 1.5 μg of purified PICK1 GST fusion protein. Fusion proteins were combined with the indicated conditions in TBS with 0.1% TX-100 for 30 min, while tilting at 4°C. Samples were rapidly washed three times with the same buffer and once with 10 mM Tris⋅Cl, pH 7.4, and then extracted with SDS sample buffer for immunoblotting as described. The IC50 value and the curve fitting for the ASIC1 peptide to inhibit this interaction were obtained from a dose response of experimental data performed in prism software (Ver. 3.02, GraphPad Software, San Diego).
Fluorescence Imaging.
COS-7 cells were transfected by electroporation, and rat E18 hippocampal neurons (G. Brewer, Southern Illinois University School of Medicine, Springfield) in primary culture for 2–3 days were transfected as described (8) with hASIC1a subcloned into pEGFP-C2 (CLONTECH). Site-directed mutagenesis of ASIC1 was performed by PCR according to the QuikChange Mutagenesis Kit protocol (Stratagene). hPICK1-DsRed was obtained by subcloning into pDsRed-N1 (CLONTECH) as described (8). All constructs were driven by a cytomegalovirus promoter and were confirmed by sequencing. After 24–48 h, cells were washed once with the recording medium (in mM: NaCl 140/KCl 5/MgCl2 1/CaCl2 1.5/glucose 5/Hepes 10, pH 7.4) and were mounted in the recording chamber. Optical recordings were performed on a Nikon Eclipse TE200 microscope with an I-PentaMAX 512 × 512 charge-coupled device camera (Princeton Instruments, Trenton, NJ). A ×100 oil SuperFluor objective (Nikon), a dichroic Quad 84000 filter set (Chroma Technology, Brattleboro, VT), and excitation filters 490/20 for enhanced GFP and 555/28 for DsRed were used for all recordings. Recordings and image analysis including region measurements were performed by using metamorph software (Universal Imaging, Media, PA).
Results
The Intracellular C Termini of ASIC1 and ASIC3 Are Substrates for PKA.
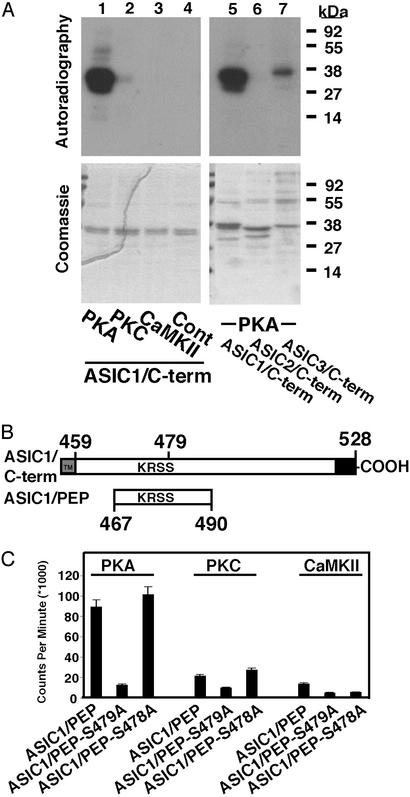
To determine whether ASIC1 is a substrate for phosphorylation, we examined the affinity-purified C terminus of ASIC1 (ASIC1/C-term). PKA phosphorylated the protein (Fig. 1A) with a stoichiometry of almost 1 mol of phosphate per mol of protein. In contrast, there was little phosphorylation by PKC and none with CaMKII.
Figure 1.
The C termini of ASIC1 and ASIC3 are substrates for PKA. (A) Affinity-purified ASIC1/C-term was immobilized on Ni2+ agarose beads for in vitro kinase assays with [32P]ATP and PKA, PKC, or CaMKII (lanes 1–3), or with kinase omitted (lane 4). The C termini of ASIC2 (ASIC2/C-term) and ASIC3 (ASIC3/C-term) were also tested with PKA (lanes 6 and 7). Autoradiography (Upper) and Coomassie staining (Lower) are shown. (B) The intracellular C terminus of ASIC1 contains the conserved PKA phosphorylation site Ser-479 and the PICK1 interaction site at the extreme C terminus. The second transmembrane domain is indicated. The location of the 24-aa ASIC1/PEP is indicated. (C) For in vitro kinase assays with PKA, PKC, or CaMKII, 24-aa peptides were synthesized with Ser-478 (ASIC1/PEP-S478A) or Ser-479 (ASIC1/PEP-S479A) mutated to alanine. Peptides were biotinylated at the N terminus and immobilized on streptavidin-coated plates for the in vitro kinase assays. After quantification of 32P incorporation through scintillation counting, results were plotted as absolute cpm (n = 9, error bars are SD).
