Abstract
Neuronal exocytosis is mediated by Ca2+-triggered rearrangements between proteins and lipids that result in the opening and dilation of fusion pores. Synaptotagmin I (syt I) is a Ca2+-sensing protein proposed to regulate fusion pore dynamics via Ca2+-promoted binding of its cytoplasmic domain (C2A-C2B) to effector molecules, including anionic phospholipids and other copies of syt. Functional studies indicate that Ca2+-triggered oligomerization of syt is a critical step in excitation–secretion coupling; however, this activity has recently been called into question. Here, we show that Ca2+ does not drive the oligomerization of C2A-C2B in solution. However, analysis of Ca2+⋅C2A-C2B bound to lipid monolayers, using electron microscopy, revealed the formation of ring-like heptameric oligomers that are ≈11 nm long and ≈11 nm in diameter. In some cases, C2A-C2B also assembled into long filaments. Oligomerization, but not membrane binding, was disrupted by neutralization of two lysine residues (K326,327) within the C2B domain of syt. These data indicate that Ca2+ first drives C2A-C2B⋅membrane interactions, resulting in conformational changes that trigger a subsequent C2B-mediated oligomerization step. Ca2+-mediated rearrangements between syt subunits may regulate the opening or dilation kinetics of fusion pores or may play a role in endocytosis after fusion.
Keywords: C2 domain‖oligomerization‖calcium‖membrane
In neurons, excitation–secretion coupling is mediated by Ca2+ (1). Ca2+ binds to Ca2+-sensing proteins that regulate the opening and dilation of exocytotic fusion pores, releasing neurotransmitters from synaptic vesicles (SV) into the synaptic cleft (2, 3). After fusion, SVs are recycled via endocytosis, refilled with transmitters, and reused for subsequent rounds of fusion (4). Over the last decade, a number of proteins that play essential roles in exocytosis and endocytosis have been identified. One family of related proteins, the synaptotagmins (syt), have been proposed to function at multiple steps in vesicle trafficking pathway (5).
In vertebrates, 13–19 isoforms of syt have been identified (6). The members of this gene family have a single membrane-spanning domain and a short intravesicular tail. The cytoplasmic domain is composed of tandem C2 domains connected by a short linker (7, 8). The membrane-proximal C2 domain is C2A, and the membrane-distal C2 domain is C2B; both C2 domains function as Ca2+-sensing modules (9–11). The most abundant and best-studied isoform is syt I. The ability of syt I to bind Ca2+, coupled to functional studies demonstrating that syt I plays a critical postdocking function in exocytosis, suggests that syt I is a major Ca2+ sensor that regulates release (12, 13). Recent studies established that changes in the ratio of syt isoforms can alter fusion pore kinetics, placing syt action in the final events of the fusion reaction (14).
To understand how Ca2+⋅syt I regulates membrane fusion, a number of putative effector molecules have been identified, including anionic phospholipids (9), components of the soluble N-ethymaleimide-sensitive factor (NSF) attachment protein receptor (SNARE) complex (15–18), and other copies of syt (i.e., oligomerization) (11, 19–22). This latter property, the ability of syt to assemble into homo- and hetero-oligomers, supports genetic studies indicating that syt I functions in a multimeric complex with separate functional domains (23, 24). syt exhibits two modes of oligomerization; Ca2+-independent oligomerization is mediated by the N-terminal region (9, 25), and Ca2+-dependent oligomerization is mediated by the C2 domains (19–22). In syt VII, both C2A and C2B oligomerize in response to Ca2+ (26); in syt I, C2B fulfills this function (11). Perturbation of Ca2+-triggered oligomerization is correlated with inhibition of exocytosis, suggesting that oligomerization is crucial for syt function during release (11, 13, 27).
Virtually nothing is known concerning the structure or stoichiometry of syt I oligomers. Furthermore, in two recent studies, Ca2+-triggered oligomerization activity of the cytoplasmic domain of syt I (C2A-C2B) was not observed, and it was suggested that oligomerization was due to tightly bound contaminants and was not an intrinsic property of this syt fragment (28, 29). We have therefore reevaluated the Ca2+-triggered oligomerization activity of C2A-C2B. We confirm that Ca2+-triggered oligomerization of highly purified C2A-C2B does not occur in solution; however, using negative stain electron microscopy (EM), we observed that C2A-C2B assembles into heptameric barrel-like structures on the surface of lipid monolayers. Assembly of syt oligomers is abolished by mutations in the C2B domain, suggesting that C2B drives multimerization. Consistent with this interpretation, we show that Ca2+ and weak interactions with anionic lipids (30–32) drive oligomerization, and thus sedimentation, of the isolated C2B domain. We propose that Ca2+ and membrane-dependent assembly of syt I into oligomeric structures regulates membrane dynamics during exocytosis and/or during endocytosis (5).
Materials and Methods
Reagents and Chemicals.
For EM studies, egg–phosphatidylcholine (PC) and phosphatidylserine (PS) were purchased from Sigma. For fluorescence studies and radio-labeled liposome-binding assays, synthetic 1,2-dioleoyl-sn-glycero-3-[phospho-l-serine] (PS), 1,2-dioleoyl-sn-glycero-3-phosphocholine (PC), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(5-dimethylamino-1-naphthalenesulfulfonyl) (dansyl-PE) were obtained from Avanti Polar Lipids. l-3-phosphatidyl[N-methyl-[3H]choline-1,2-dipalmitoyl ([3H]-PC) was purchased from Amersham Pharmacia Biotech.
cDNA encoding rat syt I (7) [the G374 form (11)], III (33), and VII (22) was kindly provided by T. C. Südhof (Dallas), S. Seino (Chiba, Japan) and M. Fukuda (Saitama, Japan), respectively. Sequences were: C2A-C2B-I, 96–421; C2A-I, 96–265; C2B-I, 248–421; C2A-C2B-II, 139–423; C2A-III, 290–421; C2A-VII, 134–262. All syt fragments were subcloned into pGEX-2T (Amersham Pharmacia) and confirmed by DNA sequencing.
