Abstract
Kinetochore proteins contribute to the fidelity of chromosome transmission by mediating the attachment of a specialized chromosomal region, the centromere, to the mitotic spindle during mitosis. In budding yeast, a subset of kinetochore proteins, referred to as the outer kinetochore, provides a link between centromere DNA-binding proteins of the inner kinetochore and microtubule-binding proteins. Using a combination of chromatin immunoprecipitation, in vivo localization, and protein coimmunoprecipitation, we have established that yeast Chl4p and Iml3p are outer kinetochore proteins that localize to the kinetochore in a Ctf19p-dependent manner. Chl4p interacts with the outer kinetochore proteins Ctf19p and Ctf3p, and Iml3p interacts with Chl4p and Ctf19p. In addition, Chl4p is required for the Ctf19p-Ctf3p and Ctf19p-Iml3p interactions, indicating that Chl4p is an important structural component of the outer kinetochore. These physical interaction dependencies provide insights into the molecular architecture and centromere DNA loading requirements of the outer kinetochore complex.
INTRODUCTION
To maintain a high fidelity of chromosome transmission during mitosis, the genetic material must be segregated accurately to daughter cells. Failures in this process lead to aneuploidy and may contribute to the development of cancer (Lengauer et al., 1998). To ensure proper chromosome segregation, duplicated chromosomes must attach to microtubules that will guide them toward opposite poles of the mitotic spindle. This attachment occurs at a specific region of the chromosome called the centromere (reviewed in Sullivan et al., 2001). The budding yeast Saccharomyces cerevisiae centromere is 125 base pairs (bp) in length and contains three conserved DNA elements (CDEs). CDEI and III are conserved sequences, whereas CDEII is an intervening A/T-rich region (Hyman and Sorger, 1995). A large multiprotein complex, the kinetochore, assembles on centromere DNA (CEN DNA) and provides a link to spindle microtubules (Dobie et al., 1999; Pidoux and Allshire, 2000).
The CBF3 complex (or inner kinetochore), which contains Ndc10p, has been shown to bind CDEIII directly (Lechner and Carbon, 1991). The outer kinetochore is comprised of several protein complexes that associate with CEN DNA chromatin via the CBF3 complex (reviewed in Ortiz and Lechner, 2000) and serve as probable links to microtubules, motor proteins, or regulatory proteins. To date, there are four known outer kinetochore protein complexes: the Ctf19 complex (Ortiz et al., 1999), the Ctf3 complex (Measday et al., 2002), the Ndc80 complex (Janke et al., 2001; Wigge and Kilmartin, 2001), and the Dam1 complex (Cheeseman et al., 2001; Janke et al., 2002; Li et al., 2002). Several other proteins have been proposed to function at the kinetochore (reviewed in Kitagawa and Hieter, 2001), including Cse4p, a modified histone H3 that is part of a specialized centromeric nucleosome (Meluh et al., 1998), and Mif2p, a homologue of human CENP-C that may bind centromeric A/T-rich regions (Meluh and Koshland, 1997). All outer kinetochore complexes contain proteins that localize to the kinetochore, interact specifically with CEN DNA, and contribute to the fidelity of chromosome transmission. Some components of the Dam1 complex bind microtubules (Hofmann et al., 1998), placing this complex at the periphery of the outer kinetochore. However, the exact physical interactions and molecular architecture of complexes within the kinetochore and the pattern of assembly of individual proteins in this supramolecular complex still remain to be elucidated.
Hallmarks for the identification of kinetochore proteins include their ability to cross-link to CEN DNA, as assayed by chromatin immunoprecipitation (ChIP), and to localize next to the nuclear side of the spindle pole body (SPB). These criteria have been used to assess candidate genes that potentially encode kinetochore proteins (He et al., 2001), including genes required for faithful chromosome transmission identified by genetic screens. Several independent screens have led to the isolation of mutants that lose chromosomes at a higher rate than a wild-type strain (the chl, mcm, ctf, and cin mutants) (Maine et al., 1984; Kouprina et al., 1988; Hoyt et al., 1990; Spencer et al., 1990). CHL4/CTF17/MCM17 was identified in three of these mutant collections (Kouprina et al., 1988; Spencer et al., 1990; Kouprina et al., 1993b; Roy et al., 1997). A strain deleted for CHL4 is viable (Roy et al., 1997), and several secondary phenotypes suggest that chl4 mutants have a compromised kinetochore: chl4 is able to maintain a dicentric plasmid with the same fidelity as a monocentric plasmid (Doheny et al., 1993; Kouprina et al., 1993a); chl4 weakens the transcription block that occurs when a CEN sequence is placed between a promoter and a reporter construct (Doheny et al., 1993); and two mutant alleles of chl4 become inviable upon increased dosage of CTF13 or NDC10 (Kroll et al., 1996; Measday et al., 2002). Recently, Chl4p was shown to have a two-hybrid interaction with Iml3p/Mcm19p (Ghosh et al., 2001). iml3 mutants exhibit an increased rate of centromere plasmid loss (Roy et al., 1997; Entian et al., 1999) as well as a number of phenotypes suggesting that IML3 encodes a previously uncharacterized kinetochore protein.
Herein, we establish Chl4p and Iml3p as bona fide kinetochore proteins, and we examine their physical juxtaposition and CEN DNA loading requirements within the kinetochore complex. Chl4p cross-links to CEN DNA chromatin, localizes to the kinetochore, and interacts with two known outer kinetochore proteins, Ctf19p and Ctf3p. In addition, Chl4p interacts with a new kinetochore component, Iml3p, that displays a Chl4p-dependent CEN DNA interaction and a kinetochore localization pattern. Using a combination of biochemical and in vivo localization techniques, we determine specific requirements and dependencies for Chl4p, Iml3p, Ctf19p, and Ctf3p interactions and kinetochore localization, providing insights into the molecular architecture of the kinetochore complex.
MATERIALS AND METHODS
Yeast Strains and Media
Strains used in this study are listed in Table 1. Media for growth and sporulation were described previously (Rose et al., 1990). To visualize chromosome fragment loss, strains were first grown on SC medium lacking uracil (selecting for the chromosome fragment) and then streaked onto YPD medium. In strains with a high rate of chromosome loss, a colony will consist of cells containing the chromosome fragment (white), and cells that have lost it (red), resulting in a white and red sectored phenotype. For the microtubule-depolymerizing drug sensitivity assay, benomyl from DuPont (Wilmington, DE) was added at the indicated concentration to YPD media; dimethyl sulfoxide was used in the control plate (0 μg/ml benomyl). Epitope tagging and gene deletions were made directly at their endogenous loci according to Longtine et al. (1998). Yeast transformations were done according to Gietz and Schiestl (1995).
Table 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| YCTF30 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 CFIII (CEN3.L. YPH278) URA3 SUP11 ctf13-30 | (Spencer et al., 1990) |
| YCTF42 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 CFIII (CEN3.L. YPH278) URA3 SUP11 ctf14-42 | (Spencer et al., 1990) |
| YPH499 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 | P. Hieter |
| YPH500 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 | P. Hieter |
| YPH1027 | MATa ura3-52 leu2-3, 112 ndc10-2 | T. Huffaker |
| YPH1124 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 CFIII (CEN3.L. YPH982) URA3 SUP11 | P. Hieter |
| YPH1315 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 ctf19Δ::HIS3 | (Hyland et al., 1999) |
| YPH1316 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 ctf19Δ::TRP1 | (Hyland et al., 1999) |
| YPH1534 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 chl4Δ::His3MX6 CFIII (CEN3.L. YPH982) URA3 SUP11 | This study |
| YPH1535 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 chl4-20 CFIII (CEN3.L. YPH982) URA3 SUP11 | This study |
| YPH1536 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 chl4-61 CFIII (CEN3.L. YPH982) URA3 SUP11 | This study |
| YPH1537 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 chl4Δ::His3MX6 | This study |
| YPH1538 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 chl4Δ::kanMX6 | This study |
| YPH1539 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 chl4Δ::kanMX6 | This study |
| YPH1540 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 chl4-20 | This study |
| YPH1541 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 chl4-61 | This study |