Both ASIC2 and ASIC3 complex with ASIC1 to form heteromultimeric channels (6, 16–18). We found that ASIC2/C-term was not a substrate for PKA, whereas ASIC3/C-term showed significantly less phosphorylation (Fig. 1A). Thus, ASIC1/C-term showed a high substrate specificity for PKA.
PKA Phosphorylates Ser-479 of ASIC1.
ASIC1/C-term contains two potential PKA consensus phosphorylation sites, Ser-478 and -479 (Fig. 1B). To identify the PKA phosphorylated residues, we synthesized 24-aa peptides (residues 467–490, Fig. 1B) with the wild-type sequence or with the candidate Ser replaced by Ala. PKA phosphorylated the wild-type peptide (Fig. 1C). Changing Ser-479 to an Ala, ASIC1/PEP-S479A, eliminated phosphorylation, whereas replacing Ser-478 had no effect. These data suggest that Ser-479 is the PKA phosphorylation site. In parallel experiments, we found little PKC or CaMKII-dependent phosphorylation (Fig. 1C), confirming that this region is predominantly a PKA substrate.
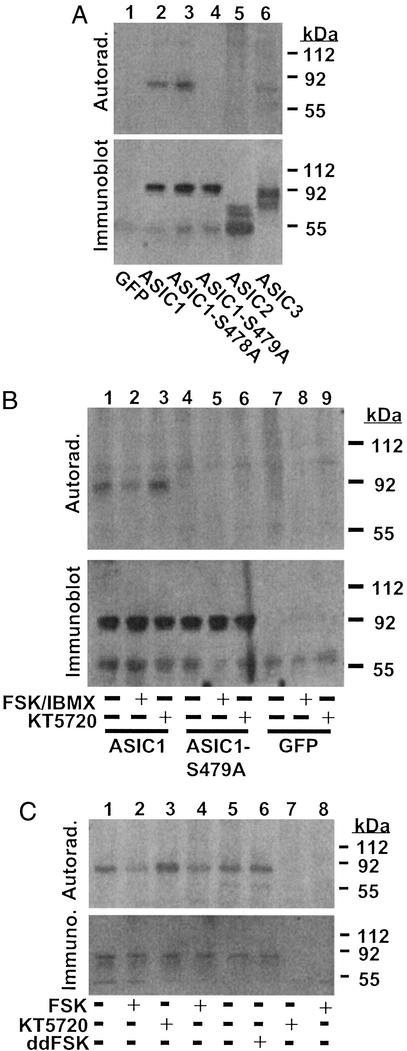
To determine whether PKA phosphorylates full-length ASIC1, we expressed the channel in COS-7 cells, immunopurified it, and tested PKA phosphorylation in vitro. Fig. 2A shows that ASIC1 was a PKA substrate. Mutation of Ser-479 but not Ser-478 abolished phosphorylation. ASIC3, but not ASIC2, showed slight PKA-dependent phosphorylation. These data indicate that full-length ASIC1 is a PKA substrate and that Ser-479 is the phosphorylation site.
Figure 2.