Purification of Recombinant Proteins.
GST-tagged proteins were purified using glutathione-Sepharose beads (Amersham Pharmacia Biotech) as described (19). The bead-immobilized proteins were washed with cleaning buffer (50 mM Hepes⋅NaOH, pH 7.4/1.1 M NaCl) plus 1 mM MgCl2/10 μg/ml DNase I (Roche Applied Science), and 10 μg/ml RNase (Sigma) followed by washes with Hepes buffer (50 mM Hepes⋅NaOH, pH 7.4/0.1 M NaCl). For EM and FPLC experiments, proteins bound to glutathione-Sepharose beads were incubated with Hepes buffer containing 20 μg/ml DNase I and 4 μg/ml RNase A, for 6 h at 4°C. The beads were then washed as described above. syt fragments were freed from the GST moiety by using thrombin, and the lack of contaminants was confirmed by measuring the UV spectra of all samples (29).
EM and Lipid Monolayers.
EM on lipid monolayers was carried out as described (34–36). Droplets (≈15 μl) of C2A-C2B (0.1 mg/ml in Tris-buffered saline; 50 mM Tris⋅HCl/100 mM NaCl, pH 7.4) were placed in Teflon wells (4 mm diameter, 0.5 mm depth) until the liquid surface bulged out. The surface of the droplet was coated with 0.5–1 μl of the indicated phospholipid mixture in chloroform/methanol (3:1, vol/vol). Samples were incubated in a sealed humidified chamber for 12 h at 4°C. For some experiments, samples were also prepared so that C2A-C2B was not exposed to organic solvents. This was achieved by placing buffer droplets (≈80 μl) in Teflon wells, followed by the spread of 0.5–1 μl of lipid mixture in chloroform/methanol (3:1, vol/vol) onto the buffer surface. The organic solvent was allowed to evaporate by incubation for 30 min at room temperature, then C2A-C2B (≈4 μl) was added into the well and the samples incubated as described (both methods of sample preparation yielded similar results). Lipid monolayers at the air/water interface were picked up with hydrophobic carbon-coated grids. After washing with incubation buffer, the grids were blotted up and negatively stained with uranyl acetate solution (1%, wt/vol). The negatively stained samples were examined in a Philips (Eindhoven, The Netherlands) CM120 transmission EM under the accelerating voltage of 100 kV. We note that the same structures were observed when samples were prepared in the presence or absence of 1 mM DTT.
Single-Particle Method.
Best films were digitized by using an Agfa Duoscan camera system. The numeric images were changed to spider format and processed by spider image processing software. Nine hundred seventy rectangular particles were aligned by using reference-free alignment. The first alignment gives an average map, which is used as the reference for a second round of alignment. The aligned particles are masked by a mask made from the reference map and were analyzed by using multivariance statistical analysis. By hierarchical classification, the particles were classified into four clusters; the main cluster included 533 particles. Five hundred and five ring-like particles were processed in the same way, and three clusters were obtained; the main cluster had 321 ring-like particles. The resolutions of the average rectangular and ring-like particle maps were 3 and 3.4 nm, respectively, according to the Differential Phase Residual method (37).
C2B⋅Liposome Cosedimentation Assays.
Large (≈100 nm) unilamellar liposomes were prepared as described (38). Three micromolar WT or K326,327A mutant C2B was incubated with 22 nM liposomes composed of 25% PS/75% PC (2 mM total lipids) in 100 μl of Hepes buffer for 5 min at room temperature, in the presence of 0.2 mM Ca2+ or 2 mM EGTA. Samples were then centrifuged at 100,000 × g for 30 min in an airfuge (Beckman Coulter), and the supernatants and pellets were separated. Pellets were washed once with 200 μl of Hepes buffer and collected again via centrifugation. Equal fractions of the supernatants and pellets were subjected to SDS/PAGE, and proteins were stained with Coomassie blue.
Results and Discussion
Reevaluation of the Oligomerization and Lipid-Binding Properties of C2A-C2B and C2B Derived from syt I.
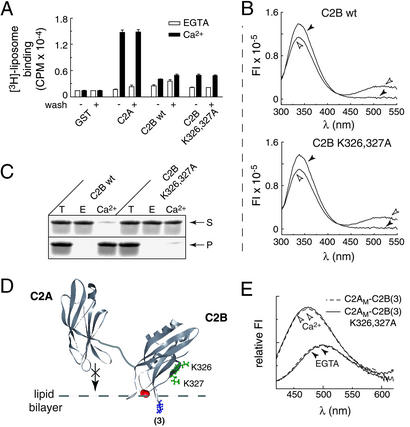
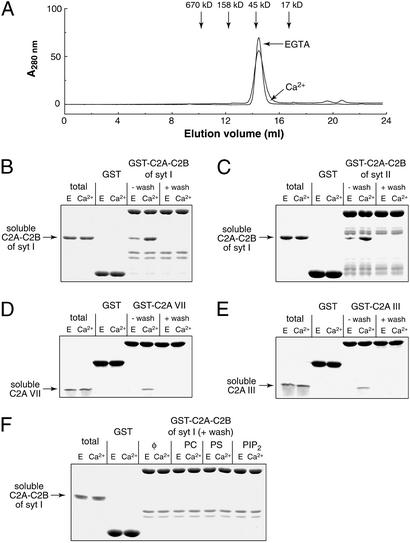
It was recently reported that the recombinant C2B domain of syt I contains tightly bound bacterial contaminants with an absorbance at 260 nm (29). Removal of these contaminants was proposed to change the biochemical properties of C2B in two ways: it abolished Ca2+-triggered oligomerization activity (29), and it reportedly enhanced binding of isolated C2B to membranes containing PS/PC (32). In our studies, removal of the contaminant did not affect the interaction of syt with membranes (Fig. 2A, detailed below). However, removal of the contaminant abolished Ca2+-triggered oligomerization activity of purified C2A-C2B. This finding is shown in Fig. 1A, where C2A-C2B eluted from a sizing column as a monomer, even in the presence of 1 mM Ca2+. This result was confirmed by using C2A-C2B immobilized as a GST fusion protein. Before removal of the contaminant, soluble C2A-C2B bound, in a Ca2+-dependent manner, to immobilized C2A-C2B; binding was abolished on removal of the contaminant (Fig. 1B). The contaminant appears to be RNA, because, under our conditions, RNase/high salt, but not DNase/high salt, effectively removed the A260 absorbing species. RNase alone was not sufficient, and high salt was required to effectively remove the contaminant. These data suggest that oligomerization is not due to the binding of multiple copies of C2A-C2B to long strands of RNA. Rather, it is likely that short fragments of RNA are tightly bound to C2B to influence the properties of syt.
