Abstract
Macroautophagy is a catabolic membrane trafficking phenomenon that is observed in all eukaryotic cells in response to various stimuli, such as nitrogen starvation and challenge with specific hormones. In the yeast Saccharomyces cerevisiae, the induction of autophagy involves a direct signal transduction mechanism that affects membrane dynamics. In this system, the induction process modifies a constitutive trafficking pathway called the cytoplasm-to-vacuole targeting (Cvt) pathway, which transports the vacuolar hydrolase aminopeptidase I, from the formation of small Cvt vesicles to the formation of autophagosomes. Apg1 is one of the proteins required for the direct signal transduction cascade that modifies membrane dynamics. Although Apg1 is required for both the Cvt pathway and autophagy, we find that Apg1 kinase activity is required only for Cvt trafficking of aminopeptidase I but not for import via autophagy. In addition, the data support a novel role for Apg1 in nucleation of autophagosomes that is distinct from its catalytic kinase activity and imply a qualitative difference in the mechanism of autophagosome and Cvt vesicle formation.
INTRODUCTION
When eukaryotic cells are challenged by nitrogen starvation or specific hormonal stimuli, they respond with a set of physiological changes that allow them to adapt to these new conditions. An important aspect of these responses is autophagy, a catabolic membrane trafficking event that is induced by signal transduction mechanisms (reviewed in Abeliovich and Klionsky, 2001; Kim and Klionsky, 2000). During autophagy, intracellular membranes, in some cases identified as originating from the endoplasmic reticulum (Dunn, 1990a, 1990b), engulf and sequester cytoplasmic material in unique double bilayer vesicles called autophagosomes. The autophagosomes then acquire endosomal and lysosomal characteristics, resulting in the degradation of the cytosolically derived material back into cellular building blocks (Dunn, 1990a, 1990b). This response is conserved in the yeast Saccharomyces cerevisiae, where autophagosomes (300–900 nm in diameter) can be seen to fuse with the yeast lysosome, the vacuole, releasing the inner vesicle (called the autophagic body) into the vacuolar lumen (Takeshige et al., 1992). In yeast, autophagy is essential for survival during nitrogen starvation and for sporulation (Tsukada and Ohsumi, 1993). A related pathway, the cytoplasm-to-vacuole targeting (Cvt) pathway is active in yeast cells under normal growth conditions. The Cvt pathway acts as a biosynthetic conduit (Klionsky et al., 1992), carrying a yeast vacuolar protease, aminopeptidase I (Ape1), into the lumen of the vacuole. In this pathway the cytosolic precursor of Ape1, prApe1 (61 kDa), is synthesized on soluble ribosomes and specifically is sequestered in double bilayer vesicles called Cvt vesicles (140–160 nm diameter). The outer bilayer of the Cvt vesicle then fuses with the limiting membrane of the yeast vacuole, leading to the release of the inner vesicle (the Cvt body) into the lumen of the vacuole. The Cvt body is then degraded, allowing the maturation of prApe1 to the active 50-kDa form (Kim and Klionsky, 2000; Abeliovich and Klionsky, 2001).
Genetic studies on the Cvt pathway and autophagy over the last several years have uncovered genes required for these processes (Tsukada and Ohsumi, 1993; Thumm et al., 1994; Harding et al., 1995). These studies found that the molecular machinery of autophagy and the Cvt pathway largely overlaps, reflecting the high degree of similarity between the two processes (Harding et al., 1996; Baba et al., 1997; Scott et al., 1997). Indeed, prApe1 is a specific cargo of both Cvt vesicles and autophagosomes (Scott et al., 1996); this specificity is mediated by the binding of the receptor/adaptor protein Cvt19 to prApe1 (Scott et al., 2001). Thus, current thinking postulates that nonspecific cargo is randomly trapped through the autophagic sequestration process, whereas specific cargoes, exemplified by prApe1, are physically recruited by their interaction with proteins such as Cvt19. Although most of the genes required for autophagy and the Cvt pathway are shared, there is a group of Cvt pathway-specific genes (Harding et al., 1996; Abeliovich et al., 1998; Scott et al., 2000) as well as a small but growing list of autophagy-specific genes (Kamada et al., 2000; Ishihara et al., 2001).
The induction of autophagy involves a direct signal transduction step that induces the formation of autophagosomes as well as a secondary expansion step that depends on de novo protein synthesis (Abeliovich et al., 2000). This second step is required for the formation of correctly sized, physiologically competent autophagosomes (Abeliovich et al., 2000). The direct signal transduction mechanism that initiates this process is not fully understood. In yeast, autophagy can be induced by the small macrolide antibiotic rapamycin, which mimics starvation by inhibiting Tor, a highly conserved protein kinase (Noda and Ohsumi, 1998). Tor kinases control a panoply of nutritional responses and cell growth decisions in all eukaryotes (reviewed in Klionsky and Emr, 2000). In yeast, inhibition of Tor by rapamycin as well as nitrogen starvation results in a rapid dephosphorylation of Apg13, a protein normally hyperphosphorylated in rich medium (Kamada et al., 2000; Scott et al., 2000). Like the induction of autophagosomes, this event is also independent of de novo protein synthesis (Abeliovich et al., 2000). Apg13 associates with a protein kinase required for the Cvt pathway and autophagy called Apg1, and it has been suggested that the dephosphorylation of Apg13 causes changes in Apg1 activity (Kamada et al., 2000), leading to a shift from the Cvt pathway to autophagic trafficking.
