Abstract
LEAFY COTYLEDON1 (LEC1) is a central regulator that is required for many aspects of Arabidopsis embryogenesis and sufficient to induce embryo development in vegetative cells when expressed ectopically. We previously showed that LEC1 encodes an HAP3 subunit of the CCAAT binding factor and that the 10 Arabidopsis HAP3 (AHAP3) subunits can be divided into two classes, the LEC1-type and the non-LEC1-type, based on sequence similarity within their B domains. By analyzing the ability of chimeric HAP3 subunits containing different combinations of domains from LEC1 and a non-LEC1-type AHAP3 subunit to suppress genetically the lec1 mutation, we show that the B domain of LEC1 is necessary and sufficient within the context of the protein for its activity in embryogenesis. Moreover, we identify one amino acid residue, Asp-55, specific to the LEC1-type B domain that is required for LEC1 activity in embryogenesis and sufficient to confer partial LEC1 activity to a non-LEC1-type B domain. Based on structural similarities between the HAP3 B domain and histone fold motif, we discuss how the Asp-55 residue may functionally differentiate LEC1 from the non-LEC1-type AHAP3 subunits.
Embryogenesis is a critical period of the flowering plant life cycle during which the single-celled zygote proliferates and undergoes a series of differentiation events, resulting in the formation of a mature embryo. During the early phase of embryogenesis, the polarity of the plant is expressed as the shoot-root axis and the embryonic tissue and organ systems are formed (1–4). Late in embryogenesis, the embryo acquires the ability to withstand desiccation, the seed accumulates storage reserves that serve as food sources after germination, and the embryo becomes quiescent metabolically as a result of seed desiccation (5, 6). Embryo development has been analyzed extensively, and many genes involved in events that characterize embryogenesis have been identified (7). However, little is known at a mechanistic level about the processes that control and coordinate these developmental events.
LEAFY COTYLEDON1 (LEC1) is a central regulator of Arabidopsis embryogenesis that controls many different aspects of embryo development (reviewed in 8). Early in embryogenesis, LEC1 is required to maintain the fate of embryonic cells that constitute the suspensor and to specify the identity of cotyledons, the embryonic leaves (9–11). During late embryogenesis, LEC1 is needed for many events involved in seed maturation, including the acquisition of desiccation tolerance and the accumulation of storage reserves (11–14). LEC1 also plays an essential role by preventing immature seeds from germinating prematurely. Significant insight into the role of LEC1 in embryo development came with the finding that ectopic LEC1 expression was sufficient to confer embryonic characteristics to seedlings and to induce somatic embryo development from vegetative cells (10). Thus, LEC1 plays multiple essential roles both early and late in embryo development and establishes a cellular environment that promotes embryo development.
LEC1 shares significant sequence identity with the HAP3 subunit of the CCAAT binding factor (CBF, also known as NF-Y) (10). CBFs are evolutionary conserved oligomeric transcription factors that contain three nonidentical subunits called HAP2 (CBF-B, NF-YA), HAP3 (CBF-A, NF-YB), and HAP5 (CBF-C, NF-YC) that interact to form a complex that binds the CCAAT DNA motif (reviewed in refs. 15 and 16). Yeast possess a fourth subunit, HAP4, that provides a transcriptional activation domain to the complex (17) whereas in mammals, this domain has been incorporated in the CBF-B and CBF-C subunits (18). Although a functional, plant CBF complex has not yet been isolated, several lines of evidence suggest its existence. First, activities that bind CAAT and CCAAT DNA sequences have been identified in plant nuclear extracts (19, 20). Second, genes encoding proteins with significant sequence identity to the functional domains of HAP2, HAP3, and HAP5 subunits have been identified in plants, though no HAP4 subunit has been identified (19–23, 34). Third, an Arabidopsis HAP2 (AHAP2) subunit suppresses a hap2 mutation of yeast, suggesting that the plant protein interacts functionally in the CBF complex (23). Thus, LEC1 is likely to regulate embryogenesis through its role as a subunit of a transcription factor that modulates the activity of genes required for embryo development.