Figure 2.
Isolated C2B does not bind PS/PC membranes with high affinity, but cosediments with liposomes via a secondary mechanism. (A) C2A-C2B, C2A, and C2B domains from syt I were immobilized as GST-fusion proteins. Samples with (+wash) or without (−wash) nuclease/salt treatment (6 μg of protein per data point) to remove bacterial contaminants were assayed for binding 3H-labeled liposomes, (≈0.24 mM 25% PS/75% PC) in 100 μl of Tris-buffered saline buffer as described (38), in either 2 mM EGTA or 0.2 mM Ca2+. Bound liposomes were quantified by liquid scintillation counting. (B) Wild-type and K326,327A mutant C2B bind PS-containing membranes to the same extent as measured by using fluorescence resonance energy transfer. Liposomes composed of 5% 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(5-dimethylamino-1-naphthalenesulfulfonyl) (dansyl-PE)/25% PS/70% PC were prepared as described (38). Native tryptophan (Trp) residues in C2B (amino acids 390 and 404) served as the energy donors and were excited at 285 nm. The emission spectra of the Trp residues (emission λMAX ≈ 337 nm) and the dansyl-PE acceptor (emission λMAX ≈ 514 nm) were collected from 300 to 550 nm. Proteins (2 μM) were mixed with 11 nM liposomes in either 2 mM EGTA (filled arrowheads) or 0.2 mM Ca2+ (open arrowheads). Fluorescence resonance energy transfer occurred in response to Ca2+ (Upper), indicating some degree of binding (32); however, the Ca2+-binding loops of isolated C2B do not insert into lipid bilayers (31). The K326,327A mutation had no effect on the weak interaction of C2B with membranes (Lower). (C) Cosedimentation of C2B with liposomes is not directly mediated by C2B⋅membrane interactions. Sedimentation assays were carried out as described in Materials and Methods. Thirteen percent of the supernatant (S), pellet (P), and total (T) reaction were subjected to SDS/PAGE and stained with Coomassie blue. Wild-type isolated C2B translocated from the supernatant to the pellet fraction in response to Ca2+; neutralization of two lysine residues (K326,327A) abolished sedimentation without affecting membrane binding. (D) Molecular model depicting the Ca2+-triggered penetration of C2B, in the context of C2AM-C2B, into lipid bilayers. C2B penetrates lipid bilayers when tethered to C2A or C2AM (31). The subscript “M” corresponds to D230,232N substitutions that disrupt C2A⋅membrane penetration (31, 58). A 5-[[2-(acetylamino)ethyl]amino] napthalene-1-sulfonic acid (AEDANS) probe was placed in the Ca2+-binding loop 3 of C2B [indicated as (3)] as described (31) to monitor membrane penetration of C2B in the context of C2AM-C2B (31). Lysines 326 and 327 are shown in green; solution structures of C2A (46) and C2B (32) from syt I were rendered in WEBLAB VIEWER LITE (Molecular Simulations)]; the flexible linker that connects them (8) was added by using a drawing program. (E) Liposomes (11 nM; 25% PS/75% PC) and C2AM-C2B (3) (0.5 μM; dashed line) were incubated in Hepes buffer in the presence of 0.1 mM EGTA. AEDANS fluorescence was excited at 336 nm, and the emission spectra were collected from 420 to 600 nm. Ca2+ was then added to a final free concentration of 0.5 mM, and spectra were obtained again. Neutralization of K326,327A (solid line) abolishes oligomerization (Fig. 4D) but does not alter the membrane penetration activity of C2B in the context of C2AM-C2B.
Figure 1.
Highly purified fragments of syt I–III and VII fail to oligomerize in response to Ca2+. (A) Size-exclusion FPLC was performed using an AKTA Explorer (Amersham Pharmacia Biotech) with a Superdex 200 column (Pharmacia). One-hundred-microliter samples of C2A-C2B from syt I (1.0 μg/μl, after nuclease/salt treatment) plus 1 mM Ca2+ or 1 mM EGTA in Tris-buffered saline were run through the column at a flow rate of 0.5 ml/min. (B) Ten-microgram GST or GST-C2A-C2B from syt I, with (+ wash) or without (−wash) nuclease/salt treatment, was immobilized on glutathione-Sepharose beads. Beads were incubated with 2 μM soluble C2A-C2B syt I for 1.5 h in 150 μl of Hepes buffer plus 0.5% Triton X-100 and either 2 mM EGTA or 1 mM Ca2+. Beads were washed three times with binding buffer and boiled in SDS sample buffer. Seven percent of the total (left two lanes) and 30% of the bound material (remaining lanes) were subjected to SDS/PAGE and visualized by staining with Coomassie blue. (C) Binding of soluble C2A-C2B from syt I (3 μM) to immobilized C2A-C2B from syt II was assayed as described in B. (D) Binding assays were carried out as in B but using C2A-VII. (E) Binding assays were carried out as in B but using C2A-III. (F) Lipid/detergent micelles containing 70 μM PC or PS or PIP2 in 1% Triton X-100 (≈98 μM micelles) were included in the binding assay described in A; rescue of oligomerization was not observed under these conditions.
