Abstract
Classical cell biology teaches that exocytosis causes the membrane of exocytic vesicles to disperse into the cell surface and that a cell must later retrieve by molecular sorting whatever membrane components it wishes to keep inside. We have tested whether this view applies to secretory granules in intact PC-12 cells. Three granule proteins were labeled with fluorescent proteins in different colors, and two-color evanescent-field microscopy was used to view single granules during and after exocytosis. Whereas neuro-peptide Y was lost from granules in seconds, tissue plasminogen activator (tPA) and the membrane protein phogrin remained at the granule site for over 1 min, thus providing markers for postexocytic granules. When tPA was imaged simultaneously with cyan fluorescent protein (CFP) as a cytosolic marker, the volume occupied by the granule appeared as a dark spot where it excluded CFP. The spot remained even after tPA reported exocytosis, indicating that granules failed to flatten into the cell surface. Phogrin was labeled with GFP at its luminal end and used to sense the pH in granules. When exocytosis caused the acidic granule interior to neutralize, GFP–phogrin at first brightened and later dimmed again as the interior separated from the extracellular space and reacidified. Reacidification and dimming could be reversed by application of NH4Cl. We conclude that most granules reseal in <10 s after releasing cargo, and that these empty or partially empty granules are recaptured otherwise intact.
Keywords: PC-12 cells‖evanescent-field microscopy‖endocytosis‖kiss and run
When vesicles undergo exocytosis, their membrane must be retrieved by endocytosis to maintain a constant cell surface area. Classical work holds that exocytic vesicles flatten into the cell surface and allow their components to mix with the plasma membrane. These membrane components then are retrieved by molecular sorting, as occurs during clathrin-mediated endocytosis. In contrast, for the stimulated exocytosis of secretory granules it has been proposed that granules release their contents through a fusion pore and then reseal again once cargo has been released (1, 2). Aside from the loss of cargo and some membrane components, the granule remains intact and may be reused or degraded. This mechanism may be referred to as granule cavity recapture, “cavicapture” (1, 2), or granule recapture. A related mechanism was proposed decades ago for synaptic vesicles referred to as “kiss-and-run” exocytosis (3).
Three experimental approaches support granule recapture. In the first, exocytosis of single granules was detected as step increases in membrane capacitance, an assay of cell surface area. Such studies showed that exocytosis can be reversible in mast cells (4, 5). When the release of catecholamine from chromaffin cells was simultaneously detected as an amperometric spike, the spike occurred almost precisely while exocytosis increased the cell surface area in a step (6, 7). Sometimes, however, a normal spike was accompanied by only a transient increase in membrane capacitance, indicating that the connection between granule lumen and the outside, or fusion pore, had opened and closed again (6, 7). The second approach explored how the kinetics of amperometric spikes changed in intact cells while stimulus patterns varied (8), or in permeabilized cells while they overexpressed proteins involved in exo- and endocytosis (9). In particular, treatments expected to interfere with the endocytic protein dynamin caused amperometric spikes to broaden and their charge to increase, suggesting that dynamin caused granules to reseal before they discharged all their catecholamine (10). The third approach was based on imaging single granules (11–13). Sea urchin eggs retrieved large vesicles after fertilization triggered exocytosis (11). In lactotropes, exocytosis caused granule matrices to bind the externally applied dye FM1-43, and some granules retained the dye after it was washed from the cell (12). In PC-12 cell membrane patches, exocytosis caused some granules to take up and sequester an extracellular marker, indicating that the interior of the granule had transiently connected to the external space (14). In chromaffin cells, horseradish peroxidase appeared in organelles indistinguishable from granules soon after the stimulation. Once again this was interpreted as granules transiently connecting their lumen with the external space (15).
The above results provide strong evidence for granule recapture but questions remain. In the first approach, cases of fusion-pore closure were too rare at normal external [Ca2+] (6, 7) to support a significant role of this mechanism under physiologic conditions. This could have been the result of observing only spontaneous fusion events. In the second approach, the observed effects on catecholamine release were subtle, and it was not clear when changes in the amount released reflected changes in the degree of emptying rather than in the catecholamine content of the granule. Finally, in some imaging experiments it was not clear whether the large vesicles retrieved were empty granules or vesicles that were newly formed through invagination of the plasma membrane (11). Other imaging studies explored a time scale of 15–30 min and carried little information on how long granule cavities remained open except that closure was unlikely to occur on the millisecond time scale implied by the duration of amperometric spikes (10).
Here we explore the postexocytic fate of granules when intact and unperturbed PC-12 cells were stimulated by membrane potential changes. Our results confirm three predictions of granule recapture in PC-12 cells: that a granule membrane protein fails to disperse, that granules keep their shape, and that they reseal after exocytosis.
Methods
Constructs and Cells.