Apg1 is a large protein of 897 amino acids with an N-terminal kinase domain that spans amino acids 24–325. There are putative Apg1 homologues in human, mouse, worm, and other genomes, suggesting that its function is conserved. The precise way in which Apg1 participates in the induction of autophagosomes and in the Cvt pathway has been under some debate. It is known that in apg1Δ mutants prApe1 accumulates at a state before sequestration by Cvt vesicles or autophagosomes, implying a role for Apg1 in early events of autophagosome/Cvt vesicle formation (Harding et al., 1996). Matsuura et al. (1997) found that Apg1 undergoes autophosphorylation and that Apg1 kinase activity, as measured by autophosphorylation, is inhibited under autophagic conditions. In contrast, Kamada et al. (2000) reported that, upon induction of autophagy, an increase is observed in both association of Apg1 with Apg13 as well as in Apg1 kinase activity, measured in vitro using myelin basic protein as a substrate.
In this study we have undertaken to further elucidate the way in which changes in Apg1 affect the shift into autophagic trafficking. To this end, we have utilized the analog-sensitive mutation approach (Bishop et al., 2000). In this technique, the active site of a protein kinase is mutated so that it is able to accommodate a cell-permeable analog of a nonspecific kinase inhibitor. Although the nonspecific inhibitor PP1 (4-amino-1-tert-butyl-3-phenylpyrazolo[3,4-d]pyrimidine) is able to bind a wide spectrum of ATP-binding sites in protein kinase active sites, the bulky analog 1-NA-PP1 cannot displace ATP from the active site of wild-type protein kinases. Replacement of the wild-type gene with the analog-specific allele creates a situation where the designed ATP competitive inhibitor is a highly specific reagent that only affects cellular events that depend on the particular kinase activity under examination (reviewed in Bishop et al., 2001). We find that inhibition of Apg1 kinase activity abrogates the Cvt pathway but not autophagy. Our findings suggest a nonkinase related role for Apg1 in the induction of autophagosomes. In addition, we find that Apg1 undergoes conformational changes in response to inhibition of Tor and that these changes depend on amino acid residues at the C terminus of the protein.
MATERIALS AND METHODS
Strains, Plasmids, and Growth Conditions
Yeast were grown in YPD medium (1% yeast extract, 2% peptone, 2% glucose) or synthetic minimal medium (SD; 0.67% yeast nitrogen base, 2% glucose, and auxotrophic amino acids and vitamins as required). Starvation medium was SD-N (0.17% yeast nitrogen base without ammonium sulfate or amino acids containing 2% glucose). Strain HAY395 was previously described (Abeliovich et al., 2000). Strain HAY75 (MATα, leu2–3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9) was a MATα segregant from strain HAY70 (Abeliovich et al. 1999). Strain HAY437 (MATα, pep4Δ::LEU2 leu2–3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 APG1-protein A::HIS5 S.p.) was constructed by integrating the protein A (prA) cassette (see below) into the APG1 reading frame in strain TVY1 (Gerhardt et al., 1998) to create a full-length APG1-prA fusion reading frame. Strains HAY512 (MATa, leu2–3,112 ura3-52 his3-Δ200 trp1-Δ901 APG1-protein A::HIS5 S.p. pep4Δ::LEU2) and HAY524 (MATa, leu2–3,112 ura3-52 his3-Δ200 trp1-Δ901 pep4Δ::LEU2) were MATa progeny from a cross between HAY437 and SEY6210.1. Strains HAY453, HAY454, HAY455, HAY518, and HAY 370 were constructed by integrating the prA cassette into HAY75 so as to fuse the prA tag to codons 800, 850, 880, 886, and 897 (full-length), respectively. Strain HAY478 was constructed by integrating the prA cassette into strain TVY1 so as to fuse the prA tag to codon 880 of the APG1 reading frame. Correct integration of the tags was verified by PCR and Western blotting with anti-prA antibodies. Strain HAY571 (apg1Δ::URA3) was constructed by disruption of the APG1 gene in TDY27 (MATα, leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 vam3ts) cells (Abeliovich et al., 1999). Strain HAY572 (apg1Δ::URA3) was constructed by disrupting the APG1 gene in strain TN124 (MATa leu2–3,112 trp1 ura3-52 pho8::pho8Δ60 pho13Δ::LEU2) (Noda et al., 1995). Strain HAY595 (MATa, leu2–3,112 ura3-52 his3-Δ200 trp1-Δ901 apg1Δ::URA3 S.p. pep4Δ::LEU2) was haploid progeny from a cross between HAY395 and HAY524. Strain HAY603 (MATα, apgΔ::URA3 apg13Δ::KanR leu2–3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9) was a haploid progeny from a cross between strains HAY595 and BY4742 apg13Δ::KanR (ResGen Corp., Carlsbad, CA) that was backcrossed to the HAY75 background. Strain HAY487 was constructed by integrating the prA cassette after codon 880 of APG1 in the BY4742 apg13Δ::KanR mutant to generate a truncated APG1 gene fusion. Strain HAY591 was progeny from a cross between strains HAY512 and BY4742 cvt9Δ::KanR (ResGen) that was backcrossed to the HAY75 background .