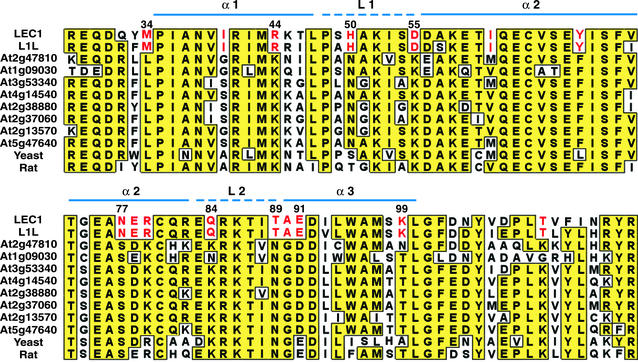
HAP3 subunits, including LEC1, consist of three regions, the A, B, and C domains, with the central B domain being conserved throughout eukaryotic evolution. The B domain possesses amino acid residues required for interaction of HAP3 with other CBF subunits and for DNA-binding activity of the CBF complex (24–26). Unlike yeast and mammals, plants possess families of genes that encode each CBF subunit (20, 22, 23). We showed previously that the 10 AHAP3 subunits could be divided into two classes on the basis of sequence similarity in the B domain: the LEC1-type, consisting of LEC1 and LEC1-LIKE (L1L), and the non-LEC1-type comprising the remaining subunits (22). Specifically, LEC1-type B domains possess 16 shared residues that differ from conserved residues found at equivalent positions in non-LEC1 type AHAP3 B domains, as shown in Fig. 1. We also showed that L1L, but not genes encoding non-LEC1-type AHAP3 subunits, could suppress the lec1 mutation when expressed ectopically, suggesting that the two LEC1-type proteins are functionally equivalent. Thus, LEC1-type AHAP3 subunits appear to function differently than their non-LEC1-type counterparts.
Figure 1.
Amino acid sequence alignment of the B domains of HAP3 subunits from Arabidopsis, yeast, and rat. Identical residues are shaded in yellow boxes. Residues specific to LEC1 and L1L are highlighted in red. The positions of α-helices and loops in the histone fold motif are indicated as continuous and dashed blue lines, respectively. Numbers indicate amino acid position within LEC1.
To begin to understand how LEC1 differs from non-LEC1-type AHAP3 subunits at a mechanistic level, we identified amino acid residues required for LEC1's specific function in embryogenesis. We show that the B domain of LEC1 is required for its function in embryo development and identify a specific B domain residue critical for its activity. Implications of these studies for the mechanism by which the CBF containing LEC1 regulates genes required for embryogenesis are discussed.
Materials and Methods
Plant Material.
Wild-type and lec1-1 mutant plants of Arabidopsis thaliana (L.) Heynh ecotype Wassilewskija lines were grown as described (11).
Constructs Encoding Chimeric Proteins.
PCR methods were used to make the domain-swap constructs. For example, the LA-LB-NCconstruct was prepared by amplifying separately regions encoding the A and B domains of LEC1 and the C domain of At4g14540 by using primer combinations LEC1A-F (5′-CCCGGGGAGATCTATGACCAGCTCAGTCATAGTAGC-3′)/LEC1B-R (5′-TTGTCTCCCTGCCGTAGTAGTACGATCGGTCTCTATCTCACG-3′) and At4gC-F (5′-CGTGAGATAGAGACCGATCGTACTACTACGGCAGGGAGACAA-3′)/At4gC-R (5′-CCCGGCCTAGGTTAAGAAAAATGATGGGAAAATTGATGTCC-3′), respectively. Primers LEC1B-R and At4gC–F contain complementary regions that facilitated the joining of the fragments by combining, annealing, and extending the two PCR products. The ORF containing the entire LA-LB-NCcDNA was then amplified by using the end primers, LEC1A-F and At4gC-R, that each contain restriction enzyme sites for cloning. After nucleotide sequencing to confirm the correct ORF, constructs were inserted into the LEC1 expression cassette (22). Specific details for preparation of other constructs are available on request.
Site-Directed Mutagenesis.