Neuropeptide Y (NPY)–enhanced GFP (EGFP) and NPY–cyan fluorescent protein (CFP) were made by excising the ORF of human pro-NPY by restriction digestion from the HindIII and EcoRI sites in NPY–GFP (16) and by subcloning the fragment into the pEGFP-N1 and pECFP-N1 parent vectors (CLONTECH/BD Bioscience, Palo Alto, CA). To fuse rat tissue plasminogen activator (tPA) to the yellow fluorescent protein, Venus (tPA–Venus), we removed the coding sequence of EGFP from tPA–EGFP (17) by digestion with AgeI and BsrGI and replaced it with the ORF of Venus (18). The predicted molecular mass of tPA–EGFP is 97 kDa, three times that of processed NPY–EGFP (31 kDa). Phogrin–Discosoma coral red fluorescent protein (DsRed) was made by excising the phogrin ORF from the phogrin-EGFP plasmid (19) and subcloning it into the EcoRI/AgeI site of the DsRed-N1 vector (CLONTECH). The ORF of rat syntaxin 1A was amplified with the Expand high-fidelity PCR system (Roche Molecular Biochemicals) using the forward primer 5′-GAAGATCTCGAGGGAAGCTTGCCACCATGAAGGACCGAACCCAGGAG- and reverse primer 5′-CCATCGGGGGCATCTTTGGAGG- GGTACCCGGGATCCGCG. The PCR product was subsequently ligated into the HindIII/KpnI site of pEGFP-N1 vector (CLONTECH). We obtained tPA–EGFP from B. Scalettar (Lewis and Clark College, Portland, OR); Venus from A. Miyawaki (RIKEN, Saitama, Japan); Phogrin-EGFP from G. Rutter (University of Bristol, Bristol, England); and syntaxin 1A from R. Scheller (Stanford University, Palo Alto, CA).
PC-12-GR5 cell stocks (provided by R. Nishi, University of Vermont, Burlington) were maintained in T80 flasks (Nalgen/Nunc) at 37°C/10% CO2 in DMEM high glucose (Invitrogen) supplemented with 5% new calf serum and 5% horse serum. For imaging, cells were replated onto poly(L-lysine) (Sigma)-coated high refractive-index glass coverslips (n488 = 1.80, Plan Optik, Elsoff, Germany) and allowed to adhere overnight. Cells then were transfected with 1 μg of plasmid DNA by using Lipofectamine 2000 (Invitrogen) according to manufacturer instructions.
Experiments were performed 24–48 h after transfection. Cells were placed in imaging buffer (130 mM NaCl/2.8 mM KCl/5 mM CaCl2/1 mM MgCl2/10 mM Hepes/10 mM glucose, pH 7.4, 300 milliosmolal), and each coverslip was imaged for up to 2 h. To stimulate secretion, individual cells were locally perfused through a micropipette (4-μm tip diameter) with a solution of elevated [K+] (105 mM KCl/50 mM NaCl/2 mM CaCl2/0.7 mM MgCl2/1 mM NaH2PO4/10 mM Hepes, as pH 7.4, 330 milliosmolal). In some experiments, cells were locally perfused with a solution in which 50 mM NaCl was replaced by 50 mM NH4Cl. All chemicals were obtained from Sigma. Experiments were carried out at room temperature (28°C). Means are given ± SE.
Fluorescence Microscopy.
To selectively illuminate the plasma membrane and its associated granules, we used evanescent-field illumination (20, 21). Cells were grown on high refractive-index glass coverslips and viewed with an inverted microscope (IX-70, Olympus America, Melville, NY) with a 1.65-numerical-aperture objective (Apo ×100 O HR, Olympus) as described (22). Fluorescent proteins of different colors were imaged simultaneously with an image splitter (Multispec MicroImager, Optical Insights, Santa Fe, NM) that separated the emission components of two fluorescent proteins into two channels that then were projected as side-by-side images on the back-illuminated chip of a charge-coupled device camera (Micromax, Roper Scientific, Trenton, NJ). Images were acquired at 2 Hz at 150-ms exposure by using METAMORPH software (Universal Imaging, Downingtown, PA). The two images were brought into focus in the same plane by adding weak lenses to one channel, and they were brought into register by careful adjustment of the mirrors in the image splitter. To correct for any residual misregistration or small differences in magnification, we took pictures in both colors before each experimental session of scattered 300-nm beads (2-FY-300, Interfacial Dynamics, Portland, OR) fluorescing at both wavelengths. Beads in the two images were brought into superposition to within 1 pixel by shifting, stretching, or shrinking one image by using a program written in MATLAB. Optical magnification was to 119 nm per pixel except in Fig. 3 (67 nm per pixel).
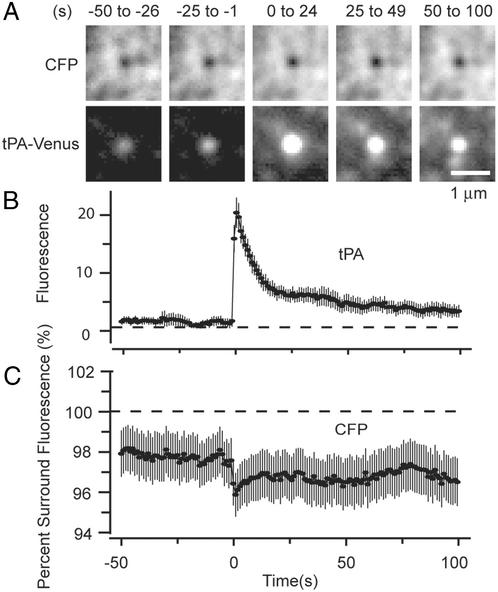
Figure 3.