PKA phosphorylates Ser-479 of ASIC1. (A) The indicated full-length ASIC1 proteins were solubilized from transfected COS-7 cells, immunopurified, tested as a substrate for PKA with [32P]ATP, and separated by SDS/PAGE. GFP served as an additional control. (A–C Upper) Autoradiograms. (A–C Lower) immunoblots with ASIC-specific antibodies. (B) Cells transfected with indicated constructs were treated with 10 μM forskolin and 100 μM IBMX, 1 μM KT5720, or vehicle and assayed via back-phosphorylation to detect PKA-dependent phosphorylation (n = 3). (C) ASIC1 was immunopurified from mouse brain sections that were treated with vehicle, 10 μM forskolin, and 100 μM IBMX, 1 μM KT5720, or 10 μM dideoxy-forskolin and assayed via back-phosphorylation to detect PKA-dependent phosphorylation. Control immunoprecipitations used preimmune sera (lanes 7–8) (n = 3).
PKA-Dependent Phosphorylation of ASIC1 in COS Cells and Brain Slices.
To test ASIC1 phosphorylation in cells, COS cells were treated with cAMP agonists (10 μM forskolin and 100 μM IBMX) or a specific PKA inhibitor (1 μM KT5720) (19) (Fig. 2B). To detect phosphorylation, immunopurified ASIC1 was assayed via back-phosphorylation, which is a measure of in vivo phosphorylation through protection of sites in vitro (15). PKA activation increased ASIC1 phosphorylation in vivo, as indicated by decreased in vitro phosphorylation, whereas PKA inhibition reduced phosphorylation in vivo (Fig. 2B). These results indicate that ASIC1 is a PKA substrate in cells, and that the channel is phosphorylated under basal conditions. There was no PKA-dependent phosphorylation of ASIC1-S479A, even after application of KT5720 to reduce the possibility that an additional PKA site was constitutively phosphorylated in cells (Fig. 2B). Control immunoprecipitations from GFP-transfected cells and immunoblotting with anti-ASIC1 antibody indicate that the results were specific.
To determine whether ASIC1 is a PKA substrate in intact neurons, we treated mouse brain slices with agents to activate or inhibit PKA, immunopurified ASIC1, and used back-phosphorylation to assess phosphorylation status. The cAMP agonist forskolin, but not the inactive isomer 1,9-dideoxyforskolin, increased ASIC1 phosphorylation (Fig. 2C). Inhibiting PKA with KT5720 had the opposite effect, indicating that some ASIC1 is phosphorylated in brain under basal conditions.
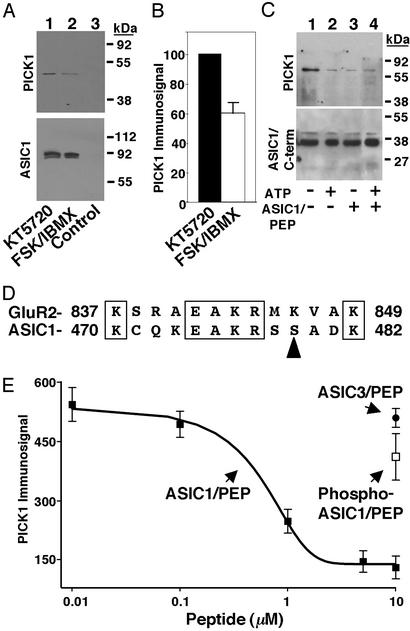
PKA Phosphorylation of ASIC1 Reduces PICK1 Binding.
Previous studies showed that the PDZ domain of PICK1 interacts with the extreme C terminus of ASIC1 (8, 9). We tested the hypothesis that phosphorylation of ASIC1 would alter this interaction. We coexpressed PICK1 and ASIC1 and treated the cells with KT5720 to reduce the basal level of ASIC1 phosphorylation. Immunoprecipitating ASIC1 specifically coprecipitated PICK1 (Fig. 3 A and B; see also ref. 8). However, treating cells with cAMP agonists reduced coprecipitation by 40%. To ensure that it was the phosphorylation of ASIC1 that influenced PICK1 binding, we used the affinity-purified ASIC1/C-term and examined binding of purified GST-tagged PICK1. When ASIC1/C-term was prephosphorylated with PKA, binding of PICK1 fell (Fig. 3C, lanes 1 and 2). These findings suggest a novel mechanism in which phosphorylation regulates PICK1 binding.
Figure 3.