Amino acid substitution mutants of LEC1 were created by using a modification of the method described above. For example, to replace Met 34 of LEC1 with Leu, LEC1 cDNA was amplified with the primer pairs LEC1 A-F (5′-CCCGGGGAGATCTATGACCAGCTCAGTCATAGTAGC-3′)/LEC1M34L-R (5′-CGTTTGCGATTGGAAGGTATTGGTCTTGC-3′) and LEC1 M34L-F (5′-AGCAAGACCAATACCTTCCAATCGCAAACG-3′)/LEC1 C-R (5′-GGGCCCCTAGGTCTTCACTTATACTGACCATAATGG-3′), where the positions of the substitution is underlined. The two PCR products were annealed and amplified by using LEC1 A-F and LEC1 C-R, and inserted into the LEC1 expression cassette after the nucleotide sequence was confirmed. Additional details are available on request.
Genetic Suppression Assay.
Constructs were transferred into homozygous lec1-1 mutants by using Agrobacterium tumefaciens strain GV3101 and in planta transformation procedures (27). T1 seeds from T0 plants were dried for 2 weeks at 28°C and plated on selection media containing 20 μg/ml hygromycin or 60 μg/ml glufosinate ammonium (Finale, AgrEvo Environmental Health, Montvale, NJ). The number of viable seedlings was counted, and their genotypes were verified in PCR amplification experiments.
Results
Domain-Swap Experiments Reveal that the LEC1 B Domain Is Critical for Its Function.
Given that LEC1 and non-LEC1-type AHAP3 subunits serve different roles during plant growth and development (22), we sought initially to identify the regions that distinguish the two proteins functionally. All HAP3 proteins, including LEC1, consist of three domains, an N-terminal A domain, a central B domain, and a C-terminal C domain, with the B domain being required for DNA binding and interaction with other CCAAT binding factor subunits (25, 26). LEC1 does not share significant sequence identity with any other AHAP3 subunit in the A or C domains (22).
To identify the domain that underlies LEC1 activity in embryogenesis, we took advantage of the modular structure of AHAP3 subunits and constructed six fusion proteins that possessed all possible combinations of A, B, and C domains from LEC1 and the non-LEC1-type AHAP3 subunit encoded by At4g14540. This non-LEC1-type AHAP3 subunit was chosen because previous studies showed that it is distantly related to LEC1 and it cannot substitute for LEC1 when expressed ectopically (22). Constructs encoding these chimeric proteins were inserted into a LEC1 expression cassette that contains 1,997 bp of 5′ and 774 bp of 3′ untranslated and flanking regions of the LEC1 gene (22). To determine which of the chimeric proteins possessed LEC1 activity, the constructs were tested for their ability to suppress the lec1 mutation in planta by transferring the constructs into lec1-1 null mutant plants in which the LEC1 gene has been deleted (10). Homozygous lec1-1 mutant seeds do not germinate because their embryos are intolerant of desiccation and die on drying, though immature seeds can be rescued before silique drying and germinated to produce vegetatively growing plants (9, 11). Thus, genetic suppression is indicated by the ability of transgenic lec1 mutant seeds to germinate after desiccation and give rise to viable seedlings.
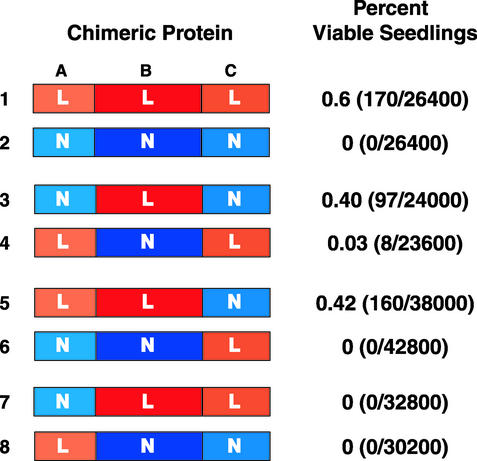
Control experiments showed that lec1 mutant plants transformed with the LEC1 gene by using the infiltration method produced an average of 0.6% viable seeds (Fig. 2, construct 1). This value reflects both the transformation efficiency of the infiltration method (27) and the ability of the LEC1 transgene to suppress the lec1 mutation efficiently. By contrast, plants transformed with At4g14540 in the LEC1 expression cassette did not give rise to viable seeds (Fig. 2, construct 2), confirming that this non-LEC1-type AHAP3 subunit cannot suppress the lec1 mutation and that the suppression strategy is valid (22).
Figure 2.