Failure of granules to collapse into the plasma membrane. (A) Two-color imaging of single granules from cells cotransfected with CFP and tPA–Venus. Cyan (Upper) and yellow (Lower) images are averages of 18 fusion events in three cells lacking extended dark regions in the CFP channel that may have resulted from failure of cells to adhere uniformly. Movies showing the small regions were aligned to the moment of fusion as reported by tPA–Venus and then averaged. The panels show averages of 25 successive frames each. (B) Fluorescence signals at the sites of tPA–Venus-labeled granules (including those in A) were plotted against time, aligned to the moment of fusion, and averaged (31 events, five cells). (C) CFP fluorescence at granule sites shown in B. No background was subtracted; instead, the CFP fluorescence at the granule site was calculated as a percentage of the intensity in a concentric annulus of 3.5-μm outer diameter. The downward spike at the time of fusion and the apparent increase in the fluorescence deficit were not consistently observed and their origins are not understood.
EGFP and DsRed were both excited by the 488-nm laser line as described (22). Images were acquired for 3.3 min at 2 Hz. To excite CFP and Venus, an acousto-optic modulator (Neos, Melbourne, FL) alternated the laser beam between 458 and 514 nm. The beam then passed through a custom dual-wavelength filter passing the 450–464- and 504–520-nm bands. Light was directed into the objective with a custom dichroic mirror that reflected from 425 to 462 and 507 to 537 nm and transmitted from 463 to 506 and 538 to 614 nm. A 520-nm dichroic mirror (Chroma 520DCLP) in the image splitter separated cyan and yellow fluorescence. Cyan passed through a 25-nm band-pass filter centered at 480 nm, and yellow passed through a 565 long-pass filter. Because 458- and 514-nm excitation alternated, the resulting stack had to be deinterleaved off line into 458- and 514-nm stacks. For analysis we used only the cyan image of the 458-nm stack and the yellow image of the 514-nm stack. Images were acquired at 1 image pair(s). Aside from being imaged with a camera, cells could also be observed by eye through dual-emission filters placed into the eyepieces. Each transmitted from 470 to 496 nm and beyond 533 nm. All filters and mirrors were from Chroma Technology (Brattleboro, VT).
Image Analysis.
Evanescent-field illumination selectively images the “footprint” of a cell where the cell adheres closely to the coverslip. Footprints were scanned by eye for individual exocytic events. The coordinates and time of occurrence of each event were marked, and a 3.6 × 3.6-μm-square region was centered on the brightest pixel of the first frame showing exocytosis. Onset of exocytosis was defined as the first frame showing a significant fluorescence increase of the granule. The 3.6 × 3.6-μm square was also placed on all other frames in the sequence and, in two-color experiments, were transferred to the corresponding point in the other-color image. The 3.6 × 3.6-μm squares thus defined were excised from the movies and stored as ministacks. A 1.2-μm-diameter circle was centered on the fusing granules, and the average fluorescence therein was measured. The circle was also transferred into the corresponding location in the other-color image, and the fluorescence was measured. The local background was determined as the average fluorescence in a concentric annulus with a 3.6-μm outer diameter and subtracted. Because the 3.6-μm annulus often contained other granules, this method systematically subtracts too much background.
Results
Imaging Exocytosis in PC-12 Cells by Evanescent-Field Fluorescence Microscopy.
Because we wished to track the fate of single granules after exocytosis, first experiments were carried out to establish that we could reliably stimulate exocytosis in PC-12 cells and detect it by imaging. Fig. 1A shows a PC-12 cell expressing a 4-kDa granule protein, NPY, fused to GFP (NPY–EGFP) (16). The evanescent field selectively illuminated fluorophores within approximately <100 nm of the plasma membrane (20) and thus imaged only the footprint where the cell adhered tightly to the coverslip. Single NPY–EGFP-containing granules were visible as small fluorescent dots that were stationary for tens of seconds to minutes. Footprints as shown in Fig. 1A were outlined, and their areas were measured. The average area was 310 μm2 (n = 5) and contained 0.358 ± 0.05 labeled granules per μm2.
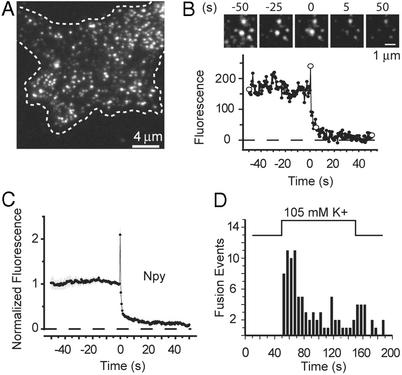
Figure 1.
Exocytosis of single NPY–EGFP-labeled granules. (A) Evanescent-field micrograph of the footprint of a live PC-12 cell (dashed line) expressing NPY–EGFP. (B) A single NPY–EGFP-labeled granule undergoing exocytosis (Upper) and the fluorescence at the granule site (Lower). Open circles refer to the images shown. There is a strong fluorescence increase lasting for a single frame; this transient increase defines the moment of fusion and the time origin. Traces as shown in B were aligned to the moment of fusion and normalized to the intensity at the granule site during the last 2 s before fusion. (C) The results then were averaged (43 events, four cells). (D) The number of fusion events in 5-s intervals is plotted against time after raising [K+] (96 events, eight cells).