It was previously shown that native syt I and II heterooligomerize in response to Ca2+; this property was recapitulated by using recombinant C2A-C2B versions of these proteins (39). However, as shown in Fig. 1C, removal of the contaminant abolished Ca2+ triggered binding of recombinant C2A-C2B from syt I with GST-C2A-C2B from syt II. Similar observations have been made for syt I (11, 19). These data suggest that other factors, associated with native syt, allow them to oligomerize in response to Ca2+. This idea is further supported by data showing that the ability of the C2A domain of syt VII, first reported to oligomerize in extracts from transfected fibroblasts (26), also depends on the presence of contaminants (Fig. 1D). We have extended these findings to the C2A domain of syt III, which also self associates in the presence of Ca2+; again, this activity is abolished on removal of the contaminant (Fig. 1E).
As noted above, the ability of native or transfected syt fragments to oligomerize in response to Ca2+ is well established (22, 26, 39). Our studies using recombinant C2A-C2B from syt I and II, and C2A domains from syt VII and III, indicate that oligomerization requires the presence of another factor. In the case of recombinant proteins, bacterial RNA appears to fulfill this requirement. Addition of either PC, PS, or PIP2 in the form of Triton X-100 micelles failed to restore oligomerization of C2A-C2B (Fig. 1F); however, as detailed below, C2A-C2B-I was able to oligomerize on monolayers composed of PS and PC.
We next addressed the effects of contaminant removal on C2B⋅PS/PC interactions. It was suggested that removal of the contaminant enhanced the ability of C2B to bind PS/PC liposomes (32). However, using radiolabeled liposome-binding assays, the degree of binding was not significantly affected by removal of the contaminant (Fig. 2A), confirming previous reports (30, 31, 40). Thus, isolated C2B exhibits only a trace of PS/PC-binding activity, regardless of whether the contaminant is bound. As a positive control, the isolated C2A domain exhibited robust Ca2+-triggered binding to radiolabeled PC/PS liposomes.
Importantly, claims that contaminant-free isolated C2B binds tightly to membranes were based on a cosedimentation assay; C2B efficiently sediments with PS/PC liposomes in the presence of Ca2+ (32, 41, 42). However, the data shown in Fig. 2 demonstrate that sedimentation does not report binding of C2B to PS/PC liposomes. Instead, Ca2+ and weak interactions with membranes trigger oligomerization and precipitation of C2B. This was demonstrated by using the K326,327A mutant version of C2B. This mutation has no effect on syt⋅PS/PC interactions monitored using fluorescence resonance energy transfer (Fig. 2B), radio ligand-binding assays (Fig. 2A), and membrane penetration assays (Fig. 2 D and E), yet this mutation completely abolished the Ca2+/membrane-induced sedimentation of C2B (Fig. 2C). Thus, the ability of C2B to cosediment with PS/PC liposomes is secondary to some other process that involves the lysine residues (e.g., oligomerization, described below). Clearly, cosedimentation data for C2 domains that are oligomerization-competent should be interpreted with caution (32).
C2A-C2B Oligomerization Requires Both Ca2+ and an Anionic Phospholipid Surface.
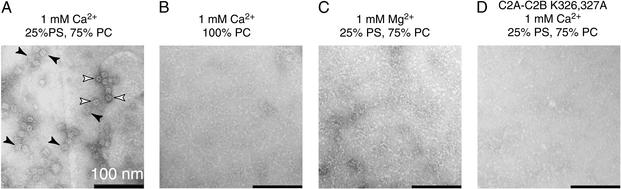
To further explore syt⋅syt and syt⋅membrane interactions, we used a negative-stain EM approach to image C2A-C2B bound to lipid monolayers (35, 36, 43, 44). Consistent with the data in Fig. 1 A and B, purified C2A-C2B failed to form oligomeric structures on hydrophilic-treated carbon-coated grids (data not shown). However, when C2A-C2B was imaged on lipid monolayers (25% PS/75% PC), dispersed particles with regular structures were observed in the presence of 50 μM − 1 mM Ca2+ (Figs. 3 and 4A; Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). At least two distinct forms of dispersed particles are visible. The most frequently observed form is rectangular in appearance; the less frequently observed particle is ring-like (Fig. 3). These particles, which correspond to C2A-C2B oligomers, were not observed when PS was not included in the monolayer (Fig. 4B), or when Ca2+ was substituted with Mg2+ (Fig. 4C). These data demonstrate that oligomerization requires both Ca2+ and anionic phospholipids; an anionic lipid surface is able to reconstitute Ca2+-triggered C2A-C2B oligomerization after removal of the contaminant. Interestingly, neutralization of the lysines 326 and 327, by substitution with alanines, disrupted assembly of the oligomers (Fig. 4D) but does not inhibit C2A-C2B⋅membrane interactions (Fig. 2 A and B). This result indicates that in response to Ca2+, C2A-C2B first penetrates bilayers (31) then assembles into oligomers. However, we cannot rule out some degree of weak oligomerization before membrane insertion.