LEC1 B domain is necessary and sufficient for its activity in embryogenesis. The diagrammatic representation shows LEC1 (construct 1), the non-LEC1-type AHAP3 subunit, At4g14540 (construct 2), and chimeric proteins containing A, B, and C domains from LEC1 (L) or At4g14540 (N). Constructs encoding these chimeric proteins were transformed into lec1-1 null mutants and, after drying of the T1 seeds, the number of viable seedlings generated from seeds tested (shown in parentheses) was determined. Transformation experiments were repeated multiple times with similar results, and the total values for all experiments are reported.
The most striking results were obtained with chimeric proteins that had their B domains exchanged. The construct encoding the chimeric protein consisting of Non-LEC1-type A and C domains and the LEC1 B domain, designated NA-LB-NC (Fig. 2, construct 3), suppressed the lec1 mutation and produced viable seedlings with an efficiency (0.4%) close to that of the LEC1 control (0.6%). T1 plants segregated T2 embryos resembling either wild-type or lec1 mutants at ratios suggesting the presence of one or two transgenes (data not shown). These results suggest that the NA-LB-NC construct behaved like the wild-type LEC1 gene. By contrast, the frequency of suppression by the construct encoding its counterpart, LA-NB-LC, was significantly lower (0.03%, Student's t test, P < 0.005) (Fig. 2, construct 4). Control experiments showed that all domain-swap constructs were competent to transform wild-type plants at a frequency similar to a vector-only control and that wild-type plants containing these constructs germinated with near 100% efficiency. These results indicate that the constructs were transformation competent and that the chimeric proteins did not have a negative effect on embryo or seedling growth (data not shown). Although a small number of lec1 mutant embryos containing the LA-NB-LC construct survived desiccation, these plants segregated progeny that resembled wild-type and lec1 mutant embryos and those with morphological phenotypes intermediate between the two, as shown in Fig. 3. Thus, LA-NB-LC was only minimally effective in rescuing morphological defects and desiccation intolerance caused by the lec1 mutation, suggesting that the chimeric protein exhibits attenuated LEC1 function. Together, these results suggest that the B domain of LEC1 within the context of the A and C domains is both necessary and sufficient for LEC1 activity.
Figure 3.
Partial rescue of morphological defects caused by the lec1 mutation. Representative T2 embryos of lec1-1 mutants transformed with the LA-NB-LC construct that display a range of morphological phenotypes intermediate between those of lec1 mutants and wild type. All embryos were dissected from siliques containing wild-type embryos at the mature green stage. (Bar = 300 μm.)
Suppression assays were also done to address the roles of the LEC1 A and C domains. The LA-LB-NC construct suppressed the lec1 mutation (Fig. 2, construct 5), but the NA-NB-LC construct did not (Fig. 2, construct 6), indicating that the LEC1 C domain is dispensable for its function. Neither the NA-LB-LC nor the LA-NB-NC constructs suppressed the lec1 mutation (Fig. 2, constructs 7 and 8). Unlike the B and C domain-swap experiments described above, no positive result showing sufficiency was obtained with this pair of constructs to corroborate the loss-of-function result showing necessity, making interpretation of these results difficult. For example, it is not clear whether the A domain influences LEC1 activity or whether the chimeric proteins do not fold or accumulate properly. The finding that the LEC1 A and C domains are not sufficient within an AHAP3 subunit to confer LEC1 activity is consistent with the conclusion that the LEC1 B domain primarily underlies its activity in embryogenesis.
Identification of B Domain Residues Required for LEC1 Function.
LEC1 and non-LEC1-type AHAP3 subunits can be distinguished on the basis of specific amino residues within the B domain that are highlighted in red in Fig. 1 (22). To determine which of these residues are important for LEC1 function, we substituted residues specific for LEC1 with those conserved at corresponding positions in non-LEC1-type AHAP3 subunits. For example, the Met at position 34 of LEC1 was replaced with the Leu present at the corresponding position in all non-LEC1-type B domains to create LEC1 (M34L). We focused on those residues that significantly changed the chemical properties of the LEC1-specific amino acid residues. cDNA clones encoding mutant forms of LEC1 with amino acid substitutions were inserted into the LEC1 expression cassette and transferred into lec1-1 null mutants to assess their ability to suppress the lec1 mutation.