PKA phosphorylation of ASIC1 reduces PICK1 binding. (A) Full-length ASIC1 and PICK1 were coexpressed in COS-7 cells and treated with 1 μM KT5720 or 10 μM forskolin and 100 μM IBMX. ASIC1 was immunoprecipitated and then gels were immunoblotted for PICK1 (Upper) or ASIC1 (Lower). Control immunoprecipitations were performed with preimmune sera. (B) Quantification of immunosignal for PICK1 (n = 3, error bars are SD). (C) Affinity-purified ASIC1/C-term (2 μg) was incubated with PKA in the absence (−) or presence (+) of 1 mM ATP and then tested for binding to 1 μM purified PICK1. ASIC1/PEP (10 μM) was included in the PICK1-binding assay, as indicated. Samples were immunoblotted with a PICK1 (Upper) or ASIC1 (Lower) antibody. (D) Sequence alignment between GluR2 and ASIC1. Boxed regions show conserved amino acids, and arrowhead indicates PKA phosphorylation site S479. (E) The affinity-purified ASIC1/C-term was immobilized for in vitro binding assays to purified PICK1. ASIC1/PEP was added as an inhibitor of PICK1 binding (filled squares). As a control, ASIC1/PEP was prephosphorylated with PKA and the PKA inactivated and then added at 10 μM (open square). As an additional control, ASIC3/PEP was added at 10 μM (filled circle).
Recent studies demonstrate that the coiled-coiled domain of PICK1 contributes to its binding to α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors in a juxtamembrane region of GluR2 (amino acids 842–853) (20). This sequence in GluR2 shares some sequence similarity with the juxtamembrane region of ASIC1 (Fig. 3D); importantly, this portion of ASIC1 includes the PKA phosphorylation site, Ser-479. To test whether this juxtamembrane region of ASIC1 binds PICK1, we assayed the ability of an ASIC1 wild-type peptide (ASIC1/PEP, Fig. 1B) to inhibit binding. Adding 10 μM ASIC1/PEP reduced the ability of ASIC1/C-term to pull down PICK1 (Fig. 3C). Prephosphorylating ASIC1/C-term did not further reduce PICK1 binding, suggesting that these interventions were not additive (Fig. 3C). Varying the concentration of ASIC1/PEP showed that inhibition was saturable with an IC50 of ≈0.5 μM (Fig. 3E). If ASIC1/PEP was phosphorylated with PKA, it lost the ability to inhibit PICK1 binding to ASIC1/C-term (Fig. 3E). The specificity of the effect was confirmed by the inability of an ASIC3 peptide (ASIC3/PEP) from the corresponding location to inhibit the interaction. These data suggest that two regions in the ASIC1 C terminus bind PICK1 and offer a mechanism for how phosphorylation regulates this interaction.
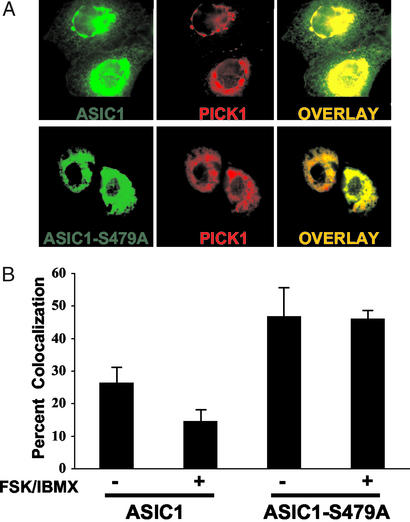
PKA Phosphorylation of ASIC1 Reduces Colocalization with PICK1.
When coexpressed in heterologous cells, PICK1 and ASIC1 colocalize in intracellular clusters (8, 9). Our finding that ASIC1 phosphorylation reduced PICK1 binding suggested that the phosphorylation status of ASIC1 would influence colocalization. To test this hypothesis, we cotransfected COS-7 cells with PICK1 and ASIC1 or ASIC1-S479A and examined the protein distribution in the presence and absence of cAMP agonists. Fig. 4A shows that ASIC1 and PICK1 colocalized in clusters, but ASIC1 was also present in the periphery of the cells not colocalized with PICK1. In contrast, ASIC1-S479A colocalized almost completely with PICK1. A quantitative analysis is shown in Fig. 4B; colocalization with PICK1 was greater with ASIC1-S479A than with wild-type ASIC1, and cAMP agonists further reduced PICK1 colocalization with ASIC1 but not with ASIC1-S479A. When expressed alone, the localization of ASIC1 and ASIC1-S479A are diffuse and unaffected by PKA activation (data not shown).