| YPH1542 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 CHL4-13Myc-TRP1 | This study |
| YPH1543 | MATα ura3-52 lys2-801 ade2-101 leu2 trp1Δ63 ndc10-2 CHL4-13Myc-TRP1 | This study |
| YPH1544 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 ctf19Δ::HIS3 CHL4-13Myc-TRP1 | This study |
| YPH1545 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 ctf3Δ::HIS3 CHL4-13Myc-TRP1 | This study |
| YPH1546 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 CHL4-13Myc-TRP1 CTF3-3HA-His3MX6 | This study |
| YPH1547 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 CHL4-13Myc-TRP1 CTF3-3HA-kanMX6 | This study |
| YPH1548 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 ctf19Δ::HIS3 CHL4-13Myc-TRP1 CTF3-3HA-kanMX6 | This study |
| YPH1549 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 iml3Δ::kanMX6 CHL4-13Myc-TRP1 | This study |
| YPH1550 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 CTF19-13Myc-TRP1 | This study |
| YPH1551 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 chl4Δ::His3MX6 CTF19-13Myc-TRP1 | This study |
| YPH1552 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 chl4Δ::His3MX6 CTF3-13Myc-TRP1 | This study |
| YPH1553 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 CTF3-3HA-His3MX6 | This study |
| YPH1554 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 ctf19Δ::HIS3 CTF3-3HA-kanMX6 | This study |
| YPH1555 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 NCD10-13Myc-kanMX6 | This study |
| YPH1556 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 chl4Δ::His3MX6 NCD10-13Myc-kanMX6 | This study |
| YPH1557 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 IML3-3HA-kanMX6 | This study |
| YPH1558 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 chl4Δ::His3MX6 IML3-3HA-kanMX6 | This study |
| YPH1559 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 ctf19Δ::HIS3 IML3-3HA-kanMX6 | This study |
| YPH1560 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 CHL4-13Myc-TRP1 IML3-3HA-kanMX6 | This study |
| YPH1561 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 ctf19Δ::HIS3 CHL4-13Myc-TRP1 IML3-3HA-kanMX6 | This study |
| YPH1562 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 IML3-13Myc-His3MX6 | This study |
| YPH1563 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 chl4Δ::His3MX6 IML3-13Myc-kanMX6 | This study |
| YPH1564 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 ctf19Δ::HIS3 IML3-13Myc-kanMX6 | This study |
| YPH1565 | MATa/α ura3-52/ura3-52 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 trp1-901/trp1-901 gal4Δ/gal4Δ gal80Δ/gal80Δ GAL2-ADE2/GAL2-ADE2 LYS2::GAL1-HIS3/LYS2::GAL1-HIS3 met2::GAL7-LacZ/met2::GAL7-LacZ pOBD2-CHL4 pOAD-MIF2 | This study |
| YPH1566 | MATa/α ura3-52/ura3-52 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 trp1-901/trp1-901 gal4Δ/gal4Δ gal80Δ/gal80Δ GAL2-ADE2/GAL2-ADE2 LYS2::GAL1-HIS3/LYS2::GAL1-HIS3 met2::GAL7-LacZ/met2::GAL7-LacZ pOBD2-CHL4 pOAD | This study |
| YPH1567 | MATa/α ura3-52/ura3-52 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 trp1-901/trp1-901 gal4Δ/gal4Δ gal80Δ/gal80Δ GAL2-ADE2/GAL2-ADE2 LYS2::GAL1-HIS3/LYS2::GAL1-HIS3 met2::GAL7-LacZ/met2::GAL7-LacZ pOBD2 pOAD-MIF2 | This study |
| YPH1568 | MATa/α ura3-52/ura3-52 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 trp1-901/trp1-901 gal4Δ/gal4Δ gal80Δ/gal80Δ GAL2-ADE2/GAL2-ADE2 LYS2::GAL1-HIS3/LYS2::GAL1-HIS3 met2::GAL7-LacZ/met2::GAL7-LacZ pOBD2 pOAD | This study |
| YPH1569* | MATa/α ura3-1/ura3-1 ade2-loc/ade2-loc his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 CHL4-YFP-His3MX6/CHL4-YFP-His3MX6 SPC29-CFP-kanMX6/SPC29-CFP-kanMX6 | This study |
| YPH1570* | MATa/α ura3-1/ura3-1 ade2-loc/ade2-loc his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/TRP1 ctf19Δ::kanMX6/ctf19Δ::kanMX6 CHL4-YFP-His3MX6/CHL4-YFP-His3MX6 SPC29-CFP-kanMX6/SPC29-CFP-kanMX6 | This study |
| YPH1571* | MATa/α ura3-1/ura3-1 ade2-loc/ade2-loc his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 chl4Δ::His3MX6/chl4Δ::kanMX6 CTF19-YFP-His3MX6/CTF19-YFP-His3MX6 SPC29-CFP-kanMX6/SPC29-CFP-kanMX6 | This study |
| YPH1572* | MATa/α ura3-1/ura3-1 ade2-loc/ade2-loc his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ctf19Δ::kanMX6/ctf19Δ::kanMX6 NDC10-YFP-His3MX6/NDC10-YFP-His3MX6 SPC29-CFP-kanMX6/SPC29-CFP-kanMX6 | This study |
| YPH1573* | MATa/α ura3-1/ura3-1 ade2-loc/ade2-loc his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/TRP1 chl4Δ::kanMX6/chl4Δ::kanMX6 NDC10-YFP-His3MX6/NDC10-YFP-His3MX6 SPC29-CFP-kanMX6/SPC29-CFP-kanMX6 | This study |
| YPH1574* | MATa/α ura3-1/ura3-1 ade2-loc/ade2-loc his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/TRP1 ctf19Δ::kanMX6/ctf19Δ::kanMX6 CTF3-YFP-His3MX6/CTF3-YFP-His3MX6 SPC29-CFP-kanMX6/SPC29-CFP-kanMX6 | This study |
| YPH1575* | MATa/α ura3-1/ura3-1 ade2-loc/ade2-loc his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 chl4Δ::His3MX6/chl4Δ::kanMX6 CTF3-YFP-His3MX6/CTF3-YFP-His3MX6 SPC29-CFP-kanMX6/SPC29-CFP-kanMX6 | This study |
| YPH1576* | MATa/α ura3-1/ura3-1 ade2-loc/ade2-loc his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/TRP1 IML3-YFP-His3MX6/IML3-YFP-His3MX6 SPC29-CFP-kanMX6/SPC29-CFP-kanMX6 | This study |
| YPH1577* | MATa/α ura3-1/ura3-1 ade2-loc/ade2-loc his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/TRP1 chl4Δ::His3MX6/chl4Δ::His3MX6 IML3-YFP-His3MX6/IML3-YFP-His3MX6 SPC29-CFP-kanMX6/SPC29-CFP-kanMX6 | This study |
| YPH1578* | MATa/α ura3-1/ura3-1 ade2-loc/ade2-loc his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ctf19Δ::kanMX6/ctf19Δ::kanMX6 IML3-YFP-His3MX6/IML3-YFP-His3MX6 SPC29-CFP-kanMX6/SPC29-CFP-kanMX6 | This study |
| YPH1579* | MATa/α ura3-1/ura3-1 ade2-loc/ade2-loc his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 MIF2-YFP-His3MX6/MIF2-YFP-His3MX6 SPC29-CFP-kanMX6/SPC29-CFP-kanMX6 | This study |
| YPH1580* | MATa/α ura3-1/ura3-1 ade2-loc/ade2-loc his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 CHL4-YFP-His3MX6/CHL4-YFP-His3MX6 MIF2-CFP-kanMX6/MIF2-CFP-kanMX6 | This study |
| YPH1582 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 iml3Δ::kanMX6 CTF19-13Myc-TRP1 | This study |
| YPH1583* | MATa/α ura3-1/ura3-1 ade2-loc/ade2-loc his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 iml3Δ::kanMX6/iml3Δ::kanMX6 CTF19-YFP-His3MX6/CTF19-YFP-His3MX6 SPC29-CFP-kanMX6/SPC29-CFP-kanMX6 | This study |
| YPH1602 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 iml3Δ::kanMX6 | This study |
| YPH1603 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 iml3Δ::kanMX6 CHL4-13Myc-TRP1 | This study |
| YVM111 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 ctf3Δ::HIS3 | V. Measday |
| YVM112 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 ctf3Δ::HIS3 | V. Measday |
| YVM218 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 trp1Δ63 CTF3-13Myc-TRP1 | (Measday et al., 2002) |
| YVM1176* | MATa/α ura3-1/ura3-1 ade2-loc/ade2-loc his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/TRP1 NDC10-YFP-His3MX6/NDC10-YFP-His3MX6 SPC29-CFP-kanMX6/SPC29-CFP-kanMX6 | (Measday et al., 2002) |
| DHY201* | MATa/α ura3-1/ura3-1 ade2-loc/ade2-loc his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 CTF19-YFP-His3MX6/CTF19-YFP-His3MX6 SPC29-CFP-kanMX6/SPC29-CFP-kanMX6 | (Measday et al., 2002) |
| DHY202* | MATa/α ura3-1/ura3-1 ade2-loc/ade2-loc his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 CTF3-YFP-His3MX6/CTF3-YFP-His3MX6 SPC29-CFP-kanMX6/SPC29-CFP-kanMX6 | (Measday et al., 2002) |
| 1cAS281 | MATa ura3 lys2 ade2 his3 leu2 trp1 cep3-1 | (Strunnikov et al., 1995) |
| 2bAS282 | MATa ura3 lys2 ade2 his3 leu2 trp1 cep3-2 | (Strunnikov et al., 1995) |
| PMY1002-3A | MATα ura3 leu2 trp1 mif2-3 | P. Meluh |
His3MX6 in these strains is the his5 gene of Schizosaccharomyces pombe. In all other strains His3MX6 is the HIS3 gene from Saccharomyces kluveri. Both complement the S. cerevisiae his3 mutation.
ChIP Assay and Coimmunoprecipitations
ChIP assays and coimmunoprecipitation from yeast lysates were performed as in Measday et al. (2002) with the following changes. For ChIP analysis, multiplex polymerase chain reaction (PCR) was performed, with three sets of primers added to a single PCR reaction. The primer pairs used to amplify specific regions of DNA are described in Meluh and Koshland (1997). The expected sizes of PCR products are 302 bp (CEN1), 288 bp (PGK1), and 243 bp (CEN3). To equilibrate the amount of PCR products obtained, 0.6 mM primer was added for each of the CEN3 and PGK1 pairs, whereas 0.8 mM primer was used for the CEN1 pair. The amount of total chromatin added varied from 1/1500 to 1/600 of the available template, whereas that of immunoprecipitated chromatin template varied from 1/10 to 1/30 of the available template, depending on the linear range for PCR.