As summarized in Table 1, constructs encoding mutagenized LEC1 proteins with single amino acid substitutions of Met at position 34 for Leu (M34L), Arg for Lys (R44K), His for Asn (H50N), Asn for Ser (N77S), or Gln for Lys (Q84K) suppressed the lec1 mutation, resulting in the production of between 0.4% and 0.8% viable seedlings. Replacement of the Thr (T)–Ala (A)–Glu (E) residues at positions 89–91 with Asn (N)–Gly (G)–Asp (D) resulted in the production of 0.4% of viable seedlings. These results suggested that the amino acid residues at these positions are not absolutely required for LEC1 function.
Table 1.
Identification of a LEC1 B domain residue required for LEC1 activity
| Amino acid substitution in LEC1* | Percent viable seedlings† |
|---|---|
| M34L | 0.8 (110 of 14,400) |
| R44K | 0.6 (82 of 12,800) |
| H50N | 0.7 (176 of 24,000) |
| D55K | 0.01 (5 of 55,600) |
| N77S | 0.4 (48 of 12,000) |
| Q84K | 0.4 (75 of 20,800) |
| TAE89-91NGD | 0.4 (73 of 16,800) |
| K99T | 0.4 (95 of 22,080) |
Mutant forms of LEC1 are designated with the wild-type amino acid and its position within LEC1, followed by the amino acid that was inserted. cDNA clones encoding LEC1 with the indicated single or triple amino acid substitutions were fused with LEC1 5′ and 3′ flanking sequences and transferred into lec1-1 mutants.
T1 lec1-1 seeds were collected and dried for 2 weeks at 28°C before germination tests were performed. Values obtained reflect the ability of the construct to suppress the lec1 mutation and the transformation efficiency. For the wild-type LEC1 gene, this value was 0.6 (see Fig. 2). The total number of viable seedlings obtained and seeds tested in all experiments are given in parentheses. Independent replicates of transformation experiments gave values similar to the percentages reported here.
By contrast, substitution of Asp at position 55 with lysine (D55K) severely compromised the ability of the LEC1 construct to suppress the lec1 mutation, resulting in a 40-fold decrease in the recovery of viable seedlings (0.01%; Table 1). Experiments showed that this construct transformed wild-type plants at a frequency similar to that of a control plasmid, indicating that it was not defective. This result suggests that the Asp-55 residue is critically required for LEC1 function.
Single Amino Acid Change Within Non-LEC1-Type B Domain Confers LEC1 Activity.
To confirm the importance of this Asp-55 residue, we determined whether the complementary change would confer LEC1 activity to a non-LEC1-type B domain. Specifically, we mutagenized the construct encoding LA-NB-LC (see Fig. 2) such that the Lys at position 55 was replaced with the Asp characteristic of the LEC1 B domain (K55D). Table 2 shows that unlike the LA-NB-LC domain-swap construct that suppressed the lec1 mutation only at a low frequency (0.03%), LA-NB-LC (K55D) resulted in an 8-fold increase in the number of viable seedlings (0.23%). Thus, this single amino acid substitution permitted a non-LEC1-type B domain to rescue the desiccation-intolerance defect caused by the lec1 mutation.
Table 2.
A single amino acid substitution confers LEC1 activity to a non-LEC1-type B domain
| Construct* | Percent viable seedlings† |
|---|---|
| LA-NB-LC | 0.03 (8 of 23,600)‡ |
| LA-NB-LC (K55D) | 0.23 (70 of 30,400) |
LA-NB-LC consists of A and C domains from LEC1 and the B domain from the non-LEC1-type AHAP3, At4g14540 (see Fig. 2).
Percent viable seedlings was determined as described. Consistent values were obtained from independently repeated experiments. The numbers of seedlings and seeds represent the sum of replicate experiments.
Data from Fig. 2.
Analysis of progeny provided additional information about the effectiveness of LA-NB-LC (K55D) to substitute for LEC1. Progeny from these transgenic plants segregated embryos with wild-type, lec1 mutant, and intermediate morphological phenotypes similar to those observed with LA-NB-LC plants (see Fig. 3). Because LA-NB-LC (K55D) significantly enhanced the ability of the chimeric protein to confer desiccation tolerance to lec1-1 mutant embryos but did not completely rescue morphological defects, we conclude that this single amino acid substitution only partially restores LEC1 activity. These results suggest that the Asp residue at position 55 plays an important role in LEC1 function.