Figure 4.
PKA phosphorylation of ASIC1 reduces colocalization with PICK1. (A) ASIC1 (Upper) or ASIC1-S479A (Lower) were coexpressed with PICK1 in COS-7 cells and imaged live by using fluorescence microscopy. (B) Percent colocalization was calculated before (−) and after (+) 20 min treatment with 10 μM forskolin and 100 μM IBMX in the same cell. Cells were chosen by a blinded observer and percent colocalization was determined by METAMORPH software with parameters set for overlapping area (n = 50, error bars are SD).
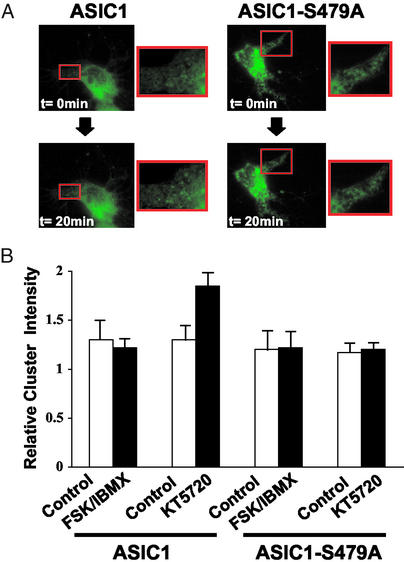
PKA Phosphorylation of ASIC1 Reduces Clustering in Hippocampal Neurons.
In hippocampal neurons, ASIC1 immunostaining is detected in a synaptic pattern and colocalizes with PSD95 (8, 11). To test the effect of phosphorylating ASIC1, we compared live fluorescent images of ASIC1 to that of ASIC1-S479A. Consistent with previous results, we found ASIC1 in a punctate and diffuse pattern (Fig. 5A Upper Left). Because our results indicated that ASIC1 showed substantial phosphorylation under baseline conditions, we applied KT5720 and 20 min later found an increase in the punctate distribution of ASIC1 (Fig. 5A Lower Left). ASIC1-S479A showed a similar appearance to ASIC1 with a punctate and diffuse appearance in the cell soma and proximal dendrites, but KT5720 did not alter its distribution (Fig. 5A Right). A quantitative analysis indicated that inhibiting PKA increased the intensity of ASIC1 in clusters (Fig. 5B). cAMP agonists had no measurable effect, consistent with significant basal phosphorylation. In contrast, neither stimulating nor inhibiting PKA altered the distribution of ASIC1-S479A.
Figure 5.
PKA phosphorylation of ASIC1 reduces clustering in hippocampal neurons. (A) Rat hippocampal neurons were transfected with ASIC1 (Left) or ASIC1-S479A (Right). Neurons were imaged live with fluorescence microscopy before (Upper) and 20 min after (Lower) addition of 1 μM KT5720. (Insets) Enlargements of boxed areas. (B) Relative cluster intensity was calculated before (−) and 20 min after (+) adding 1 μM KT5720 in the same cell. A total of 30 clusters per neuron were analyzed with METAMORPH software (n = 3, error bars are SD). Images were thresholded to define clusters. Relative cluster intensity was a ratio of cluster intensity and immediate background.
To test whether phosphorylating ASIC1 influences H+-gated currents, we recorded whole-cell currents from cultured hippocampal neurons and COS-7 cells expressing ASIC1 and PICK1. We applied cAMP agonists (forskolin and IBMX) and PKA inhibitor (KT5720); neither intervention significantly altered acid-evoked currents (data not shown). Thus, PKA-dependent phosphorylation appeared to have little direct effect on H+-gated ASIC1 current, as expected from previous results characterizing the channel's interaction with PICK1 (8, 9).