Fluorescence Microscopy
Proteins were tagged at their C terminus with yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP) as described in Hailey et al. (2002). In all cases, the tag was integrated into the genome to maintain gene expression from the endogenous promoter. To ensure that wild-type and mutant strains expressed similar amounts of YFP or CFP fusion proteins, 50 μg of yeast lysate was run on an SDS-PAGE gel and Western blotted with anti-green fluorescent protein (GFP) antibody from Roche Diagnostics (Indianapolis, IN) for all strains used in fluorescence imaging. Cells were imaged using a DeltaVision microscopy system from Applied Precision (Issaquah, WA). The system incorporates an IL-70 microscope (Olympus, Tokyo, Japan), a u-plan-apo 100× oil objective (1.35 numerical aperature), a CoolSnap HQ digital camera from Roper Scientific (Tucson, AZ) and optical filter sets from Omega Optical (Battleboro, VT). Live cells were imaged on a thin pad of media containing 1% agarose (Hailey et al., 2002). Images were analyzed using SoftWoRx software. To quantify the image intensities at the kinetochore and in the nucleus, the image intensity values in a 5 × 5 pixel square centered either on the kinetochore or within the nucleus, respectively, were summed. A background value from a 5 × 5 pixel square either adjacent to the kinetochore (within the nucleus), or a 5 × 5 pixel square within the cytoplasm was subtracted from the summed values for the kinetochore or nucleus, respectively. For the conversion of the image files to the TIFF format, the output was 8-bit grayscale, and all images of a particular protein in different mutant backgrounds were scaled with the same set minimum and maximum values.
Genome-Wide Two-Hybrid Assay
CHL4 was cloned into pOBD2 as described in Cagney et al. (2000). The Chl4p-DNA binding domain fusion was functional as judged by rescue of the chl4Δ sectoring phenotype described in Figure 1. Two-hybrid screens were performed as described in Uetz et al. (2000). To confirm positive two-hybrid interactions, strains containing the DNA binding domain fusion plasmid (or pOBD2 vector alone) and the activation domain fusion plasmid (or pOAD vector alone) were mated. Diploid strains containing both plasmids were grown to log phase in media selecting for the plasmids, and fivefold dilutions of 5 × 106 cells were plated on media selecting for the two-hybrid interaction as indicated in Figure 7.
Figure 1.
Phenotypes of chl4 mutants. (A) Chromosome loss phenotype visualized using the SUP11 system. A nonessential chromosome fragment carrying SUP11 can suppress the ade2-101 mutation that leads to accumulation of a red pigment in the cells (Koshland and Hieter, 1987). Thus, cells containing the chromosome fragment are white, whereas those that have lost it are red. Wild-type (YPH1124), chl4Δ (YPH1534), chl4-20 (YPH1535), and chl4-61 (YPH1536) strains containing a nonessential chromosome fragment marked with URA3 were grown in SC-uracil medium and then plated on YPD plates. (B) Benomyl sensitivity. Wild-type (YPH500), chl4Δ (YPH1539), chl4-20 (YPH1540), and chl4-61 (YPH1541) strains were spotted in fivefold dilutions on YPD plates containing the amount of benomyl indicated on the left.
Figure 7.
Chl4p interacts with the kinetochore protein Mif2p. Growth of PJ694a/α strains containing the Chl4p-DNA–binding domain fusion in plasmid pOBD2 (or vector alone as control) and the Mif2p-activation domain fusion in plasmid pOAD (or vector alone as control) on selective media. SC medium lacking tryptophan and leucine only selects for the vectors, whereas medium lacking tryptophan, leucine, and histidine selects for the vectors and the two-hybrid interaction. 3-Aminotriazole (3AT) is added to prevent growth due to leaky expression from the HIS3 promoter. Strains are pOBD2-CHL4 pOAD-MIF2 (YPH1565), pOBD2-CHL4 pOAD (YPH1566), pOBD2 pOAD-MIF2 (YPH1567), and pOBD2 pOAD (YPH1568).
RESULTS
chl4Δ Displays Phenotypes Commonly Observed in Kinetochore Mutants
To confirm the chromosome loss phenotype observed in chl4 point mutants, we deleted CHL4 from a strain containing a nonessential marked chromosome fragment, and scored chromosome missegregation in the deletion mutant by a colony sectoring assay (Koshland and Hieter, 1987). chl4Δ sectored heavily, similar to two chl4 alleles isolated in the ctf mutant collection (chl4-20 and chl4-61), indicating a high rate of chromosome fragment loss (Figure 1A) (Spencer et al., 1990).
Many chromosome segregation and spindle integrity mutants have heightened sensitivity to microtubule-destabilizing drugs, perhaps due to the synergistic effect of a mutation affecting a microtubule-dependent process together with compromised microtubule networks (Poddar et al., 1999). Several outer kinetochore mutants have been shown to be sensitive to sublethal doses of the microtubule-depolymerizing drug benomyl (Roy et al., 1997; Hyland et al., 1999; Poddar et al., 1999). We tested the ability of chl4Δ, chl4-20, and chl4-61 to grow on benomyl-containing media at 25°C (Figure 1B). All mutant alleles exhibited increased sensitivity to 15 μg/ml benomyl and were inviable at 20 μg/ml benomyl. The chromosome loss and benomyl sensitivity phenotypes are consistent with chl4Δ having a defect in kinetochore function.
chl4Δ Genetically Interacts with Kinetochore Mutants
To test for genetic interactions with kinetochore mutants, we mated chl4Δ to strains carrying individual mutations in three of the CBF3 subunits (ndc10, cep3, and ctf13) (Spencer et al., 1990; Strunnikov et al., 1995; Kopski and Huffaker, 1997) or to a mif2 mutant strain (Meluh and Koshland, 1995). Dissection of the heterozygous diploids showed that chl4Δ was synthetically lethal with mif2-3 and ndc10-42 and that chl4Δ lowered the permissive temperature of ctf13-30, cep3-1, and cep3-2 mutants (Table 2). The synthetic interaction observed with ndc10-42 was allele dependent, because ndc10-2 was not synthetically lethal with chl4Δ (Table 2). We also tested for genetic interaction between chl4Δ and two outer kinetochore deletion mutations, ctf19Δ (Hyland et al., 1999) and ctf3Δ (Measday et al., 2002), and observed no synthetic effect on growth, even when the three deletion mutations were present in a single strain (Table 2). Thus, a chl4 deletion mutation genetically interacts with mutations in genes of the inner kinetochore, but not with mutations in two outer kinetochore genes.
Table 2.
Genetic interactions between chl4Δ and kinetochore mutants
| Straina | Genetic Interactionb |
|---|---|
| chl4Δ ndc10-42 | SLc |
| chl4Δ ndc10-2 | Viable |
| chl4Δ cep3-1 | CSLd |
| chl4Δ cep3-2 | CSLd |
| chl4Δ ctf13-30 | CSLd |
| chl4Δ mif2-3 | SLc |
| chl4Δ ctf19Δ | Viable |
| chl4Δ ctf3Δ | Viable |
| chl4Δ ctf19Δ ctf3Δ | Viable |
| iml3Δ chl4Δ | Viable |
| iml3Δ ctf19Δ | Viable |
| iml3Δ ctf3Δ | Viable |
| iml3Δ chl4Δ ctf3Δ | Viable |
| iml3Δ ctf19Δ ctf3Δ | Viable |
The strains used in the crosses were chl4Δ (YPH1538 or YPH1539), ndc10-42 (YCTF42), ndc10-2 (YPH1027), cep3-1 (1cAS281), cep3-2 (2bAS282), ctf13-30 (YCTF30), mif2-3 (PMY1002-3A), ctf19Δ (YPH1316), ctf3Δ (YVM111), and iml3Δ (YPH1602).
Strains were incubated at 25°C.
SL, synthetic lethality. Individual mutations do not cause inviability on their own, but a strain containing both mutations is inviable.
CSL, conditional synthetic lethality. Spores are viable at 25°C but die at a lower temperature than the restrictive temperature of the single temperature-sensitive mutant. chl4Δ ctf13-30 double mutants died at 32°C, whereas the nonpermissive temperature of the ctf13-30 single mutant is 35°C; chl4Δ cep3-1 and chl4Δ cep3-2 double mutants were inviable at 31°C and 30°C, respectively, whereas the nonpermissive temperature of the cep3-1 and cep3-2 single mutants is 34°C.
Chl4p Associates with CEN DNA and Localizes to the Kinetochore
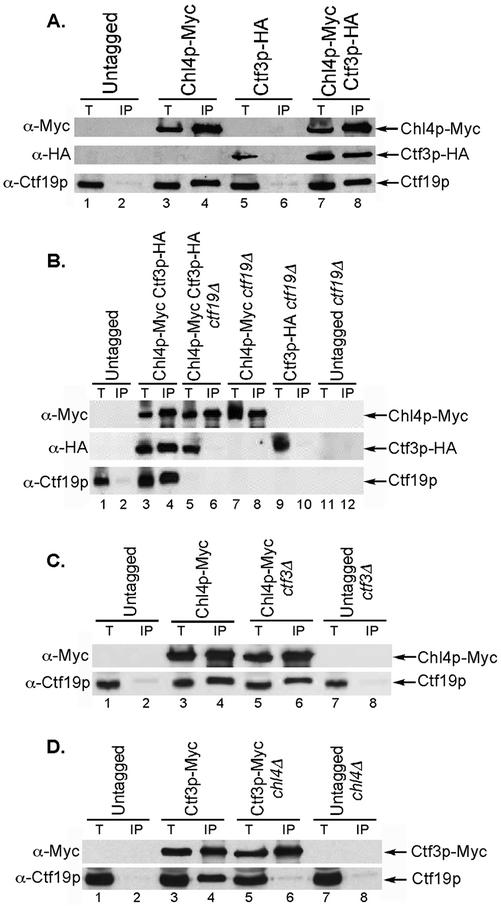
Genetic and phenotypic evidence strongly suggested that the CHL4 gene product was a candidate kinetochore protein. To obtain biochemical evidence that Chl4p is located at the centromere, we used ChIP to assay whether Chl4p could interact with CEN DNA. Myc-tagged Chl4p was immunoprecipitated from formaldehyde cross-linked extracts with anti-Myc–conjugated beads. The coprecipitated DNA was analyzed by PCR with primer pairs specific to centromeric regions of chromosomes I and III (CEN1 and CEN3) and to a noncentromeric region (PGK1) as a control for binding specificity. Chl4p interacted specifically with CEN DNA but not with a non-CEN locus, similar to Ctf19p and Ndc10p, two known kinetochore proteins (Figure 2A, lanes 4, 6, and 8).