Discussion
LEC1 Defines a Specialized Class of HAP3 Subunit.
Our previous studies showed that AHAP3 subunits can be divided into at least two classes, LEC1-type and non-LEC1-type, based primarily on sequence differences in their B domains (22). Here we provide two lines of evidence that strongly support the hypothesis that the B domain is primarily responsible for differentiating LEC1 functionally from non-LEC1-type AHAP3 subunits. First, the ability of NA-LB-NC to rescue desiccation intolerance and morphological defects of lec1 mutant embryos strongly suggests that the LEC1 B domain is sufficient for LEC1 activity within the context of the A and C domain (Fig. 2). Consistent with this conclusion, all constructs without the LEC1 B domain did not rescue lec1 mutant defects efficiently, suggesting that the LEC1 B domain is necessary for its function.
L1L, the only other AHAP3 subunit with a LEC1-type B domain, but not two non-LEC1-type AHAP3 subunits suppresses the lec1 mutation when expressed from the LEC1 expression cassette and confers embryonic characteristics to seedlings when driven by the cauliflower mosaic virus 35S promoter (22). Because LEC1 and L1L do not exhibit sequence identity in their A and C domains, this finding provides additional evidence that the B domain differentiates LEC1 and non-LEC1-type AHAP3 subunits functionally.
We note that neither the LA-NB-NC nor the NA-LB-LC chimeric proteins rescued lec1 mutant embryos, making interpretation of this set of results difficult. Because the A domains of LEC1 and At4g14540 differ substantially (46 residues vs. 14 residues, respectively) (22), it is formally possible that the length of the A domain may be important for LEC1 function. However, because the NA-LB-NC and LA-LB-NC constructs suppressed the lec1 mutation, a more likely explanation is that the NA-LB-LC chimeric protein adopted an abnormal confirmation, thereby negating its activity and/or accumulation.
Second, our results show that a single amino acid specific to the LEC1 B domain, Asp-55, is required to confer desiccation tolerance to lec1 mutant embryos and that substitution of the non-LEC1-type B domain residue, Lys-55, with Asp to create LA-NB-LC (K55D) confers partial LEC1 activity to the non-LEC1-type B domain (Tables 1 and 2). These results strongly support the importance of Asp-55 in conferring LEC1 function and suggest that the inability of LA-NB-LC and LEC1 (D55K) to suppress the lec1 mutation efficiently does not result from instability of the mutagenized proteins. Because LA-NB-LC (K55D) does not completely suppress the lec1 mutation, it is likely that other LEC1-specific B domain residues also play important roles in conferring LEC1 activity.
Thus, the B domain differentiates LEC1 functionally from non-LEC1-type AHAP3 subunits. Because yeast and mammalian B domains are more closely related to non-LEC1-type than LEC1-type B domains (22), these results suggest that LEC1 represents a previously undescribed class of HAP3 subunit.
Role of Asp-55 in Determining LEC1 Activity.
Results of the domain-swap and site-directed mutagenesis studies show that the B domain differentiates LEC1 functionally from non-LEC1-type AHAP3 subunits. The B domain is required for the interaction of yeast and mammalian HAP3 subunits with the other CBF subunits, HAP2 and HAP5, and for the DNA binding activity of the CBF complex (24–26). Several alternatives could explain how the LEC1 B domain confers unique activity to the CBF (22). First, the LEC1 B domain may affect the DNA binding specificity or binding affinity of the CBF complex. Second, because Arabidopsis possesses 10 and 9 genes encoding AHAP2 and AHAP5 subunits, respectively, the LEC1 B domain may bind with a specific combination of these other two subunits to form a CBF that activates genes required for embryogenesis. Third, the LEC1 B domain may recruit another transcription factor to confer a unique activity to the CBF complex. AHAP2, -3, and -5 subunits do not possess obvious transcriptional activation domains, as do the mammalian HAP2 and HAP5 subunits (18). A different subunit may be necessary to provide a transcriptional activation function to the CBF complex or to alter its binding specificity.