To stimulate exocytosis, external [K+] was elevated by superfusing single cells through a micropipette. The elevated [K+] made the plasma membrane potential less negative and thereby opened voltage-gated calcium channels. Fig. 1B shows a still image of a granule undergoing exocytosis; the fluorescence at the granule site is plotted beneath it. After fusion, the granule briefly lit up and then dimmed. The transient brightening is not well resolved at the image-acquisition frequency used here, but faster recordings (data not shown) show NPY–EGFP emerging from a single granule in a cloud and rapidly diffusing away, as in bovine chromaffin cells (20) and with acridine orange in INS-1 cells (23). Brightening occurs both because the EGFP in the acidic granule encounters the neutral pH of the external medium and because release against the coverslip moves NPY–EGFP closer into the evanescent field. After rising briefly in Fig. 1B, the fluorescence fell as NPY–EGFP was released. Sometimes measurable punctate fluorescence remained at the granule site even after exocytosis, indicating that some granules failed to release all their NPY–EGFP.
Fig. 1C shows averaged results. Fluorescence rose for <1 s and then fell as NPY–EGFP was released. In differentiated PC-12 cells, stimulation was reported to raise the pH in granules and cause EGFP to brighten already before exocytosis (24), but any such effect in Fig. 1C must have either been too small to detect or preceded the release of NPY–EGFP by <1 s. Fig. 1D shows a latency histogram of exocytic events. Elevated [K+] caused vigorous exocytosis, the rate of which rose to a peak and then fell to lower levels. However, fusion events occurred rarely or never before depolarization, hence the exocytosis observed here is a triggered event.
Other Proteins Leave Granules More Slowly.
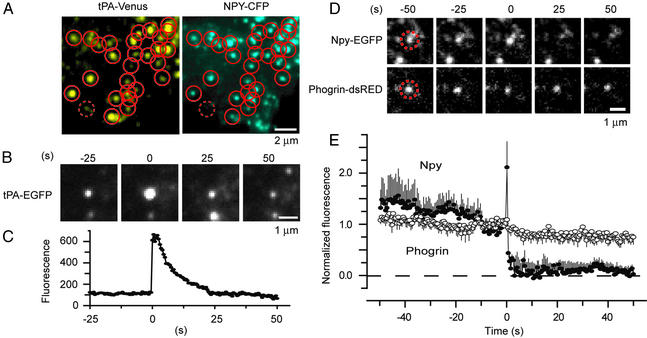
Although NPY–EGFP is well suited to report exocytosis, it leaves granules too fast to be useful as a marker for postexocytic granule components. Two proteins were found to leave granules much more slowly: tPA and phogrin. tPA is a 70-kDa serine protease normally found in PC-12 cell and chromaffin granules (25). To verify that tPA is targeted to granules, we fused tPA to the yellow fluorescent protein Venus (18) and coexpressed it with NPY–CFP. Fig. 2A shows a fluorescence image separated into its cyan and yellow components. Small circles were drawn around the tPA–Venus-labeled structures in the yellow image and transferred into the cyan image. Nearly all tPA–Venus-positive spots (86 ± 2% of 126 granules in two cells) colocalized with NPY–CFP-positive granules. At least near the cell surface, therefore, most or all tPA–Venus-labeled structures were granules. Next, exocytosis was stimulated in a cell expressing tPA–EGFP. A representative tPA–EGFP fusion event (Fig. 2B) is shown together with the time course of fluorescence at the granule site (Fig. 2C). After brightening, the granule dimmed slowly because either the tPA is released and/or the granule resealed and reacidified (see below). Clearly tPA–EGFP was retained in granules much longer than NPY–EGFP (for similar results with tPA–Venus, see Fig. 3B).
Figure 2.
Other granule proteins disperse more slowly after exocytosis. (A) Colocalization of tPA–Venus and NPY–CFP. Circles were drawn around fluorescent dots in the tPA–Venus image and transferred into the NPY–CFP image. The dotted circle shows an example where no NPY–CFP was detected at the site of a tPA–Venus granule and colocalization was scored negative. (B) tPA–EGFP-labeled granule undergoing exocytosis. Here and in other figures the apparent widening of the fluorescent spot at 0 s artifactually resulted from printing all four panels at the same contrast; the effect is not seen if the panel is displayed such that saturation of gray levels is avoided (not shown). (C) Fluorescence at the granule site. (D) Two-color imaging of a single NPY–EGFP- and phogrin–DsRed-labeled granule undergoing exocytosis. The granule position is outlined by a dashed circle. (E) Average fluorescence trace of granules containing both NPY–EGFP and phogrin–DsRed (17 events, three cells). Fluorescence traces were temporally aligned to the moment of fusion as reported by NPY and then normalized to the last 2 s before fusion and finally averaged as shown in Fig. 1C. Throughout, time is relative to the moment of fusion.