Figure 2.
Chl4p interacts with CEN DNA in an Ndc10p- and Ctf19-dependent, but Ctf3p-independent manner. (A) Chl4p interacts with CEN DNA. ChIP assay performed by immunoprecipitation of Myc-tagged Chl4p, Ctf19p, or Ndc10p followed by multiplex PCR analysis. Lanes 9–12, dilutions (2.5-fold) of one of the total templates, showing that PCR is in the linear range. Strains are untagged (YPH499), Chl4p-Myc (YPH1542), Ctf19p-Myc (YPH1550), and Ndc10p-Myc (YPH1555). (B) The interaction of Chl4p with CEN DNA depends on Ndc10p. ChIP assay performed by immunoprecipitation of Myc-tagged Chl4p or Ndc10p from wild-type, ndc10-2, or chl4Δ strains, followed by multiplex PCR analysis. For the temperature-sensitive assay, wild-type and ndc10-2 strains were grown to log phase at 25°C, and half of the culture was then shifted to 37°C for 3 h. Strains are untagged (YPH499), Chl4p-Myc (YPH1542), Chl4p-Myc ndc10-2 (YPH1543), Ndc10p-Myc (YPH1555), and Ndc10p-Myc chl4Δ (YPH1556). (C) The interaction of Chl4p with CEN DNA depends on Ctf19p. ChIP assay performed by immunoprecipitation of Myc-tagged Chl4p or Ctf19p from wild-type, ctf19Δ, or chl4Δ strains, followed by multiplex PCR analysis. Strain are untagged (YPH499), Chl4p-Myc (YPH1542), Chl4p-Myc ctf19Δ (YPH1544), Ctf19p-Myc (YPH1550), and Ctf19p-Myc chl4Δ (YPH1551). (D) The interaction of Chl4p with CEN DNA is independent of Ctf3p. ChIP assay performed by immunoprecipitation of Myc-tagged Chl4p or Ctf3p from wild-type, ctf3Δ, or chl4Δ strains, followed by multiplex PCR analysis. Strains are untagged (YPH499), Chl4p-Myc (YPH1542), Chl4p-Myc ctf3Δ (YPH1545), Ctf3p-Myc (YVM218), and Ctf3p-Myc chl4Δ (YPH1552). In A–D, T, total lysate; IP, immunoprecipitated fraction.
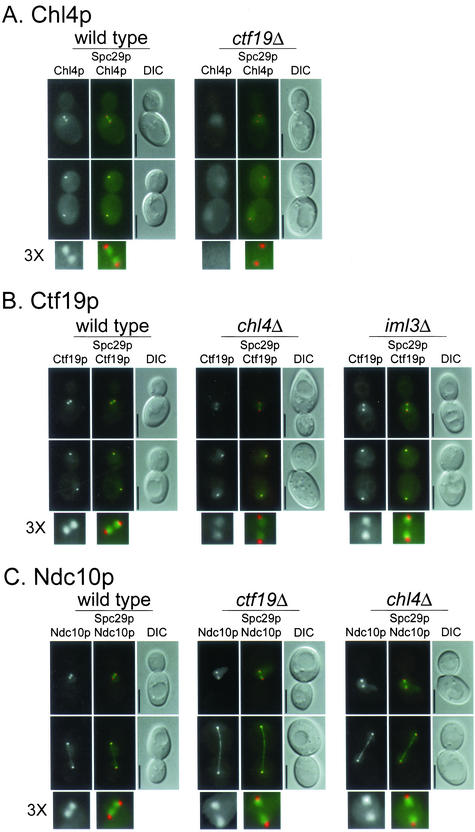
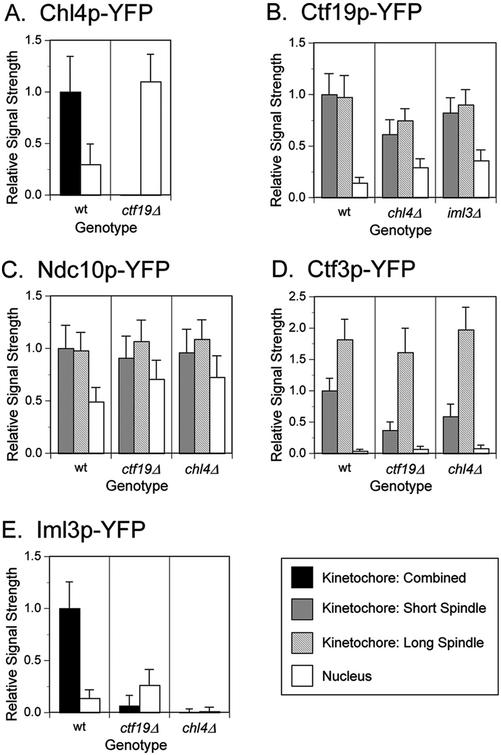
The localization of several kinetochore proteins tagged with GFP or its variants YFP and CFP has recently been determined in yeast cells by fluorescence microscopy. The resulting images have shown that kinetochore proteins reside next to the nuclear side of the SPB in cells with short spindles and colocalize with the SPB in late anaphase cells (He et al., 2001; Pearson et al., 2001; Measday et al., 2002). To visualize the localization of Chl4p, we tagged it with YFP and imaged Chl4p-YFP in a strain containing a tagged SPB protein, Spc29p-CFP (Figure 3A, wild-type panel). We found that Chl4p-YFP had a pattern of localization similar to Ctf19p-YFP and Ctf3p-YFP (Figure 3, B and D, wild-type panels) and other kinetochore proteins in cells with short and long spindles. The specific interaction of Chl4p with CEN DNA chromatin and its cellular localization suggest that Chl4p is part of the budding yeast kinetochore complex.
Figure 3.
Interdependence of kinetochore protein localization. (A–E) Indicated proteins were tagged with YFP and imaged as described under MATERIALS AND METHODS. Spc29p-CFP was included in the genetic background as an SPB marker, which allowed determination of the relative location of the kinetochore. In all panels, upper images are cells with short spindles and lower images are cells with long spindles. The left frame has the YFP signal, the middle frame has merged YFP and CFP signals, and the right frame is the corresponding differential interference contrast image. In the color images, YFP is pseudocolored green and CFP is pseudocolored red. A threefold enlargement of the YFP and CFP signals in cells with short spindles, indicated by 3X, is shown at the bottom of each panel. The relevant genetic background (wild-type or kinetochore mutant) is indicated on top of each panel. (F) For colocalization, Chl4p was tagged with YFP and Mif2p was tagged with CFP in a wild-type strain and imaged as in A–E. All strains are homozygous diploids of (A) Chl4p-YFP in wt (YPH1569) and ctf19Δ (YPH1570); (B) Ctf19p-YFP in wt (DHY201), chl4Δ (YPH1571), and iml3Δ (YPH1583); (C) Ndc10p-YFP in wt (YVM1176), ctf19Δ (YPH1572), and chl4Δ (YPH1573); (D) Ctf3p-YFP in wt (DHY202), ctf19Δ (YPH1574), and chl4Δ (YPH1575); (E) Iml3p-YFP in wt (YPH1576), chl4Δ (YPH1577), and ctf19Δ (YPH1578); and (F) Mif2p-YFP (YPH1579) and Chl4p-YFP Mif2p-CFP (YPH1580). All strains (except YPH1580) contain Spc29p-CFP. wt, wild-type. Bar, 5 μm.
Chl4p Requires Ndc10p and Ctf19p, but Not Ctf3p, to Interact with CEN DNA
Whereas components of the CBF3 complex bind directly to CEN DNA (Lechner and Carbon, 1991), outer kinetochore complexes, such as the Ctf19 complex, interact with CEN DNA via CBF3 (Ortiz et al., 1999). In addition, the Ctf3 complex was shown to require both Ctf19p and Cse4p to interact efficiently with CEN DNA (Measday et al., 2002). The CEN DNA loading requirements of kinetochore proteins can thus be used to probe their molecular organization within the kinetochore complex. To determine whether the interaction of Chl4p with CEN DNA requires the CBF3 complex, we performed ChIP analysis with Chl4p-Myc in a strain containing a temperature-sensitive mutation in NDC10. In the ndc10-2 background, Chl4p interacted with CEN DNA at permissive temperature (25°C) (Figure 2B, lane 10), but not at restrictive temperature (37°C) (Figure 2B, lane 12). Because Chl4p is able to coimmunoprecipitate CEN DNA in the wild-type strain at 37°C (Figure 2B, lane 8), the lack of interaction in ndc10-2 is not simply due to a failure of Chl4p to associate with CEN DNA at high temperature. Western blots also demonstrated that Chl4p-Myc was immunoprecipitated efficiently in wild-type and mutant backgrounds at both temperatures (data not shown). Conversely, deletion of CHL4 did not affect the ability of Ndc10p to interact with CEN DNA (Figure 2B, lanes 14 and 16). Thus, functional Ndc10p is required for the Chl4p-CEN DNA interaction. In accordance with these data, Chl4p-GFP localization becomes diffuse in an ndc10-2 strain incubated at the restrictive temperature (K. Mythreye and K. Bloom, personal communication). Therefore, Chl4p requires an intact CBF3 complex to properly localize to and interact with the centromere.