Discussion
Our data show that PKA phosphorylated ASIC1 at Ser-479 in the intracellular C-terminal tail. In the absence of an added stimulus, there was significant phosphorylation of ASIC1 expressed in heterologous cells and endogenous ASIC1 in brain slices. cAMP agonists further increased phosphorylation. Although ASIC1 is the first member of the DEG/ENaC family demonstrated to be a direct substrate for PKA, phosphorylation of associated proteins has been shown to indirectly regulate the activity and trafficking of the ENaC family members (21–23), and ASIC1 and ASIC2 have been reported to be phosphorylated by PKC (24, 25). Whereas the C terminus of ASIC1 was an excellent PKA substrate, ASIC2 was not phosphorylated, and ASIC3 showed less phosphorylation.
Previous studies demonstrated that the last four amino acids of ASIC1 bind the PDZ domain of PICK1 (8, 9). Our results indicated that an additional site in the ASIC1 C terminus contributes to PICK1 binding. Finding that ASIC1 contains a second PICK1-binding site is similar to the observation that GluR2 binding to PICK1 involves both a PDZ interaction and an interaction between a sequence in the GluR2 C terminus and the coiled-coiled domain in PICK1 (20, 26). Interestingly, the PICK1-interacting sequence we identified in ASIC1 shows some sequence similarity to that in GluR2.
The physiologic significance of ASIC1 phosphorylation was shown by its influence on PICK1 binding. The phosphorylation site, Ser-479, lies in the sequence involved in binding to PICK1. Minimizing Ser-479 phosphorylation or mutating Ser-479 to Ala enhanced ASIC1 binding to PICK1. Conversely, phosphorylating Ser-479 reduced ASIC1 binding to PICK1. These data are consistent with other reports of phosphorylation-dependent regulation of protein binding to coiled-coiled domains (27).
Earlier studies showed that PICK1 transfected into hippocampal neurons localized at glutamatergic synapses (28). PICK1 participated in glutamate receptor trafficking by clustering the receptor to internalized pools (26, 29, 30). Previous work has also shown that ASIC1 colocalized with PICK1 at synapses (8, 11). Our present data show that this interaction is regulated by phosphorylation of ASIC1. PKA-dependent phosphorylation of ASIC1 altered the channel's subcellular distribution and reduced its colocalization with PICK1. Thus, much like the trafficking of glutamate receptors to the synapse, the delivery of ASIC1 subunits is likely regulated by a combination of protein interactions and posttranslational modifications.
The phosphorylation of ASIC1 also contributes to our understanding of these channels as signal integrators (31). ASIC channel activity is regulated by extracellular protons (2), neuropeptides (32, 33), extracellular calcium (34–36), zinc (37), temperature (38), and mechanical stimuli (39, 40). Our current data indicate that ASIC1 is also a target for an intracellular signal-transduction cascade that may connect stimuli from both external and internal sources.
Acknowledgments
A.S.L., A.H.-H., and C.C.A. are Postdoctoral Associates, and M.J.W. is an Investigator of Howard Hughes Medical Institute (HHMI). We thank the University of Iowa DNA Core Facility (supported by National Institutes of Health Grant DK25295) for assistance with sequencing and oligonucleotide synthesis and the University of Iowa Central Microscopy Research Facility. This work was supported by HHMI. J.A.W. was supported by the Veterans Administration Research Career Development Award.