The granule membrane protein phogrin is made as a 112-kDa precursor that is later shortened by intraluminal cleavage to 60 kDa (26). Phogrin was fused at its cytoplasmic end to DsRed. To verify that phogrin was targeted to granules, cells were cotransfected with phogrin–DsRed and NPY–EGFP. Phogrin–DsRed fluorescence was punctate, and 84 ± 6% of the red phogrin spots colocalized with green NPY–EGFP spots (137 granules in three cells from experiments as shown in Fig. 2A). Similar to tPA–Venus, therefore, phogrin–DsRed selectively labeled granules. Cells were stimulated with elevated [K+] as shown in Figs. 1 and 2B. Fig. 2D shows a granule labeled with both NPY–EGFP and phogrin–DsRed undergoing exocytosis. Whereas the NPY–EGFP signal vanished after exocytosis, the phogrin–DsRed signal diminished only slightly, and a fluorescent spot remained throughout. Related results have been obtained in INS-1 cells (23). Fig. 2E shows averaged fluorescence measurements from experiments as shown in Fig. 2D. Most NPY–EGFP was lost rapidly from the granule as shown in Fig. 1, but phogrin–DsRed fluorescence remained nearly constant; what changes did occur are attributable at least in part to bleaching or minor movement. Evidently, most phogrin remained in the granule and failed to migrate into the plasma membrane for >1 min after exocytosis. The persistence of phogrin and, to a lesser extent, of tPA at exocytic sites makes both proteins useful probes for monitoring the fate of granules after exocytosis.
Most Granules Do Not Collapse into the Plasma Membrane After Exocytosis.
In chromaffin cells (27) and sea urchin eggs (28), membrane-associated granule components remain as patches on the cell surface after the granules have performed exocytosis (27) and flattened into the plasma membrane (27). The following experiment shows that PC-12 cell granules do not readily flatten into the plasma membrane. Cells were cotransfected with tPA–Venus as a granule marker and with CFP as a cytoplasmic marker. Where the volume of a granule excludes cytosolic CFP, we expect diminished CFP fluorescence to provide a negative image of the granule. Exocytosis was stimulated as shown in Fig. 1, and movies were recorded as alternating images of tPA–Venus and CFP. The CFP channel showed a mottled fluorescence where tPA–Venus-labeled and unlabeled structures excluded CFP, and some cells showed extended dark regions, possibly resulting from where the cell membrane had lifted itself out of the evanescent field (not shown). Where such regions did not interfere, however, the CFP image showed a dark spot at the site of 21 of 31 granules (five cells) that later performed exocytosis. In each of the 21 granules, the spot persisted after exocytosis. Fig. 3A shows still images from a clip showing the average of movies temporally aligned to the moment of fusion. For 50 s before fusion, a dark spot in the center of the CFP image superimposed on the spot of tPA–Venus fluorescence, and the spot remained after exocytosis for the duration of the recording. although tPA–Venus fluorescence increased 12-fold and then declined, CFP fluorescence at the granule site showed a deficit that failed to diminish after fusion (Fig. 3 B and C). GFP diffuses freely in cytoplasm with a diffusion coefficient one-third that in aqueous solution (29) or ≈30 μm2/s. The same may be assumed for CFP. Thus the failure for CFP to fill the dark spot at the granule site indicates that CFP continued to be excluded. Evidently granules do not, or do not fully, collapse into the plasma membrane and instead retain their volume for >1 min after exocytosis.
Most Granule Cavities Reseal After Exocytosis and Reacidify.
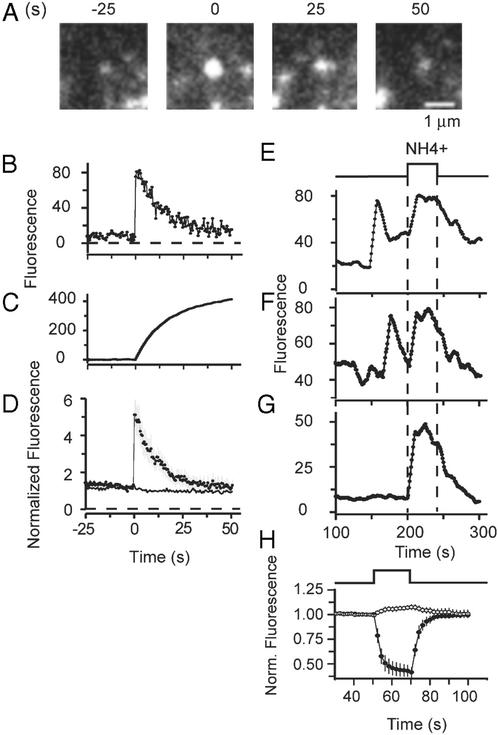
We next asked whether the granules resealed or whether their lumen remained continuous with the external space during the 100-s postexocytic interval explored in Fig. 3. Again phogrin was used as a granule marker, but this time it was conjugated with EGFP near the end of phogrin's luminal domain (EGFP–phogrin). A mutation blocked cleavage of phogrin at its intraluminal cleavage site and ensured that the EGFP remained on the protein. In addition, a C-terminal 18-kDa fragment was removed to limit the size of the construct (30). Similar to tPA–EGFP, the construct served as a pH-sensitive probe of the granule lumen. EGFP–phogrin-labeled granules brightened as exocytosis connected their lumens with the external space and neutralized the acidic pH (Fig. 4A). Later the granules dimmed again, approximately to their prefusion values. This result is analyzed in Fig. 4 B and D and is in striking contrast to the result with phogrin–DsRed. Because the main difference between the constructs was the location and pH sensitivity of the chromophore, we conclude that the dimming of EGFP–phogrin reports the reacidification of the granule after it had resealed and disconnected from the extracellular space (fission). To gain insight into the timing of fission, we plotted the time integral of Fig. 4B to remove high-frequency noise (Fig. 4C). The trace reached half its final value within 8.0 ± 1.4 s after fusion (14 events in three cells), indicating that half the fluorescence gained during exocytosis was quenched within 8 s. Clearly, fission occurred in <8 s after the average fusion event. It may have occurred in much <8 s, because much of this time may be taken up by acidification of already-fissioned granules.