Our finding that Asp-55 of LEC1 is critical for its function provides insight into how the LEC1 B domain may confer unique activity to the CBF complex. The B domain of HAP3 subunits shares sequence identity with the histone fold motif of histone H2B, a structurally conserved region consisting of three α helices (α1, α2, and α3) connected by two short loops (L1 and L2, see Fig. 1) (26, 28). The histone fold motif mediates protein–protein and DNA binding interactions (29–32). For example, the HAP3 histone fold motif mediates heterodimerization with the histone H2A-like histone fold motif of HAP5 as the first step in CBF assembly (33). Although LEC1 possesses amino acid residues that differentiates it from other AHAP3 subunits, it retains residues characteristic of the histone fold motif.
Sequence alignment of the B domain and histone H2B positions Asp-55 of LEC1 at the junction between L1 and α2-helix of the histone fold motif (Fig. 1). Based on the three-dimensional structure of the nucleosome core particle, the amino acid residue at this position is located on the surface of the histone octamer complex in a region that contacts DNA (29, 31). Assuming that Asp-55 is located similarly in the CBF containing LEC1, this result supports the hypothesis that Asp-55 of the LEC1 B domain may affect the DNA binding specificity or affinity relative to that of non-LEC1-type AHAP3 subunits. In support of this hypothesis, it has been shown that the histone fold motif of the HAP3 subunit contributes to the CCAAT specificity to the complex, though the α1 helix has been implicated to play a primary role (32). Moreover, substitution of the Lys residue at the corresponding position of the rat HAP3 subunit with Ala did not affect its ability to interact with HAP2 and HAP5 subunits but, rather, altered the DNA binding activity of the CBF complex (26). Together, these findings open the possibility that the CBF containing LEC1 may regulate genes required for embryo development by binding a different DNA consensus sequence than a CBF containing a non-LEC1-type AHAP3 subunit. Because Asp-55 is predicted to be on the surface of LEC1, we cannot formally exclude the possibility that Asp-55 acts in the LEC1 B domain to mediate specific interactions with other CBF subunits or with other transcription factors (26). Additional studies about the components of LEC1-CBF and identification of the complex's target sequence are necessary to define precisely how Asp-55 contributes to the unique function of the CBF containing LEC1 during embryogenesis.
Acknowledgments
We thank Abeba Kiros and Julie Pelletier for technical assistance, Hongyu Zhang and Nickolai Alexandrov for help in analyzing protein structure, and Raymond Kwong, Katie Dehesh, and Jack Okamuro for helpful discussions. This work was supported by grants from the Department of Energy (to J.J.H.) and Ceres, Inc.
Abbreviations
- AHAP
Arabidopsis heme-activated protein
- CBF
CCAAT binding factor
- LEC1
LEAFY COTYLEDON1
- L1L
LEC1-LIKE
Note Added in Proof.
A recent publication (35) confirms that the HAP3 (NF-YB) and HAP5 (NF-YC) subunits from humans interact through their histone fold motifs and that the residue corresponding to Asp-55 of LEC1 is located at the junction between L1 and α2.
References
- 1.Laux T, Jurgens G. Plant Cell. 1997;9:989–1000. doi: 10.1105/tpc.9.7.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldberg R B, de Paiva G, Yadegari R. Science. 1994;266:605–614. doi: 10.1126/science.266.5185.605. [DOI] [PubMed] [Google Scholar]
- 3.West M A L, Harada J J. Plant Cell. 1993;5:1361–1369. doi: 10.1105/tpc.5.10.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jurgens G. EMBO J. 2001;20:3609–3616. doi: 10.1093/emboj/20.14.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harada J J. In: Cellular and Molecular Biology of Seed Development, Advances in Cellular and Molecular Biology of Plants. Larkins B A, Vasil I K, editors. Vol. 4. Dordrecht, The Netherlands: Kluwer Academic; 1997. pp. 545–592. [Google Scholar]