Abbreviations
- ASIC1
acid-sensing ion channel-1
- ASIC1/PEP
ASIC1 peptide
- PKA
protein kinase A
- IBMX
3-isobutyl-1-methylxanthine
- ASIC/C-term
polyhistidine fusion proteins containing the C-terminal region
- CaMKII
calmodulin-dependent protein kinase II
References
- 1.Mano I, Driscoll M. BioEssays. 1999;21:568–578. doi: 10.1002/(SICI)1521-1878(199907)21:7<568::AID-BIES5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 2.Waldmann R, Lazdunski M. Curr Opin Neurobiol. 1998;8:418–424. doi: 10.1016/s0959-4388(98)80070-6. [DOI] [PubMed] [Google Scholar]
- 3.Welsh M J, Price M P, Xie J. J Biol Chem. 2002;277:2369–2372. doi: 10.1074/jbc.R100060200. [DOI] [PubMed] [Google Scholar]
- 4.Waldmann R, Champigny G, Bassilana F, Heurteaux C, Lazdunski M. Nature. 1997;386:173–177. doi: 10.1038/386173a0. [DOI] [PubMed] [Google Scholar]
- 5.García-Añoveros J, Derfler B, Neville-Golden J, Hyman B T, Corey D P. Proc Natl Acad Sci USA. 1997;94:1459–1464. doi: 10.1073/pnas.94.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lingueglia E, de Weille J R, Bassilana F, Heurteaux C, Sakai H, Waldmann R, Lazdunski M. J Biol Chem. 1997;272:29778–29783. doi: 10.1074/jbc.272.47.29778. [DOI] [PubMed] [Google Scholar]
- 7.Waldmann R, Bassilana F, de Weille J R, Champigny G, Heurteaux C, Lazdunski M. J Biol Chem. 1997;272:20975–20978. doi: 10.1074/jbc.272.34.20975. [DOI] [PubMed] [Google Scholar]
- 8.Hruska-Hageman A M, Wemmie J A, Price M P, Welsh M J. Biochem J. 2001;361:443–450. doi: 10.1042/0264-6021:3610443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duggan A, Garcia-Anoveros J, Corey D P. J Biol Chem. 2002;277:5203–5208. doi: 10.1074/jbc.M104748200. [DOI] [PubMed] [Google Scholar]
- 10.Anzai N, Deval E, Schaefer L, Friend V, Lazdunski M, Lingueglia E. J Biol Chem. 2002;277:16655–16661. doi: 10.1074/jbc.M201087200. [DOI] [PubMed] [Google Scholar]
- 11.Wemmie J A, Chen J, Askwith C C, Hruska-Hageman A M, Price M P, Nolan B C, Yoder P G, Lamani E, Hoshi T, Freeman J H J, Welsh M J. Neuron. 2002;34:463–477. doi: 10.1016/s0896-6273(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 12.Obrenovitch T P, Garofalo O, Harris R J, Bordi L, Ono M, Momma F, Bachelard H S, Symon L. J Cereb Blood Flow Metab. 1988;8:866–874. doi: 10.1038/jcbfm.1988.144. [DOI] [PubMed] [Google Scholar]
- 13.Johnson M B, Jin K, Minami M, Chen D, Simon R P. J Cereb Blood Flow Metab. 2001;21:734–740. doi: 10.1097/00004647-200106000-00011. [DOI] [PubMed] [Google Scholar]
- 14.Leonard A S, Hell J W. J Biol Chem. 1997;272:12107–12115. doi: 10.1074/jbc.272.18.12107. [DOI] [PubMed] [Google Scholar]
- 15.Leonard A S, Lim I A, Hemsworth D E, Horne M C, Hell J W. Proc Natl Acad Sci USA. 1999;96:3239–3244. doi: 10.1073/pnas.96.6.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bassilana F, Champigny G, Waldmann R, de Weille J R, Heurteaux C, Lazdunski M. J Biol Chem. 1997;272:28819–28822. doi: 10.1074/jbc.272.46.28819. [DOI] [PubMed] [Google Scholar]
- 17.Babinski K, Catarsi S, Biagini G, Seguela P. J Biol Chem. 2000;275:28519–28525. doi: 10.1074/jbc.M004114200. [DOI] [PubMed] [Google Scholar]
- 18.Benson C J, Xie J, Wemmie J A, Price M P, Henss J M, Welsh M J, Snyder P M. Proc Natl Acad Sci USA. 2002;99:2338–2343. doi: 10.1073/pnas.032678399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weisskopf M G, Castillo P E, Zalutsky R A, Nicoll R A. Science. 1994;265:1878–1882. doi: 10.1126/science.7916482. [DOI] [PubMed] [Google Scholar]