Figure 4.
Granules reseal after exocytosis and reacidify. (A) EGFP–phogrin-labeled granules brighten transiently on exocytosis. (B) Plot of the fluorescence intensity at the granule site along with its time integral (C). (D) Average of 14 events in three cells, obtained as described for Fig. 1C. Continuous line re-plots data with phogrin–DsRed from Fig. 2E. (E–G) Brightening of three granules during application of 50 mM NH4Cl. The granules in E and F had previously undergone exocytosis, whereas the granule in G had not. (E–G) Plotted as rolling averages of 10 successive measurements to reduce noise. Background was measured as the average of intensities in seven 1.2-μm circles in the image of each cell placed where granules were lacking. (H) Fluorescence changes in cells expressing syntaxin–EGFP on the cell surface. The fluorescence drops markedly when the external medium is acidified (35-μm2 membrane area, average of six cells) but changed only slightly when the NH4Cl-containing solution is applied (six cells). This change may be due to acidic-docked granules that contain small amounts of syntaxin. Because EGFP–phogrin was poorly expressed in our cells, we sought to increase the calcium influx and thus raise the frequency of exocytic events in experiments as desccribed for E and F. This was done by maintaining 50 mM CaCl2 externally both before (62.5 mM NaCl/3 mM KCl/50 mM CaCl2/1 mM MgCl2/10 mM Hepes/10 mM glucose, pH 7.4, 300 milliosmolal) and during (105 mM KCl/50 mM CaCl2/0.7 mM MgCl2/1 mM NaH2PO4/10 mM Hepes, 379 milliosmolal) stimulation. The 50 mM [Ca2+] did not significantly change the fluorescence response during exocytosis in EGFP–phogrin-expressing cells. Fluorescence rose abruptly by a factor 5.5 ± 0.8 and declined with a half-time of 7.9 ± 0.9 s (16 events, five cells).
To confirm that dimming reports reacidification of a sequestered compartment, we stimulated cells and waited for EGFP–phogrin-labeled granules to brighten during exocytosis and then to dim. We then applied external ammonium chloride to supply NH3, a membrane-permeant base that neutralizes the acidic pH of organelles (31, 32). Fig. 4 E and F shows fluorescence traces from two exocytic events. As shown in Fig. 4 A, B, and D, the granules first brightened and then dimmed. NH4Cl caused the granules to brighten again (Fig. 4 E and F) and caused brightening even in granules that had not undergone exocytosis (Fig. 4G). As a control, cells were transfected with syntaxin–EGFP, a construct carrying EGFP on the extracellular side of the plasma membrane where NH4Cl is not expected to cause pH changes. Whereas external acidification caused the strong and reversible drop in fluorescence expected from the pH dependence of EGFP (33, 34), NH4Cl caused little or no fluorescence change (Fig. 4H). We conclude that NH4Cl caused granules to brighten in Fig. 4 E–G by collapsing proton gradients and not through a direct effect on EGFP.
We analyzed 21 granules that fused <100 s before the NH4Cl perfusion. Thirteen were seen to brighten in response to NH4Cl; these granules must have resealed and reestablished a proton gradient. Five became invisible before NH4Cl was applied and could not be scored. Three granules failed to brighten and presumably lost most of their phogrin into the plasma membrane. In the majority of granules, therefore (between 62% and 86% in this data set), the dimming reported resealing.
Discussion
We have explored the release and retention of three proteins after the stimulated exocytosis of secretory granules in intact PC-12 cells. The release of NPY–EGFP was so rapid that we could not resolve it well. However, tPA–EGFP and tPA–Venus were released more slowly, and the release of the membrane protein phogrin was altogether too slow to be detected reliably during our observation period. Phogrin and tPA were used as markers of postexocytic granules or granule remnants. By imaging the volume excluded by a granule, we found that most granules failed to collapse into the cell surface and instead retained their volume for >100 s, thus offering refuge for proteins that were reluctant to leave. Within the 100 s most granules reacidified their lumen and therefore must have disconnected from the cell surface. Hence most granules in PC-12 cells reseal after exocytosis.