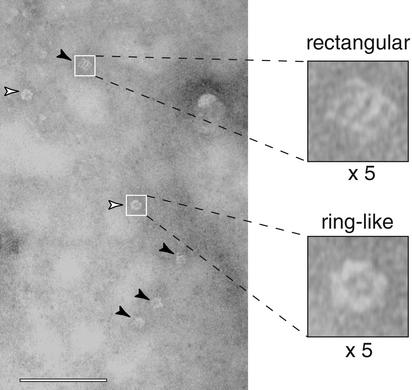
Figure 3.
Ca2+-triggered assembly of the cytoplasmic domain of syt I into oligomers on PS/PC monolayers. Rectangular C2A-C2B oligomers are marked with filled arrowheads; ring-like forms are marked with open arrowheads. Examples of individual rectangular and ring-like C2A-C2B oligomers are also shown at ×5 magnification. Lipid monolayers were composed of 25% PS/75% PC; [Ca2+] was 50 μM. (Bar = 100 nm.)
Figure 4.
Cation, lipid, and structural requirements for assembly of C2A-C2B-I into oligomers. (A) The image was obtained as described in Fig. 3, except 1 mM Ca2+ was used. (B) Conditions were as in A, except PS was omitted. (C) Conditions were as in A, except 1 mM Ca2+ was replaced with 1 mM Mg2+. (D) Conditions were as in A, except that lysines 326 and 327, which are critical for the oligomerization of C2A-C2B and C2B that harbor contaminants (Fig. 1 A and B) (11, 45), were neutralized by substitution with alanines. This mutation does not affect membrane binding (Fig. 2E) but abolished assembly of oligomeric particles in the presence of 1 mM Ca2+ and 25% PS. Monomeric C2A-C2B K326,327A bound to membranes is below the resolution of our EM assay and was not observed.
The requirement of an anionic lipid surface for C2A-C2B oligomerization suggests that Ca2+ and membranes might cooperate to drive conformational changes in syt that trigger oligomerization. NMR studies indicate that Ca2+ alone does not drive large structural rearrangements within the isolated C2 domains of syt I (32, 46). However, at present, virtually nothing is known concerning the structure of Ca2+-loaded C2 domains in the presence of effector molecules. NMR studies of C2A from syt I and syntaxin (47–49) made use of a fragment of syntaxin that is completely dispensable for binding (16, 50, 51) and thus did not reveal insights into potential conformational changes in syt. Other studies have made use of short acyl chain lipids, but as shown here (Fig. 1F), PS dispersed in detergent micelles failed to reconstitute oligomerization (52). Thus, PS in a mono- or bilayer interacts with syt I in a manner distinct from PS in micelles.
Assembly of C2A-C2B into Heptameric Oligomers on Lipid Monolayers.
Under our experimental conditions (50 μM–10 mM Ca2+; 25% PS/75% PC), most of the oligomers were rectangular; ≈20% were ring-like. The predominance of the rectangular form may be due to the simultaneous interaction of both C2A and C2B with the lipid monolayer (31); this idea is described in further detail below. By single-particle analysis, the particles with rectangular shape were picked, aligned, classified, and averaged. Their average map revealed that the rectangular particle has an overall dimension of 11 × 11 nm. Four bar-like subunits are apparent; each bar-like subunit has two high-density domains exhibiting a biconcave shape (Fig. 5A). The projection maps of the four bar-like subunits are not equally dense. The two subunits in the middle are high, whereas those on both sides are low in projection density.
Figure 5.
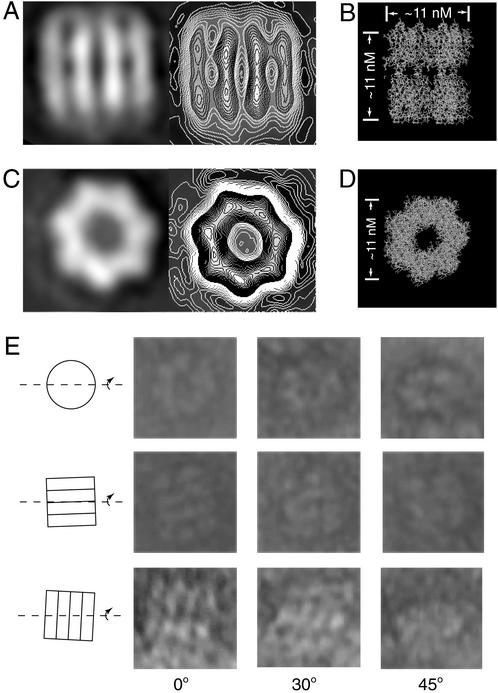
Single-particle analysis of syt oligomers. (A) Average maps of the rectangular particles before (Left) and after (Right) contouring. Within a bar, the two electron-dense regions probably correspond to C2A and C2B. At this level of resolution, we cannot discriminate between C2A and C2B, but, because C2B appears to drive oligomerization (Fig. 4D), it is likely that each syt subunit is arranged in a parallel manner. In this case, the membrane anchors of native syt would emerge from the end of the oligomer that contains C2A. (B) Molecular model in which the crystal structure of C2A-C2B from syt III (8) has been packed into a heptameric oligomer; the heptamer is viewed from the side, where four C2A-C2B subunits, vertically oriented, are visible. In this model, the structures of the C2 domains were not altered, but the tandem C2 domains were aligned along their long axes by changing the flexible linker that connects them. (C) Average map of the ring-like particles before (Left) and after (Right) contouring. (D) Top view of the model shown in B; the ends of all seven subunits are visible. (E) Tilt analysis of heptameric particles. In each set of images, a single particle is tilted through 0, 30, and 45° about the axis indicated by the line drawing on the left. (Top) A ring-like particle is analyzed. (Middle and Bottom) Two rectangular particles in the indicated orientations are analyzed.