To investigate whether Chl4p requires other kinetochore components, such as Ctf19p or Ctf3p, for its interaction with CEN DNA, we performed ChIP with Chl4p-Myc in strains lacking CTF19 or CTF3. We found that although Chl4p required Ctf19p to interact with the centromere (Figure 2C, lane 6), it did not require Ctf3p (Figure 2D, lane 6). Conversely, immunoprecipitation of Ctf19p-Myc in strains lacking CHL4 revealed that Ctf19p coimmunoprecipitated CEN DNA in the absence of Chl4p (Figure 2C, lane 10). Similarly, Ctf3p was able to interact with CEN DNA in the absence of Chl4p (Figure 2D, lane 10), although in some instances the CEN DNA coimmunoprecipitation seemed to be less efficient (data not shown). Western blots demonstrated that in all cases, efficient immunoprecipitation of the Myc-tagged protein was not affected in the deletion strain compared with the wild-type strain (data not shown; see Western blots in Figure 5 showing immunoprecipitations in wild-type and mutant strains). Thus, Chl4p, like the Ctf3 complex, requires Ctf19p to interact with CEN DNA, whereas neither Chl4p nor the Ctf3 complex are required for the Ctf19p-CEN DNA interaction (Figure 2, C and D; Measday et al., 2002). Moreover, Chl4p and Ctf3p are able to interact with the centromere independently of each other.
Figure 5.
Chl4p coimmunoprecipitates with outer kinetochore proteins. (A) Chl4p coimmunoprecipitates with Ctf19p and Ctf3p. Anti-Myc immunoprecipitations were performed in a strain containing Myc-tagged Chl4p and HA-tagged Ctf3p and control strains containing one or no tagged proteins. Total lysate (40 μg) and 15% of the immunoprecipitated fraction were loaded on SDS-PAGE gels, and Western blots were used to detect Myc- and HA-tagged proteins, and Ctf19p, with the antibodies indicated on the left. Strains are untagged (YPH499), Chl4p-Myc (YPH1542), Ctf3p-HA (YPH1553), and Chl4p-Myc Ctf3p-HA (YPH1546). (B) Ctf19p is required for the coimmunoprecipitation between Chl4p and Ctf3p. Anti-Myc immunoprecipitations and Western blots were performed as in A in strains lacking Ctf19p. Lanes 7–12 are controls. Strains are untagged (YPH499), Chl4p-Myc Ctf3p-HA (YPH1547), Chl4p-Myc Ctf3p-HA ctf19Δ (YPH1548), Chl4p-Myc ctf19Δ (YPH1544), Ctf3p-HA ctf19Δ (YPH1554), and untagged ctf19Δ (YPH1315). (C) Ctf3p is dispensable for the coimmunoprecipitation between Chl4p and Ctf19p. Anti-Myc immunoprecipitations and Western blots were performed as in A in strains lacking Ctf3p. Lanes 7 and 8 are controls. Strains are untagged (YPH499), Chl4p-Myc (YPH1542), Chl4p-Myc ctf3Δ (YPH1545), and untagged ctf3Δ (YVM112). (D) Chl4p is required for the interaction of Ctf3p with Ctf19p. Anti-Myc immunoprecipitations and Western blots were performed as in A in strains containing Myc-tagged Ctf3p in the presence or absence of Chl4p. Lanes 7 and 8 are controls. Strains are untagged (YPH499), Ctf3p-Myc (YVM218), Ctf3p-Myc chl4Δ (YPH1552), and untagged chl4Δ (YPH1537). In A–D, T, total lysate; IP, immunoprecipitated fraction.
Chl4p Requires Ctf19p for Proper Kinetochore Localization, and Lack of Chl4p or Ctf19p Affects the Localization of Ctf3p in Early Anaphase
Because Ctf19p is required for Chl4p to interact with CEN DNA, we asked whether lack of CTF19 disturbed the localization of Chl4p-YFP. We found that in a ctf19Δ strain, Chl4p-YFP did not localize to the kinetochore. Instead, the YFP signal was diffuse throughout the nucleus (Figures 3A and 4A). The intensity of the diffuse nuclear signal was fourfold greater in the ctf19Δ mutant than in the wild-type strain (Figure 4A). Conversely, the localization of Ctf19p-YFP was only modestly impaired by the absence of Chl4p (Figures 3B and 4B). In agreement with our ChIP data (Figure 2B; Measday et al., 2002), deletions of either CTF19 or CHL4 do not significantly disturb the inner kinetochore structure, because Ndc10p-YFP still localizes to the kinetochore in ctf19Δ and chl4Δ strains (Figures 3C and 4C). A slight increase in a diffuse nuclear signal in these strains is currently under further investigation (data not shown). In all cases, the wild-type and mutant strains expressed similar levels of fusion proteins (data not shown; see MATERIALS AND METHODS). In summary, our ChIP and in vivo localization data indicate that Ctf19p, which interacts with CEN DNA via CBF3 (Ortiz et al., 1999), is essential not only for the Chl4p–CEN DNA interaction but also for the proper localization of Chl4p to the kinetochore.
Figure 4.
Quantification of kinetochore localization signals. (A–E) Kinetochore protein signal strengths of the strains imaged in Figure 3, A–E, were determined as described under MATERIALS AND METHODS. For each graph, the values were normalized to the mean value of the wild-type strain. In B–D, the mean value of the kinetochore signal from cells with a short spindle was used for normalization. Bars represent the standard deviation. The mean value of the signal intensity in wild-type strains and the number of kinetochores or nuclei examined for each panel are as follows, where n1 is the sample size for kinetochore signals in cells with either short or long spindles, n2 is the sample size for kinetochore signals in cells with short spindles, and n3 is the sample size for kinetochore signals in cells with long spindles (sample sizes for determining the nuclear signal strength are equal to either n1 or n2): (A) Mean value (wt) = 2297; for wt, n1 = 78; for ctf19Δ, n1 = 45. (B) Mean value (wt) = 12368; for wt, n2 = 33 and n3 = 26; for chl4Δ, n2 = 36 and n3 = 30; for iml3Δ, n2 = 22 and n3 = 14. (C) Mean value (wt) = 12587; for wt, n2 = 86 and n3 = 62; for ctf19Δ, n2 = 64 and n3 = 38; for chl4Δ, n2 = 82 and n3 = 33. (D) Mean value (wt) = 4035; for wt, n2 = 24 and n3 = 16; for ctf19Δ, n2 = 45 and n3 = 24; for chl4Δ, n2 = 38 and n3 = 19. (E) Mean value (wt) = 5740; for wt, n1 = 74; for ctf19Δ, n1 = 33; for chl4Δ, n1 = 63. wt, wild-type.
Because Ctf3p, like Chl4p, requires Ctf19p to interact with CEN DNA (Measday et al., 2002), we examined the localization of Ctf3p in the absence of Ctf19p. Interestingly, in the ctf19Δ strain, Ctf3p-YFP showed a threefold decrease in kinetochore localization signal in cells with short spindles, whereas in cells with long spindles, Ctf3p-YFP showed normal colocalization with the SPB (Figures 3D and 4D). To investigate whether this behavior was specific to the lack of Ctf19p, we imaged Ctf3p-YFP in a chl4Δ strain. We observed a 40% decrease in the intensity of the Ctf3p-YFP kinetochore signal in chl4Δ cells with short spindles, whereas there was no decrease in Ctf3p-YFP signal intensity in chl4Δ cells with long spindles, similar to ctf19Δ (Figures 3D and 4D). These data suggest that Ctf19p, and possibly Chl4p, are important for Ctf3p localization to the kinetochore during early stages of mitosis, but not in later stages.
Chl4p Interacts with Known Outer Kinetochore Proteins
Given the interaction of Chl4p with CEN DNA and its localization to the kinetochore, we asked whether Chl4p would also interact with known outer kinetochore proteins. In a strain containing Myc-tagged Chl4p, we performed immunoprecipitation with anti-Myc–conjugated beads and were able to detect both Chl4p and Ctf19p in the immunoprecipitate (Figure 5A, lane 4). We also tested for interaction of Chl4p with Ctf3p by using a strain that contained hemagglutinin (HA)-tagged Ctf3p. Ctf3p-HA was detected in anti-Myc immunoprecipitates only when Chl4p-Myc was present (Figure 5A, lane 8). Similar results were obtained when the same strain was used in an anti-HA immunoprecipitation (data not shown). To determine whether the interaction of Chl4p with Ctf3p depended on Ctf19p, we performed immunoprecipitations in a ctf19Δ strain with both anti-Myc– (Figure 5B) and anti-HA- (data not shown) conjugated beads. We found that Chl4p no longer interacted with Ctf3p in the absence of Ctf19p (Figure 5B, lane 6). Immunoprecipitation of Chl4p-Myc was also performed in a ctf3Δ strain and revealed that Ctf19p was still present in the immunoprecipitate, indicating that the Chl4p–Ctf19p interaction is independent of Ctf3p (Figure 5C, lane 6). Conversely, immunoprecipitating Ctf3p-Myc in the absence of Chl4p disrupted the interaction between Ctf3p and Ctf19p (Figure 5D, lane 6). Thus, our immunoprecipitation data suggest that Chl4p is connected to the centromere and to the Ctf3 complex via Ctf19p. Moreover, because the Ctf3p–CEN DNA interaction depends on Ctf19p (Measday et al., 2002), and Chl4p is essential for the Ctf3p-Ctf19p interaction, Chl4p may contribute to the efficient association of the Ctf3 complex with the centromere.