- 20.Hanley J G, Khatri L, Hanson P I, Ziff E B. Neuron. 2002;34:53–67. doi: 10.1016/s0896-6273(02)00638-4. [DOI] [PubMed] [Google Scholar]
- 21.Snyder P M. Endocr Rev. 2002;23:258–275. doi: 10.1210/edrv.23.2.0458. [DOI] [PubMed] [Google Scholar]
- 22.Volk K A, Snyder P M, Stokes J B. J Biol Chem. 2001;276:43887–43893. doi: 10.1074/jbc.M108714200. [DOI] [PubMed] [Google Scholar]
- 23.Shimkets R A, Lifton R, Canessa C M. Proc Natl Acad Sci USA. 1998;95:3301–3305. doi: 10.1073/pnas.95.6.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berdiev B K, Xia J, Jovov B, Markert J M, Mapstone T B, Gillespie G Y, Fuller C M, Bubien J K, Benos D J. J Biol Chem. 2002;277:45734–45740. doi: 10.1074/jbc.M208995200. [DOI] [PubMed] [Google Scholar]
- 25.Baron A, Deval E, Salinas M, Lingueglia E, Voilley N, Lazdunski M. J Biol Chem. 2002;277:50463–50468. doi: 10.1074/jbc.M208848200. [DOI] [PubMed] [Google Scholar]
- 26.Xia J, Zhang X, Staudinger J, Huganir R L. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- 27.Szilak L, Moitra J, Vinson C. Protein Sci. 1997;6:1273–1283. doi: 10.1002/pro.5560060615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boudin H, Craig A M. J Biol Chem. 2001;276:30270–30276. doi: 10.1074/jbc.M102991200. [DOI] [PubMed] [Google Scholar]
- 29.Perez J L, Khatri L, Chang C, Srivastava S, Osten P, Ziff E B. J Neurosci. 2001;21:5417–5428. doi: 10.1523/JNEUROSCI.21-15-05417.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim C H, Chung H J, Lee H K, Huganir R L. Proc Natl Acad Sci USA. 2001;98:11725–11730. doi: 10.1073/pnas.211132798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez J C. Nat Rev Neurosci. 2002;3:763. [Google Scholar]
- 32.Askwith C C, Cheng C, Ikuma M, Benson C J, Price M P, Welsh M J. Neuron. 2000;26:133–141. doi: 10.1016/s0896-6273(00)81144-7. [DOI] [PubMed] [Google Scholar]
- 33.Catarsi S, Babinski K, Seguela P. Neuropharmacology. 2001;41:592–600. doi: 10.1016/s0028-3908(01)00107-1. [DOI] [PubMed] [Google Scholar]
- 34.Immke D C, McCleskey E W. Nat Neurosci. 2001;4:869–870. doi: 10.1038/nn0901-869. [DOI] [PubMed] [Google Scholar]
- 35.Berdiev B K, Mapstone T B, Markert J M, Gillespie G Y, Lockhart J, Fuller C M, Benos D J. J Biol Chem. 2001;276:38755–38761. doi: 10.1074/jbc.M107266200. [DOI] [PubMed] [Google Scholar]
- 36.Zhang P, Canessa C M. J Gen Physiol. 2002;120:553–566. doi: 10.1085/jgp.20028574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baron A, Schaefer L, Lingueglia E, Champigny G, Lazdunski M. J Biol Chem. 2001;276:35361–35367. doi: 10.1074/jbc.M105208200. [DOI] [PubMed] [Google Scholar]
- 38.Askwith C C, Benson C J, Welsh M J, Snyder P M. Proc Natl Acad Sci USA. 2001;98:6459–6463. doi: 10.1073/pnas.111155398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price M P, Lewin G B, McIlwrath S L, Cheng C, Xie J, Heppenstall P A, Stucky C L, Mannsfeldt A G, Brennan T J, Drummond H A, et al. Nature. 2000;407:1007–1011. doi: 10.1038/35039512. [DOI] [PubMed] [Google Scholar]
- 40.Price M P, McIllwrath S L, Xie J, Cheng C, Qiao J, Tarr D E, Sluka K A, Brennan T J, Lewin G R, Welsh M J. Neuron. 2001;32:1071–1083. doi: 10.1016/s0896-6273(01)00547-5. [DOI] [PubMed] [Google Scholar]