Fig. 5C shows the results of single particle analysis of the ring-like particles. The average map clearly revealed that each ring-like structure has seven high-density regions, suggesting it consists of seven subunits. The diameter of the ring-like structure is ≈11 nm, the same value that was obtained for the rectangular particles.
To determine the relationship between the two types of particles, we analyzed tilted views (0, 30, and 45°) of each particle. In Fig. 5E Top, tilted projections of the ring-like particle become progressively elliptical. In Fig. 5E Middle and Bottom, tilted views of rectangular particles, in two distinct orientations, are shown. In Fig. 5E Middle, the tilt angle is perpendicular to the axis of the bars; this tilt does not affect the shape of the image. In contrast, in the gallery at the bottom, the tilt angle is parallel to the axis of the bars; this tilt changes the appearance of the rectangular form so it becomes elliptical, similar to the image of the ring-like form viewed with a 45° tilt. These data indicate that the rectangular and ring-like forms represent the side and top views of the same C2A-C2B oligomer. This interpretation is supported by the higher density of the two middle bars of the rectangular map; they have a different height, compared with the lower adjacent outside bars, because the shape of the whole oligomer is similar to a barrel or cylinder.
To further establish whether the observed oligomers are composed of C2A-C2B heptamers, we generated a model using the crystal structure of C2A-C2B from syt III (Fig. 5 B and D). Seven C2A-C2B subunits were packed into a heptameric barrel (Fig. 5 B and D), resulting in a model that exhibits striking similarity to the density maps shown in Fig. 5 A and C. The existence of two high-density domains within a bar-like subunit is consistent with the individual C2 domains in C2A-C2B (8). Thus, each bar-like subunit in the density map represents the side view of a C2A-C2B monomer, and each ring-like C2A-C2B oligomer consists of seven monomers.
These findings might explain the dominance of the rectangular form; if the Ca2+ and membrane-binding loops of C2A and C2B point outward, the number of contact sites for membranes would be maximized in complexes that lie on their side. Interestingly, the subunits at the peak of the arch of the heptamer would not be in direct contact with the monolayer. This result suggests that conformational changes within membrane-bound C2A-C2B subunits might be propagated to subunits that are not in direct contact with the monolayer, to form a seven-subunit complex. It is also possible that additional lipids are bound to either the surface, or within the barrel, of the heptamer. Future studies are needed to determine the precise orientation of the Ca2+ and membrane-binding loops of the C2 domains within the oligomer.
Finally, we observed that, in addition to forming heptameric particles, Ca2+⋅C2A-C2B also assembled into filaments on lipid monolayers (Fig. 7, which is published as supporting information on the PNAS web site). The cytoplasmic domain of syt VII has also been reported to form filaments (53). The EM in the syt VII study was carried out in the absence of membranes, suggesting that bacterial contaminants, again, were able to partially substitute for an anionic lipid surface. We observed a similar phenomenon for C2A-C2B from syt I that harbored bound contaminants; filaments were formed in response to Ca2+ in the absence of PS. However, under these conditions, C2A-C2B-I did not appear to form heptamers. These findings indicate that RNA and membrane surfaces may act differently to facilitate syt oligomerization, potentially by driving distinct conformational changes in syt. An alternative possibility is that the spatial limitations imposed by RNA, as compared with membrane surfaces, allows assembly of filaments but not heptamers.
We note that fragments corresponding to C2A-III, C2A-VII, and C2A-C2B-I inhibit exocytosis from PC12 cells (11, 41, 54) and in an incompletely purified form oligomerize in response to Ca2+. Future studies are needed to determine whether these fragments disrupt secretion, at least in part, by interfering with the oligomerization of syt isoforms that are expressed in PC12 cells (55). The idea that syt oligomerization is important for release is further supported by the observation that a point mutation in the C2B domain (56) that inhibits Ca2+-triggered oligomerization of syt (27) reduces the evoked exocytosis of docked synaptic vesicles (13).
Preliminary studies in our laboratory indicate that the cytoplasmic domain of syt penetrates into the target membrane during exocytosis (J.B. and E.R.C., unpublished observations; ref. 31). The data shown here indicate that membrane-bound syt assembles into ordered multimers. The structure of these oligomers in vivo may differ from the structure observed in our in vitro studies, because syt is anchored to the vesicle membrane via its transmembrane domain. Furthermore, it is unclear whether heptameric barrels, which are 11 × 11 nm, would fit between the vesicle and target membrane during fusion. We favor a model in which copies of the cytoplasmic domain of syt subunits form lower-ordered oligomers (e.g., dimers or tetramers) on the inner leaflet of the plasma membrane. Assembly of these oligomers might represent structural rearrangements within the fusion complex to regulate the time to opening or dilation kinetics of fusion pores (14). We speculate that syt that has bound to an effector, e.g., membranes and/or SNARE proteins (30), subsequently assembles into multimers, thereby rearranging components of the fusion complex to regulate fusion (14, 57). A key question now is the structure of membrane-embedded full length syt that is bound in trans to target membranes (25).
Supplementary Material
Acknowledgments
We thank Dr. H. M. Zhou for the use of the AKTA purifier; J. Frank for providing the spider software; Z. Liu for helping to write the spider software; Z. A. Pan and W. L. Jiang for maintenance of the EM facility; H. J. Kim for preliminary data regarding syt III oligomerization; L. L. Liu for assistance with the membrane penetration experiments; and M. Jackson, M. Dong, P. Wang, and A. Bhalla for comments. This work was supported by a grant from the National Natural Science Foundation of China (to S.F.S.), the National Institutes of Health (National Institute of General Medical Sciences Grant GM 56827 and National Institute of Mental Health Grant MH61876 to E.R.C.) and the Milwaukee Foundation (to E.R.C.). E.R.C. is a Pew Scholar in the Biomedical Sciences. J.B. is supported by an American Heart Association Predoctoral Fellowship.