The advantages to a cell in adopting this type of exocytosis have been discussed (1, 2, 35). When secretory vesicles avoid mixing their membrane components with the cell surface in the first place, cells save themselves the effort of reassembling these components during endocytosis. Some of this benefit would accrue even when granule membranes flatten into the cell surface as long as their components stay together in patches, as seen after prolonged stimulation in bovine chromaffin cells (27). Retrieving intact secretory vesicle cavities as in kiss-and-run exocytosis would strongly benefit small synaptic terminals that must re-cycle a limited reservoir of synaptic vesicle membrane in repeated rounds of exo- and endocytosis (36). However, it is less clear how often recaptured granules are used again for exocytic hormone release. They lack the molecular machinery to refill with proteins but may well reaccumulate small neurotransmitters such as catecholamines. Indeed, granule components and perhaps entire recaptured granules can participate in a second round of catecholamine release when chromaffin cells are restimulated after 5 min of rest (37). Exocytosis of recaptured granules was also observed in PC-12 cell membrane patches (14).
We cannot tell exactly how rapidly granules resealed after exocytosis. However, half the exocytic brightening of EGFP–phogrin is reversed in <8 s. Granules in other cells reseal more slowly. In MIN6 pancreatic β cells, EGFP–phogrin-labeled granules remained bright for tens of seconds before they began to dim (30), and in chromaffin cells granules occasionally stay open for minutes (D.P., unpublished data). In PC-12 cells, 8 s is too short for a granule to release a protein such as tPA completely, and significant amounts of tPA remain in granules long after exocytosis. We do not know why tPA–EGFP is released so slowly, because its molecular mass is only approximately three times larger than that of NPY–EGFP. It may be bound to components of the granule membrane. Nonetheless, the results show that resealing of granules results in the differential release of cargo.
How granules reseal is unknown. In principle, the resealing process could represent the molecular reverse of the opening of the fusion pore. If fusion-pore opening results directly from the assembly of N-ethylmaleimide-sensitive fusion protein attachment protein receptor (SNARE) cis complexes, then its molecular reversal requires the partial disassembly of cis complexes, which may pose energetic puzzles (14). Nonetheless, molecular reversal has been considered for synaptic vesicles in hippocampal terminals where fusion pores are thought to stay open for <6 ms (38). Alternatively, the resealing process may require the recruitment of a dedicated fission machinery either before or after exocytosis. The endocytic protein dynamin has been proposed to serve this function (10, 14, 39). Two-color evanescent-field microscopy seems well suited to test this idea and to test for the transient recruitment of other proteins to spatially defined sites in the plasma membrane (22).
Acknowledgments
We thank A. Miyawaki, G. Rutter, B. Scalettar, and R. Scheller for constructs and S. An, S. Arch, S. Barg, and T. Blackmer for helpful comments on the manuscript. This work was supported by National Institutes of Health Grant MH60600. D.P. was supported by fellowships from the French Ministry of Foreign Affairs and the Human Frontier Science Program organization.
Abbreviations
- NPY
neuropeptide Y
- EGFP
enhanced GFP
- CFP
cyan fluorescent protein
- tPA
tissue plasminogen activator
- DsRed
Discosoma coral red fluorescent protein
References
- 1.Henkel A W, Almers W. Curr Opin Neurobiol. 1996;6:350–357. doi: 10.1016/s0959-4388(96)80119-x. [DOI] [PubMed] [Google Scholar]
- 2.Palfrey H C, Artalejo C R. Neuroscience. 1998;83:969–989. doi: 10.1016/s0306-4522(97)00453-3. [DOI] [PubMed] [Google Scholar]
- 3.Valtorta F, Meldolesi J, Fesce R. Trends Cell Biol. 2001;11:324–328. doi: 10.1016/s0962-8924(01)02058-x. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez J M, Neher E, Gomperts B D. Nature. 1984;312:453–455. doi: 10.1038/312453a0. [DOI] [PubMed] [Google Scholar]
- 5.Spruce A E, Breckenridge L J, Lee A K, Almers W. Neuron. 1990;4:643–654. doi: 10.1016/0896-6273(90)90192-i. [DOI] [PubMed] [Google Scholar]
- 6.Albillos A, Dernick G, Horstmann H, Almers W, Alvarez de Toledo G, Lindau M. Nature. 1997;389:509–512. doi: 10.1038/39081. [DOI] [PubMed] [Google Scholar]
- 7.Ales E, Tabares L, Poyato J M, Valero V, Lindau M, Alvarez de Toledo G. Nat Cell Biol. 1999;1:40–44. doi: 10.1038/9012. [DOI] [PubMed] [Google Scholar]
- 8.Elhamdani A, Palfrey H C, Artalejo C R. Neuron. 2001;31:819–830. doi: 10.1016/s0896-6273(01)00418-4. [DOI] [PubMed] [Google Scholar]