The Chl4p Two-Hybrid Interactor Iml3p Is a New Outer Kinetochore Protein
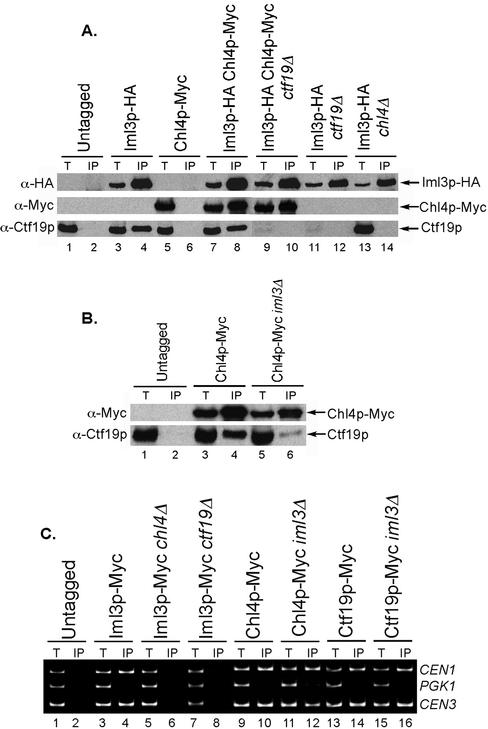
Phenotypic analyses of iml3/mcm19 mutants indicated that Iml3p was a putative kinetochore protein (Entian et al., 1999; Ghosh et al., 2001). We tested for genetic interactions between an iml3 deletion mutation and mutations in other outer kinetochore genes. We found that similar to chl4Δ, iml3Δ did not compromise growth when combined with chl4Δ, ctf19Δ or ctf3Δ, or various multiple combinations of these mutations (Table 2). Iml3p was shown to interact with Chl4p by two-hybrid assay (Ghosh et al., 2001). We confirmed this interaction biochemically by performing an anti-HA immunoprecipitation in a strain containing Iml3p-HA and Chl4p-Myc. Anti-Myc and anti-Ctf19p Western blots showed that Iml3p was able to coimmunoprecipitate both Chl4p and Ctf19p (Figure 6A, lanes 4 and 8). The Iml3p–Chl4p interaction was also observed when we used anti-Myc beads for immunoprecipitation (data not shown). Because Ctf19p is required for Chl4p to interact with Ctf3p, we tested whether Ctf19p was also required for the Iml3p–Chl4p interaction. We found that in contrast to Ctf3p, Iml3p still coimmunoprecipitated with Chl4p in a ctf19Δ strain (Figure 6A, lane 10). Thus, Iml3p and Chl4p can form a complex independently of Ctf19p. We next asked whether lack of Chl4p would disrupt the Iml3p–Ctf19p interaction. When Iml3p-HA was immunoprecipitated from a chl4Δ strain, Ctf19p was no longer present in the immunoprecipitate (Figure 6A, lane 14), indicating that Chl4p is required for the Iml3p–Ctf19p interaction. Finally, to determine whether Iml3p was required for the Chl4p–Ctf19p interaction, we immunoprecipitated Chl4p-Myc from an iml3Δ strain and found that the amount of Ctf19p in the immunoprecipitate was reduced, albeit not completely absent (Figure 6B). Thus, we conclude that Iml3p is part of the outer kinetochore complex and interacts with the Ctf19 complex in a Chl4p-dependent manner and that Iml3p contributes to the efficient interaction of Chl4p with Ctf19p.
Figure 6.
Iml3p is a new outer kinetochore protein. (A) Iml3p interacts with Chl4p and with Ctf19p in a Chl4p-dependent manner. Anti-HA immunoprecipitations and Western blots were performed as in Figure 5 in wild-type, ctf19Δ, or chl4Δ strains containing Myc-tagged Chl4p and HA-tagged Iml3p or control strains containing one or no tagged proteins. Strains are untagged (YPH499), Iml3p-HA (YPH1557), Chl4p-Myc (YPH1542), Iml3p-HA Chl4p-Myc (YPH1560), Iml3p-HA Chl4p-Myc ctf19Δ (YPH1561), Iml3p-HA ctf19Δ (YPH1559), and Iml3p-HA chl4Δ (YPH1558). (B) The interaction of Ctf19p with Chl4p is reduced in the absence of Iml3p. Anti-Myc immunoprecipitations and Western blots were performed as in Figure 5 in wild-type or iml3Δ strains containing either Myc-tagged Chl4p or no tagged protein. Strains are untagged (YPH499), Chl4p-Myc (YPH1542), and Chl4p-Myc iml3Δ (YPH1549). (C) Iml3p interacts with CEN DNA in a Chl4p- and Ctf19p-dependent manner. ChIP assay performed by immunoprecipitation of Myc-tagged Iml3p from wild-type, chl4Δ or ctf19Δ strains, or by immunoprecipitation of Myc-tagged Chl4p or Ctf19p from wild-type or iml3Δ strains, followed by multiplex PCR analysis. Strains are untagged (YPH499), Iml3p-Myc (YPH1562), Iml3p-Myc chl4Δ (YPH1563), Iml3p-Myc ctf19Δ (YPH1564), Chl4p-Myc (YPH1542), Chl4p-Myc iml3Δ (YPH1603), Ctf19p-Myc (YPH1550), and Ctf19p-Myc iml3Δ (YPH1582). In A–C, T, total lysate; IP, immunoprecipitated fraction.
Next, we asked whether Iml3p met the criteria we used to define a kinetochore protein: association with CEN DNA and a kinetochore localization pattern. We were able to specifically isolate CEN DNA from an Iml3p-Myc immunoprecipitate, similar to Chl4p (Figure 6C, lane 4). We also found that the Iml3p–CEN DNA interaction depended on Chl4p and Ctf19p, because CEN DNA no longer coprecipitated with Iml3p-Myc in chl4Δ and ctf19Δ strains (Figure 6C, lanes 6 and 8). In contrast, Iml3p was not required for the Chl4p–CEN DNA or Ctf19p–CEN DNA interactions (Figure 6C, lanes 12 and 16), although in some instances the efficiency of CEN DNA coimmunoprecipitation appeared to be slightly reduced for Chl4p (data not shown), perhaps due to the reduced interaction of Chl4p with Ctf19p in the absence of Iml3p. These data suggest that Iml3p lies more distal to the centromere than Ctf19p and Chl4p. We then imaged Iml3p-YFP in a strain containing Spc29p-CFP and observed localization of Iml3p to the kinetochore (Figure 3E). In accordance with the ChIP data described above, Iml3p-YFP failed to localize to the kinetochore in a ctf19Δ or a chl4Δ strain (Figures 3E and 4E). Interestingly, a diffuse nuclear Iml3p-YFP signal was visible in ctf19Δ, as was observed for the Chl4p-YFP signal in ctf19Δ; no Iml3p-YFP signal was detected in chl4Δ (Figure 4E). In contrast, lack of Iml3p did not disturb Ctf19p-YFP localization to the kinetochore (Figures 3B and 4B). Thus, our results strongly suggest that Iml3p is a new outer kinetochore protein that is connected to the centromere via Chl4p and Ctf19p.
A Genome-Wide Two-Hybrid Screen Reveals that Chl4p Interacts with the Kinetochore Protein Mif2p
To uncover other potential protein partners of Chl4p, we constructed a Chl4p-DNA binding domain fusion and tested for two-hybrid interactions with a genome-wide array of activation domain fusions (Cagney et al., 2000; Uetz et al., 2000). Two independent screens showed that Mif2p interacted with Chl4p (Figure 7). However, we have so far been unsuccessful in our attempts to coimmunoprecipitate the two proteins from yeast lysates by using various combinations of epitope tags. Mif2p has been shown to localize to the kinetochore by ChIP assay (Meluh and Koshland, 1997). Here, we determined by fluorescence microscopy that the localization of YFP-tagged Mif2p in relation to Spc29p-CFP was similar to that of other kinetochore proteins (Figure 3F). We also observed that YFP-tagged Chl4p colocalized with Mif2p-CFP in both short and long spindle stage cells (Figure 3F). Thus, Chl4p shows not only a genetic interaction (Table 2) but also a two-hybrid interaction and colocalization with Mif2p, an established kinetochore protein.
DISCUSSION
Our coimmunoprecipitation, ChIP, and fluorescence imaging data contribute to the mapping of protein complexes within the kinetochore, by determining requirements for protein–protein interactions, CEN DNA loading, and proper kinetochore localization of four outer kinetochore proteins (Table 3). We first demonstrate that Chl4p is a protein of the outer kinetochore, confirming previous suggestions from genetic data (Kouprina et al., 1993a), and then probe for the location of Chl4p within the kinetochore complex. Chl4p specifically coimmunoprecipitates with CEN DNA and localizes to the kinetochore in an Ndc10p- and Ctf19p-dependent manner, suggesting that Chl4p is located more distal to the DNA than the CBF3 and Ctf19 complexes. Moreover, because the Ctf19 complex itself requires CBF3 for interacting with CEN DNA (Ortiz et al., 1999), a plausible hypothesis is that Chl4p interacts with CBF3 and CEN DNA via the Ctf19 complex. Other kinetochore proteins, such as Mtw1p, have also been shown to depend on Ndc10p for interaction with the centromere (Goshima and Yanagida, 2000), suggesting that disruption of this inner kinetochore component will affect the localization and CEN DNA interaction of many kinetochore proteins. However, our data demonstrate further specificity in dependency relationships by showing that association of Chl4p with CEN DNA requires one outer kinetochore protein (Ctf19p) but not another (Ctf3p).