Abbreviations
- syt
synaptotagmins
- EM
electron microscopy
- PC
phosphatidylcholine
- PS
phosphatidylserine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Katz B. The Release of Neural Transmitter Substances. Springfield, IL: Thomas; 1969. [Google Scholar]
- 2.Lindau M, Almers W. Curr Opin Cell Biol. 1995;7:509–517. doi: 10.1016/0955-0674(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 3.Augustine G J. Curr Opin Neurobiol. 2001;11:320–326. doi: 10.1016/s0959-4388(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 4.Takei K, Haucke V. Trends Cell Biol. 2001;11:385–391. doi: 10.1016/s0962-8924(01)02082-7. [DOI] [PubMed] [Google Scholar]
- 5.Chapman E R. Nat Rev Mol Cell Biol. 2002;3:498–508. doi: 10.1038/nrm855. [DOI] [PubMed] [Google Scholar]
- 6.Craxton M. Genomics. 2001;77:43–49. doi: 10.1006/geno.2001.6619. [DOI] [PubMed] [Google Scholar]
- 7.Perin M S, Fried V A, Mignery G A, Jahn R, Sudhof T C. Nature. 1990;345:260–263. doi: 10.1038/345260a0. [DOI] [PubMed] [Google Scholar]
- 8.Sutton R B, Ernst J A, Brunger A T. J Cell Biol. 1999;147:589–598. doi: 10.1083/jcb.147.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brose N, Petrenko A G, Sudhof T C, Jahn R. Science. 1992;256:1021–1025. doi: 10.1126/science.1589771. [DOI] [PubMed] [Google Scholar]
- 10.Davletov B A, Sudhof T C. J Biol Chem. 1993;268:26386–26390. [PubMed] [Google Scholar]
- 11.Desai R C, Vyas B, Earles C A, Littleton J T, Kowalchyck J A, Martin T F, Chapman E R. J Cell Biol. 2000;150:1125–1136. doi: 10.1083/jcb.150.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez-Chacon R, Konigstorfer A, Gerber S H, Garcia J, Matos M F, Stevens C F, Brose N, Rizo J, Rosenmund C, Sudhof T C. Nature. 2001;410:41–49. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- 13.Littleton J T, Bai J, Vyas B, Desai R, Baltus A E, Garment M B, Carlson S D, Ganetzky B, Chapman E R. J Neurosci. 2001;21:1421–1433. doi: 10.1523/JNEUROSCI.21-05-01421.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C T, Grishanin R, Earles C A, Chang P Y, Martin T F, Chapman E R, Jackson M B. Science. 2001;294:1111–1115. doi: 10.1126/science.1064002. [DOI] [PubMed] [Google Scholar]
- 15.Sollner T, Bennett M K, Whiteheart S W, Scheller R H, Rothman J E. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 16.Chapman E R, Hanson P I, An S, Jahn R. J Biol Chem. 1995;270:23667–23671. doi: 10.1074/jbc.270.40.23667. [DOI] [PubMed] [Google Scholar]
- 17.Schiavo G, Stenbeck G, Rothman J E, Sollner T H. Proc Natl Acad Sci USA. 1997;94:997–1001. doi: 10.1073/pnas.94.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett M K, Calakos N, Scheller R H. Science. 1992;257:255–259. doi: 10.1126/science.1321498. [DOI] [PubMed] [Google Scholar]
- 19.Chapman E R, An S, Edwardson J M, Jahn R. J Biol Chem. 1996;271:5844–5849. doi: 10.1074/jbc.271.10.5844. [DOI] [PubMed] [Google Scholar]
- 20.Sugita S, Hata Y, Sudhof T C. J Biol Chem. 1996;271:1262–1265. doi: 10.1074/jbc.271.3.1262. [DOI] [PubMed] [Google Scholar]
- 21.Damer C K, Creutz C E. J Neurochem. 1996;67:1661–1668. doi: 10.1046/j.1471-4159.1996.67041661.x. [DOI] [PubMed] [Google Scholar]
- 22.Fukuda M, Mikoshiba K. J Biol Chem. 2000;275:28180–28185. doi: 10.1074/jbc.M001376200. [DOI] [PubMed] [Google Scholar]
- 23.DiAntonio A, Parfitt K D, Schwarz T L. Cell. 1993;73:1281–1290. doi: 10.1016/0092-8674(93)90356-u. [DOI] [PubMed] [Google Scholar]
- 24.Littleton J T, Stern M, Perin M, Bellen H J. Proc Natl Acad Sci USA. 1994;91:10888–10892. doi: 10.1073/pnas.91.23.10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai J, Earles C A, Lewis J L, Chapman E R. J Biol Chem. 2000;275:25427–25435. doi: 10.1074/jbc.M906729199. [DOI] [PubMed] [Google Scholar]
- 26.Fukuda M, Mikoshiba K. J Biol Chem. 2001;276:27670–27676. doi: 10.1074/jbc.M100851200. [DOI] [PubMed] [Google Scholar]
- 27.Fukuda M, Kabayama H, Mikoshiba K. FEBS Lett. 2000;482:269–272. doi: 10.1016/s0014-5793(00)02064-0. [DOI] [PubMed] [Google Scholar]
- 28.Garcia R A, Forde C E, Godwin H A. Proc Natl Acad Sci USA. 2000;97:5883–5888. doi: 10.1073/pnas.100127197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ubach J, Lao Y, Fernandez I, Arac D, Sudhof T C, Rizo J. Biochemistry. 2001;40:5854–5860. doi: 10.1021/bi010340c. [DOI] [PubMed] [Google Scholar]