- 6.Bewley J D. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berleth T, Chatfield S. In: The Arabidopsis Book. Somerville C R, Meyerowitz E M, editors. Rockville, MD: Am. Soc. Plant Biol.; 2002. , doi/10.1199/tab.0009. [Google Scholar]
- 8.Harada J J. J Plant Physiol. 2001;158:405–409. [Google Scholar]
- 9.Meinke D W. Science. 1992;258:1647–1650. doi: 10.1126/science.258.5088.1647. [DOI] [PubMed] [Google Scholar]
- 10.Lotan T, Ohto M, Matsudaira Yee K, West M A L, Lo R, Kwong R W, Yamagishi K, Fischer R L, Goldberg R B, Harada J J. Cell. 1998;93:1195–1205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- 11.West M A L, Matsudaira Yee K L, Danao J, Zimmerman J L, Fischer R L, Goldberg R B, Harada J J. Plant Cell. 1994;6:1731–1745. doi: 10.1105/tpc.6.12.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meinke D W, Franzmann L H, Nickle T C, Yeung E C. Plant Cell. 1994;6:1049–1064. doi: 10.1105/tpc.6.8.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parcy F, Valon C, Kohara A, Misera S, Giraudat J. Plant Cell. 1997;9:1265–1277. doi: 10.1105/tpc.9.8.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vicient C M, Bies-Etheve N, Delseny M. J Exp Bot. 2000;51:995–1003. doi: 10.1093/jexbot/51.347.995. [DOI] [PubMed] [Google Scholar]
- 15.Maity S N, de Crombrugghe B. Trends Biochem Sci. 1998;23:174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- 16.Mantovani R. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 17.Forsburg S L, Guarente L. Genes Dev. 1989;3:1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- 18.Coustry F, Maity S N, Sinha S, de Crombrugghe B. J Biol Chem. 1996;272:14485–14491. doi: 10.1074/jbc.271.24.14485. [DOI] [PubMed] [Google Scholar]
- 19.Kusnetsov V, Landsberger M, Meurer J, Oelmuller R. J Biol Chem. 1999;274:36009–36014. doi: 10.1074/jbc.274.50.36009. [DOI] [PubMed] [Google Scholar]
- 20.Gusmaroli G, Tonelli C, Mantovani R. Gene. 2001;264:173–185. doi: 10.1016/s0378-1119(01)00323-7. [DOI] [PubMed] [Google Scholar]
- 21.Albani D, Robert L S. Gene. 1995;167:209–213. doi: 10.1016/0378-1119(95)00680-x. [DOI] [PubMed] [Google Scholar]
- 22.Kwong R W, Bui A Q, Lee H, Kwong L W, Fischer R L, Goldberg R B, Harada J J. Plant Cell. 2003;15:5–18. doi: 10.1105/tpc.006973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards D, Murray J A H, Smith A G. Plant Physiol. 1998;117:1015–1022. doi: 10.1104/pp.117.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X-Y, Mantovani R, Hooft Van Huijsduijnen R, Andre I, Benoist C, Mathis D. Nucleic Acids Res. 1992;20:1087–1091. doi: 10.1093/nar/20.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xing Y, Fikes J D, Guarente L. EMBO J. 1993;12:4647–4655. doi: 10.1002/j.1460-2075.1993.tb06153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinha S, Kim I S, Sohn K-Y, de Crombrugghe B, Maity S N. Mol Cell Biol. 1996;16:328–337. doi: 10.1128/mcb.16.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bechtold N, Ellis J, Pelletier G. C R Acad Sci Paris. 1993;316:1194–1199. [Google Scholar]
- 28.Bolognese F, Imbriano C, Caretti G, Mantovani R. Nucleic Acids Res. 2000;28:3830–3838. doi: 10.1093/nar/28.19.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arents G, Moudrianakis E N. Proc Natl Acad Sci USA. 1995;92:11170–11174. doi: 10.1073/pnas.92.24.11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baxevanis A D, Arents G, Mourdianakis E N, Landsman D. Nucleic Acids Res. 1995;23:2685–2691. doi: 10.1093/nar/23.14.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 32.Zemzoumi K, Frontini M, Bellorini M, Mantovani R. J Mol Biol. 1999;286:327–337. doi: 10.1006/jmbi.1998.2496. [DOI] [PubMed] [Google Scholar]
- 33.Sinha S, Maity S N, Lu J, de Crombrugghe B. Proc Natl Acad Sci USA. 1995;92:1624–1628. doi: 10.1073/pnas.92.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gusmaroli G, Tonelli C, Mantovani R. Gene. 2002;283:41–48. doi: 10.1016/s0378-1119(01)00833-2. [DOI] [PubMed] [Google Scholar]
- 35.Romier C, Cocchiarella F, Mantovani R, Moras D. J Biol Chem. 2003;278:1336–1345. doi: 10.1074/jbc.M209635200. [DOI] [PubMed] [Google Scholar]