Table 3.
Summary of dependency relationships among kinetochore proteins
n/d, not determined.
From Measday et al. (2002).
From Ortiz et al. (1999).
K. Mythreye and K. Bloom, personal communication.
ChlP assay was done in nocodazole-arrested cells.
ss, in cells with short spindles; ls, in cells with long spindles.
Ctf19p was tagged with HA instead of Myc for this particular assay.
CEN ChIP may be less efficient in the mutant strain than in the wild-type strain.
Our studies with Chl4p have led to the identification of another component of the kinetochore complex, Iml3p. Previous genetic data suggested that the IML3 gene product might function at the kinetochore (Ghosh et al., 2001). The mutant phenotypes of iml3, which include a high rate of chromosome and plasmid loss, benomyl sensitivity, relaxation of a transcription block, and stable maintenance of a dicentric plasmid (Entian et al., 1999; Ghosh et al., 2001), are very similar to those of chl4. Iml3p also interacts with Chl4p by two-hybrid assay (Ghosh et al., 2001). We have confirmed the Iml3p–Chl4p two-hybrid interaction by coimmunoprecipitation from yeast lysates (Figure 6A). Interestingly, it was recently shown that Chl4p is present in a tandem-affinity purification of Iml3p (Gavin et al., 2002). Herein, we show that Iml3p both interacts with CEN DNA and localizes to the kinetochore in a Ctf19p- and Chl4p-dependent manner, which establishes Iml3p as a component of the outer kinetochore. Unlike the Ctf3p-Chl4p interaction, the Iml3p–Chl4p interaction does occur in the absence of Ctf19p, indicating that Chl4p and Iml3p could form a separate complex from the Ctf19 and Ctf3 complexes. It should be noted that the interactions of Chl4p and Iml3p with each other and with other kinetochore proteins may not be direct and that it is not known whether they require the presence of centromere DNA or an intact inner kinetochore.
The coimmunoprecipitation data suggest that Chl4p connects the Ctf3 complex to the Ctf19 complex. Whereas Chl4p interacts with Ctf19p and CEN DNA in the absence of Ctf3p (Figures 2D and 5C), Ctf3p does not interact with Ctf19p in the absence of Chl4p (Figure 5D). Because the interaction of Ctf3p with CEN DNA requires Ctf19p (Measday et al., 2002), a simple interpretation of this result is that Chl4p is more proximal to CEN DNA than the Ctf3 complex. However, deletion of CHL4 does not abolish the Ctf3p–CEN DNA interaction and only reduces the localization signal of Ctf3p to the kinetochore by 40% (Figures 3C and 4C). Therefore, it is likely that a more complex network of protein interactions occurs. For example, other members of the Ctf3p complex may allow for interaction with the Ctf19 complex in the absence of Chl4p, as is schematically depicted in Figure 8B. Alternatively, the lack of Ctf3p-Ctf19p coimmunoprecipitation in the absence of Chl4p could reflect a change in the overall conformation of the outer kinetochore that may result in reduced CEN DNA binding efficiency by the Ctf3 complex. Moreover, we have observed that proper in vivo localization of Ctf3p requires Ctf19p, and to some extent Chl4p, only in cells with short spindles; in cells with long spindles, Ctf3p localizes to the kinetochore even in the absence of Ctf19p and Chl4p. Because kinetochores overlap with the SPB near the end of anaphase, the SPB may provide an additional anchor for outer kinetochore proteins at this stage of the cell cycle.
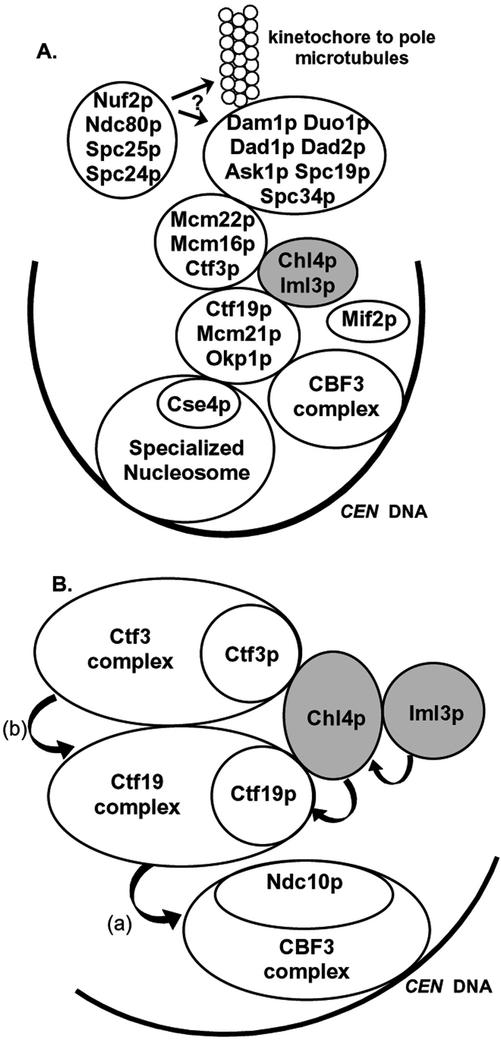
Figure 8.
Model of the budding yeast kinetochore. (A) Revision of the kinetochore model presented in Measday et al. (2002), which now includes Chl4p and Iml3p as part of the outer kinetochore. Mif2p has also been added to the diagram because of its two-hybrid interaction with Chl4p. (B) Proposed molecular architecture of the proteins that have been examined in this article, deduced from requirements for CEN DNA interaction, localization, and protein–protein interactions. Arrows represent requirements for interaction with CEN DNA, beginning with the complex/protein interacting with CEN DNA, and pointed toward the complex/protein required for the interaction. (a) From Ortiz et al. (1999). (b) From Measday et al. (2002).
In addition to the interactions of Chl4p with proteins of the outer kinetochore, we have found that Chl4p interacts with the kinetochore protein Mif2p by two-hybrid assay. The fact that we were unable to detect the Chl4p-Mif2p interaction by coimmunoprecipitation suggests that the conditions in which the experiment was performed may not have been ideal to preserve the interaction. For instance, because Mif2p has been proposed to interact with DNA at CDEII or CDEIII, owing to the presence of potential AT hooks in its structure, it is possible that a certain DNA conformation is required to position Mif2p so that it can interact with other kinetochore proteins. This particular conformation may be lost during the immunoprecipitation procedure. In fact, we have seen no report of proteins interacting with Mif2p by standard immunoprecipitation from yeast lysates, and thus the exact positioning of Mif2p within the kinetochore complex has not been determined yet. Interestingly, Mif2p was also found to interact with the CBF3 component Cep3p in our two-hybrid array (data not shown), suggesting that Mif2p lies close to the inner kinetochore. Our studies show that Mif2p has a kinetochore localization pattern and that it colocalizes with Chl4p by fluorescence microscopy (Figure 3F).
One major difficulty in understanding the function of outer kinetochore proteins is their genetic and phenotypic redundancy. Several outer kinetochore proteins, including Ctf19p, Mcm21p, Ctf3p, Mcm16p, Mcm22p, Chl4p, and Iml3p, are not essential for cell viability. Simultaneous deletion of several of these components does not seem to compromise cell viability or have a synergistic effect on the chromosome loss phenotype (Table 2; Ghosh et al., 2001; Measday et al., 2002), which may be indicative of the presence of a “mega-complex” within the outer kinetochore, encompassing the Ctf19 and Ctf3 complexes, Chl4p, Iml3p, and additional players found in affinity purifications (Gavin et al., 2002; Cheeseman et al., 2002b). Whole genome synthetic lethal screens of chl4Δ and iml3Δ are currently underway with the aim of uncovering genetic interactions and possibly other proteins performing a similar function, and may provide some insights into Chl4p and Iml3p function.
Chl4p is a 453 amino acid protein that has no recognizable domains as assessed by standard conserved domain homology searches. It has been suggested that part of Chl4p is similar to a small region of Escherichia coli recA, and some Chl4p residues have been predicted to fold into a helix-turn-helix motif that could be part of a putative DNA-binding domain (Kouprina et al., 1993a). A BLASTP analysis (Altschul et al., 1997) of the Chl4p sequence against the nr database revealed that two predicted proteins have significant similarity to Chl4p, the Neurospora crassa protein B2A19.050 “related to trfA protein” (GenBank accession no. CAB98235) and the Schizosaccharomyces pombe pi022/SPBP22H7.09c gene product (GenBank accession no. CAC37377). Additionally, a Candida albicans protein, orf6.7000 (CandidaDB CA4453), shows 27% identity to Chl4p. Although these putative homologues of Chl4p have yet to be functionally characterized, conservation of this protein in distant fungal species indicates that it probably plays an important role in the maintenance of genomic integrity.
We demonstrate herein that Chl4p is required for interactions between known outer kinetochore complexes and that it may affect their CEN DNA loading efficiency. Thus, Chl4p could be an important structural component of the outer kinetochore. It has been proposed that Chl4p is involved in the initial step of kinetochore formation rather than maintenance of preexisting ones, because de novo kinetochores are more affected by deletion of CHL4 than established ones (K. Mythreye and K. Bloom, personal communication). Thus, Chl4p could be instrumental in establishing the structure of nascent kinetochores by holding together the building blocks in the proper conformation. Additionally, our data show that chl4 strains are sensitive to a drug that disrupts microtubule networks. Thus, a kinetochore lacking CHL4 may not attain the optimal structural conformation required for efficient interaction with spindle microtubules. Whether dynamic rearrangements of the kinetochore occur during the progression of the cell cycle is unknown. Our data with Ctf3p localization in chl4 and ctf19 mutants suggest that changes do occur, because the localization of Ctf3p is disturbed only in early anaphase in the absence of Ctf19p and Chl4p. Future studies will determine whether Chl4p and other proteins of the outer kinetochore actively participate in kinetochore dynamics during mitosis.