- 9.Fisher R J, Pevsner J, Burgoyne R D. Science. 2001;291:875–878. doi: 10.1126/science.291.5505.875. [DOI] [PubMed] [Google Scholar]
- 10.Graham M E, O'Callaghan D W, McMahon H T, Burgoyne R D. Proc Natl Acad Sci USA. 2002;99:7124–7129. doi: 10.1073/pnas.102645099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whalley T, Terasaki M, Cho M S, Vogel S S. J Cell Biol. 1995;131:1183–1192. doi: 10.1083/jcb.131.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angleson J K, Cochilla A J, Kilic G, Nussinovitch I, Betz W J. Nat Neurosci. 1999;2:440–446. doi: 10.1038/8107. [DOI] [PubMed] [Google Scholar]
- 13.Williams R M, Webb W W. J Cell Sci. 2000;113:3839–3850. doi: 10.1242/jcs.113.21.3839. [DOI] [PubMed] [Google Scholar]
- 14.Holroyd P, Lang T, Wenzel D, De Camilli P, Jahn R. Proc Natl Acad Sci USA. 2002;99:16806–16811. doi: 10.1073/pnas.222677399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henkel A W, Horstmann H, Henkel M K. FEBS Lett. 2001;505:414–418. doi: 10.1016/s0014-5793(01)02861-7. [DOI] [PubMed] [Google Scholar]
- 16.Lang T, Wacker I, Steyer J, Kaether C, Wunderlich I, Soldati T, Gerdes H H, Almers W. Neuron. 1997;18:857–863. doi: 10.1016/s0896-6273(00)80325-6. [DOI] [PubMed] [Google Scholar]
- 17.Lochner J E, Kingma M, Kuhn S, Meliza C D, Cutler B, Scalettar B A. Mol Biol Cell. 1998;9:2463–2476. doi: 10.1091/mbc.9.9.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagai T, Ibata K, Park E S, Kubota M, Mikoshiba K, Miyawaki A. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 19.Pouli A E, Emmanouilidou E, Zhao C, Wasmeier C, Hutton J C, Rutter G A. Biochem J. 1998;333:193–199. doi: 10.1042/bj3330193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steyer J A, Almers W. Nat Rev Mol Cell Biol. 2001;2:268–275. doi: 10.1038/35067069. [DOI] [PubMed] [Google Scholar]
- 21.Axelrod D. J Biomed Opt. 2001;6:6–13. doi: 10.1117/1.1335689. [DOI] [PubMed] [Google Scholar]
- 22.Merrifield C J, Feldman M E, Wan L, Almers W. Nat Cell Biol. 2002;4:691–698. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- 23.Tsuboi T, Zhao C, Terakawa S, Rutter G A. Curr Biol. 2000;10:1307–1310. doi: 10.1016/s0960-9822(00)00756-9. [DOI] [PubMed] [Google Scholar]
- 24.Han W, Li D, Stout A K, Takimoto K, Levitan E S. J Neurosci. 1999;19:900–905. doi: 10.1523/JNEUROSCI.19-03-00900.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parmer R J, Mahata M, Mahata S, Sebald M T, O'Connor D T, Miles L A. J Biol Chem. 1997;272:1976–1982. doi: 10.1074/jbc.272.3.1976. [DOI] [PubMed] [Google Scholar]
- 26.Wasmeier C, Hutton J C. J Biol Chem. 1996;271:18161–18170. doi: 10.1074/jbc.271.30.18161. [DOI] [PubMed] [Google Scholar]
- 27.Wick P F, Trenkle J M, Holz R W. Neuroscience. 1997;80:847–860. doi: 10.1016/s0306-4522(97)00062-6. [DOI] [PubMed] [Google Scholar]
- 28.Smith R M, Baibakov B, Ikebuchi Y, White B H, Lambert N A, Kaczmarek L K, Vogel S S. J Cell Biol. 2000;148:755–767. doi: 10.1083/jcb.148.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swaminathan R, Hoang C P, Verkman A S. Biophys J. 1997;72:1900–1907. doi: 10.1016/S0006-3495(97)78835-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohara-Imaizumi M, Nakamichi Y, Tanaka T, Katsuta H, Ishida H, Nagamatsu S. Biochem J. 2002;363:73–80. doi: 10.1042/0264-6021:3630073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klempner M S, Styrt B. J Clin Invest. 1983;72:1793–1800. doi: 10.1172/JCI111139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sankaranarayanan S, Ryan T A. Nat Cell Biol. 2000;2:197–204. doi: 10.1038/35008615. [DOI] [PubMed] [Google Scholar]
- 33.Patterson G H, Knobel S M, Sharif W D, Kain S R, Piston D W. Biophys J. 1997;73:2782–2790. doi: 10.1016/S0006-3495(97)78307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Llopis J, McCaffery J M, Miyawaki A, Farquhar M G, Tsien R Y. Proc Natl Acad Sci USA. 1998;95:6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meldolesi J, Ceccarelli B. Philos Trans R Soc London B. 1981;296:55–65. doi: 10.1098/rstb.1981.0171. [DOI] [PubMed] [Google Scholar]
- 36.Harata N, Pyle J L, Aravanis A M, Mozhayeva M, Kavalali E T, Tsien R W. Trends Neurosci. 2001;24:637–643. doi: 10.1016/s0166-2236(00)02030-0. [DOI] [PubMed] [Google Scholar]
- 37.von Grafenstein H, Knight D E. J Physiol (London) 1992;453:15–31. doi: 10.1113/jphysiol.1992.sp019215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevens C F, Williams J H. Proc Natl Acad Sci USA. 2000;97:12828–12833. doi: 10.1073/pnas.230438697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Artalejo C R, Henley J R, McNiven M A, Palfrey H C. Proc Natl Acad Sci USA. 1995;92:8328–8332. doi: 10.1073/pnas.92.18.8328. [DOI] [PMC free article] [PubMed] [Google Scholar]