- 30.Earles C A, Bai J, Wang P, Chapman E R. J Cell Biol. 2001;154:1117–1123. doi: 10.1083/jcb.200105020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bai J, Wang P, Chapman E R. Proc Natl Acad Sci USA. 2002;99:1665–1670. doi: 10.1073/pnas.032541099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernandez I, Arac D, Ubach J, Gerber S H, Shin O, Gao Y, Anderson R G, Sudhof T C, Rizo J. Neuron. 2001;32:1057–1069. doi: 10.1016/s0896-6273(01)00548-7. [DOI] [PubMed] [Google Scholar]
- 33.Mizuta M, Inagaki N, Nemoto Y, Matsukura S, Takahashi M, Seino S. J Biol Chem. 1994;269:11675–11678. [PubMed] [Google Scholar]
- 34.Uzgiris E E, Kornberg R D. Nature. 1983;301:125–129. doi: 10.1038/301125a0. [DOI] [PubMed] [Google Scholar]
- 35.Wang H W, Sui S. J Struct Biol. 1999;127:283–286. doi: 10.1006/jsbi.1999.4161. [DOI] [PubMed] [Google Scholar]
- 36.Wang H W, Sui S. J Struct Biol. 2001;134:46–55. doi: 10.1006/jsbi.2001.4364. [DOI] [PubMed] [Google Scholar]
- 37.Frank J, Verschoor A, Boublik M. Science. 1981;214:1353–1355. doi: 10.1126/science.7313694. [DOI] [PubMed] [Google Scholar]
- 38.Davis A F, Bai J, Fasshauer D, Wolowick M J, Lewis J L, Chapman E R. Neuron. 1999;24:363–376. doi: 10.1016/s0896-6273(00)80850-8. [DOI] [PubMed] [Google Scholar]
- 39.Osborne S L, Herreros J, Bastiaens P I, Schiavo G. J Biol Chem. 1999;274:59–66. doi: 10.1074/jbc.274.1.59. [DOI] [PubMed] [Google Scholar]
- 40.Schiavo G, Gu Q M, Prestwich G D, Sollner T H, Rothman J E. Proc Natl Acad Sci USA. 1996;93:13327–13332. doi: 10.1073/pnas.93.23.13327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugita S, Shin O H, Han W, Lao Y, Sudhof T C. EMBO J. 2002;21:270–280. doi: 10.1093/emboj/21.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shin O H, Rizo J, Sudhof T C. Nat Neurosci. 2002;5:649–656. doi: 10.1038/nn869. [DOI] [PubMed] [Google Scholar]
- 43.Sui S F, Liu Z, Li W, Xiao C, Wang S, Gao Q, Zhou Q. FEBS Lett. 1996;388:103–111. doi: 10.1016/0014-5793(96)00528-5. [DOI] [PubMed] [Google Scholar]
- 44.Qin H, Liu Z, Sui S F. Biophys J. 1995;68:2493–2496. doi: 10.1016/S0006-3495(95)80431-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chapman E R, Desai R C, Davis A F, Tornehl C K. J Biol Chem. 1998;273:32966–32972. doi: 10.1074/jbc.273.49.32966. [DOI] [PubMed] [Google Scholar]
- 46.Shao X, Fernandez I, Sudhof T C, Rizo J. Biochemistry. 1998;37:16106–16115. doi: 10.1021/bi981789h. [DOI] [PubMed] [Google Scholar]
- 47.Ubach J, Zhang X, Shao X, Sudhof T C, Rizo J. EMBO J. 1998;17:3921–3930. doi: 10.1093/emboj/17.14.3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shao X, Li C, Fernandez I, Zhang X, Sudhof T C, Rizo J. Neuron. 1997;18:133–142. doi: 10.1016/s0896-6273(01)80052-0. [DOI] [PubMed] [Google Scholar]
- 49.Fernandez I, Ubach J, Dulubova I, Zhang X, Sudhof T C, Rizo J. Cell. 1998;94:841–849. doi: 10.1016/s0092-8674(00)81742-0. [DOI] [PubMed] [Google Scholar]
- 50.Kee Y, Scheller R H. J Neurosci. 1996;16:1975–1981. doi: 10.1523/JNEUROSCI.16-06-01975.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matos M F, Rizo J, Sudhof T C. Eur J Cell Biol. 2000;79:377–382. doi: 10.1078/0171-9335-00063. [DOI] [PubMed] [Google Scholar]
- 52.Chae Y K, Abildgaard F, Chapman E R, Markley J L. J Biol Chem. 1998;273:25659–25663. doi: 10.1074/jbc.273.40.25659. [DOI] [PubMed] [Google Scholar]
- 53.Fukuda M, Katayama E, Mikoshiba K. J Biol Chem. 2002;277:29315–29320. doi: 10.1074/jbc.M201697200. [DOI] [PubMed] [Google Scholar]
- 54.Sugita S, Han W, Butz S, Liu X, Fernandez-Chacon R, Lao Y, Sudhof T C. Neuron. 2001;30:459–473. doi: 10.1016/s0896-6273(01)00290-2. [DOI] [PubMed] [Google Scholar]
- 55.Zhang X, Kim-Miller M J, Fukuda M, Kowalchyk J A, Martin T F. Neuron. 2002;34:599–611. doi: 10.1016/s0896-6273(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 56.DiAntonio A, Schwarz T L. Neuron. 1994;12:909–920. doi: 10.1016/0896-6273(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 57.Tucker W C, Chapman E R. Biochem J. 2002;366:1–13. doi: 10.1042/BJ20020776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.