By combining our fluorescence imaging, ChIP, and immunoprecipitation data, we are proposing an extension of our previous model (Measday et al., 2002) of the kinetochore structure, which includes Chl4p and Iml3p as central components of the outer kinetochore (Figure 8A). Although the budding yeast centromere is much simpler than the centromere of other eukaryotes, the number of proteins comprising the budding yeast kinetochore is large and still growing (Cheeseman et al., 2002b). Our studies not only establish two new protein components in the kinetochore complex but also give a deeper understanding of the spatial relationships existing among several of its components.
ACKNOWLEDGMENTS
We thank T. Huffaker, D. Koshland, and P. Meluh for yeast strains; B. Sundin for expert help with microscopy and preliminary imaging of Chl4p and Mif2p; M. Mayer, A. Page, K. Kitagawa, and C. Boone for critical reading of the manuscript; K. Mythreye and K. Bloom for sharing data before publication and helpful discussions and comments on the manuscript; and members of the Hieter laboratory. I.P. was supported by a National Cancer Institute of Canada Research Studentship and a University of British Columbia Killam Predoctoral Fellowship. V.M. was supported by a Michael Smith Foundation for Health Research Postdoctoral Fellowship. P.H. was supported by a Canadian Institute of Health Research Senior Scientist Award. S.F. is an investigator of the Howard Hughes Medical Institute. This work was supported by a Canadian Institute of Health Research operating grant MOP-38096 (to P.H.), a National Institutes of Health grant CA-16519 (to P.H.) and National Institutes of Health grant NCRR RR11823 (to T.D. and S.F.).
Abbreviations used:
- CBF3
centromere-binding factor 3
- CDE
conserved DNA element
- CEN DNA
centromere DNA
- CFP
cyan fluorescent protein
- ChIP
chromatin immunoprecipitation
- GFP
green fluorescent protein
- SPB
spindle pole body
- YFP
yellow fluorescent protein
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–08–0517. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–08–0517.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagney G, Uetz P, Fields S. High-throughput screening for protein-protein interactions using two-hybrid assay. Methods Enzymol. 2000;328:3–14. doi: 10.1016/s0076-6879(00)28386-9. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Brew C, Wolyniak M, Desai A, Anderson S, Muster N, Yates JR, Huffaker TC, Drubin DG, Barnes G. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J Cell Biol. 2001;155:1137–1145. doi: 10.1083/jcb.200109063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Drubin DG, Barnes G. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J Cell Biol. 2002a;157:199–203. doi: 10.1083/jcb.200201052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR, III, Chan CSM, Orubin DG, Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the aurora kinase Ipl1p. Cell. 2002a;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- Dobie KW, Hari KL, Maggert KA, Karpen GH. Centromere proteins and chromosome inheritance: a complex affair. Curr Opin Genet Dev. 1999;9:206–217. doi: 10.1016/S0959-437X(99)80031-8. [DOI] [PubMed] [Google Scholar]
- Doheny KF, Sorger PK, Hyman AA, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the S. cerevisiaekinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian KD, et al. Functional analysis of 150 deletion mutants in Saccharomyces cerevisiaeby a systematic approach. Mol Gen Genet. 1999;262:683–702. doi: 10.1007/pl00013817. [DOI] [PubMed] [Google Scholar]
- Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Ghosh SK, Poddar A, Hajra S, Sanyal K, Sinha P. The IML3/MCM19 gene of Saccharomyces cerevisiaeis required for a kinetochore-related process during chromosome segregation. Mol Genet Genomics. 2001;265:249–257. doi: 10.1007/s004380000408. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. Transforming Yeast with DNA. Methods Mol Cell Biol. 1995;5:255–269. [Google Scholar]
- Goshima G, Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- Hailey DW, Davis TN, Muller EG. Fluorescence resonance energy transfer using color variants of green fluorescent protein. Methods Enzymol. 2002;351:34–49. doi: 10.1016/s0076-6879(02)51840-1. [DOI] [PubMed] [Google Scholar]
- He X, Rines DR, Espelin CW, Sorger PK. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- Hofmann C, Cheeseman IM, Goode BL, McDonald KL, Barnes G, Drubin DG. Saccharomyces cerevisiaeDuo1p and Dam1p, novel proteins involved in mitotic spindle function. J Cell Biol. 1998;143:1029–1040. doi: 10.1083/jcb.143.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Stearns T, Botstein D. Chromosome instability mutants of Saccharomyces cerevisiaethat are defective in microtubule-mediated processes. Mol Cell Biol. 1990;10:223–234. doi: 10.1128/mcb.10.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland KM, Kingsbury J, Koshland D, Hieter P. Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiaeand a potential link between the kinetochore and mitotic spindle. J Cell Biol. 1999;145:15–28. doi: 10.1083/jcb.145.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Sorger PK. Structure and function of kinetochores in budding yeast. Annu Rev Cell Dev Biol. 1995;11:471–495. doi: 10.1146/annurev.cb.11.110195.002351. [DOI] [PubMed] [Google Scholar]
- Janke C, Ortiz J, Lechner J, Shevchenko A, Magiera MM, Schramm C, Schiebel E. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 2001;20:777–791. doi: 10.1093/emboj/20.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Ortiz J, Tanaka TU, Lechner J, Schiebel E. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 2002;21:181–193. doi: 10.1093/emboj/21.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K, Hieter P. Evolutionary conservation between budding yeast, and human kinetochores. Nat Rev Mol Cell Biol. 2001;2:678–689. doi: 10.1038/35089568. [DOI] [PubMed] [Google Scholar]
- Kopski KM, Huffaker TC. Suppressors of the ndc10–2 mutation: a role for the ubiquitin system in Saccharomyces cerevisiaekinetochore function. Genetics. 1997;147:409–420. doi: 10.1093/genetics/147.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D, Hieter P. Visual assay for chromosome ploidy. Methods Enzymol. 1987;155:351–372. doi: 10.1016/0076-6879(87)55024-8. [DOI] [PubMed] [Google Scholar]
- Kouprina N, Kirillov A, Kroll E, Koryabin M, Shestopalov B, Bannikov V, Zakharyev V, Larionov V. Identification and cloning of the CHL4 gene controlling chromosome segregation in yeast. Genetics. 1993a;135:327–341. doi: 10.1093/genetics/135.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Pashina OB, Nikolaishwili NT, Tsouladze AM, Larionov VL. Genetic control of chromosome stability in the yeast Saccharomyces cerevisiae. Yeast. 1988;4:257–269. doi: 10.1002/yea.320040404. [DOI] [PubMed] [Google Scholar]
- Kouprina N, Tsouladze A, Koryabin M, Hieter P, Spencer F, Larionov V. Identification and genetic mapping of CHL genes controlling mitotic chromosome transmission in yeast. Yeast. 1993b;9:11–19. doi: 10.1002/yea.320090103. [DOI] [PubMed] [Google Scholar]
- Kroll ES, Hyland KM, Hieter P, Li JJ. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics. 1996;143:95–102. doi: 10.1093/genetics/143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Li Y, Bachant J, Alcasabas AA, Wang Y, Qin J, Elledge SJ. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 2002;16:183–197. doi: 10.1101/gad.959402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Maine GT, Sinha P, Tye BK. Mutants of S. cerevisiaedefective in the maintenance of minichromosomes. Genetics. 1984;106:365–385. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday V, Hailey DW, Pot I, Givan SA, Hyland KM, Cagney G, Fields S, Davis TN, Hieter P. Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev. 2002;16:101–113. doi: 10.1101/gad.949302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiaeencodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Ortiz J, Lechner J. The budding yeast kinetochore: less simple than expected. Protoplasma. 2000;211:12–19. [Google Scholar]
- Ortiz J, Stemmann O, Rank S, Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Maddox PS, Salmon ED, Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Allshire RC. Centromeres: getting a grip of chromosomes. Curr Opin Cell Biol. 2000;12:308–319. doi: 10.1016/s0955-0674(00)00094-6. [DOI] [PubMed] [Google Scholar]
- Poddar A, Roy N, Sinha P. MCM21 and MCM22, two novel genes of the yeast Saccharomyces cerevisiaeare required for chromosome transmission. Mol Microbiol. 1999;31:349–360. doi: 10.1046/j.1365-2958.1999.01179.x. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1990. [Google Scholar]
- Roy N, Poddar A, Lohia A, Sinha P. The mcm17 mutation of yeast shows a size-dependent segregational defect of a mini-chromosome. Curr Genet. 1997;32:182–189. doi: 10.1007/s002940050264. [DOI] [PubMed] [Google Scholar]
- Spencer F, Gerring SL, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov AV, Kingsbury J, Koshland D. CEP3 encodes a centromere protein of Saccharomyces cerevisiae. J Cell Biol. 1995;128:749–760. doi: 10.1083/jcb.128.5.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Blower MD, Karpen GH. Determining centromere identity: cyclical stories and forking paths. Nat Rev Genet. 2001;2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kilmartin JV. The Ndc80p complex from Saccharomyces cerevisiaecontains conserved centromere components and has a function in chromosome segregation. J Cell Biol. 2001;152:349–360. doi: 10.1083/jcb.152.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]