The GFP-AUT7(414) plasmid was made by ligation of the EcoRI/XhoI fragment from pAUT7(416) (Huang et al., 2000) into the EcoRI/XhoI sites of pRS414 to generate pAUT7(414). The BglII restriction site was then introduced just after the initiation codon of the AUT7 gene on pAUT7(414) using a QuikChange Site-directed Mutagenesis kit (Stratagene, La Jolla, CA), to generate pAUT7BglII(414). The DNA fragment encoding GFP (S65T) with BamHI sites on both sides was then ligated to the BglII site of pAUT7BglII(414) to generate pGFP-AUT7(414). The APG1 gene was cloned into the SpeI and SalI sites of pRS415 (Sikorsky and Hieter, 1989) by amplification of the open reading frame plus 500 bases upstream of the initiation codon and 300 bases downstream of the termination codon using primers containing SpeI and SalI sites.
Reagents
Kinase inhibitors PP1 and 1-NA-PP1 were synthesized as described previously (Bishop et al., 1999). Antiserum to Ape1 has been described (Klionsky et al., 1992). CompleteTM EDTA-free protease inhibitor tablets were from Roche Molecular Biochemicals (Indianapolis, IN). Other reagents were from Sigma-Aldrich (St. Louis, MO) unless specified.
Anti-Apg1 Antibody
Codons 1–250 of the Apg1 reading frame were cloned in frame into the NdeI and BamHI sites of plasmid pET-14b (Novagen, Madison, WI). The recombinant 6xHis-tagged protein was purified on Ni-NTA agarose according to the Novagen manual and polyclonal rabbit antiserum was generated by Covance Research Products, Inc. (Denver, PA).
In Vitro Mutagenesis
Introduction of the M102A mutation in the APG1 reading frame was done by site-directed PCR mutagenesis (gene SOEing; Pogulis et al., 1996), and the mutation was verified by DNA sequencing of the reading frame. The K54A, L886G, and N884A mutations were introduced using the Stratagene QuickChange kit (Stratagene Inc.).
Integrated Tagging of Apg1 with 2X IgG Binding Domain from Protein A
Plasmid pHAB102 was constructed by excising a PacI-AscI fragment of pFA6a-GFP(S65T)-His3MX6 (Longtine et al., 1998) and replacing it with a PacI-AscI PCR fragment carrying a 2× repeat of the Staphylococcus aureus prA IgG binding domain (gift of Dr. Peter Rehling). To generate an integration module, DNA primers were designed to amplify the prA/His5 region with 5′ and 3′ 40-base overhangs that correspond to the targeted integration site.
Immunoprecipitation and Protein Kinase Assays
A logarithmically growing cell culture (40 A600 units) was harvested and resuspended in 300 μl of lysis buffer (50 mM HEPES, pH7.5, 1 M NaCl, 1 mM EGTA, 1 mM MgCl2, 30% glycerol, 30 mM sodium pyrophosphate, 0.2 mM Na3VO4, 50 mM NaF, 2.5 mM EDTA, 1 μg each of leupeptin and pepstatin, and 1 mM PMSF, supplemented with Complete EDTA-free protease inhibitor tablets). Acid-washed glass beads, 500 μl, were added, and the cells were vortexed at 4°C for 5 min. The cell extract was diluted with 1.5 ml of IP buffer (50 mM HEPES, pH 7.5, 100 mM NaCl, 2.5 mM EDTA, 0.5% Tween-20, 10% glycerol, 0.2 mM Na3VO4, 1 μg each of leupeptin and pepstatin, and 1 mM PMSF) and clarified twice by centrifugation for 10 min at 13,000 × g. The extract (8 mg of total protein in 0.7 ml) was further diluted twofold in IP buffer and incubated with gentle agitation for 2 h at 4°C with 10 μl of anti-Apg1 antibody. Three hundred microliters of a 10% suspension of prA sepharose was added and incubated a further 1.5 h. Immune complexes were washed once in IP buffer, three times in wash buffer (50 mM HEPES, pH 7.5, 0.5 M NaCl, 2.5 mM EDTA, 10% glycerol, and 0.5% Tween 20), and three times in kinase buffer (50 mM HEPES, pH 7.5, 1 mM EGTA, 2 mM MgCl2, 0.1% Tween-20, and 10% glycerol). The immunoprecipitate was divided equally into four tubes and resuspended in 100 μl of kinase buffer containing 0 or 20 μM 1-NA-PP1 for 5 min. Precipitates were collected by centrifugation and resuspended in 30 μl kinase buffer containing the appropriate inhibitor concentration plus 5 μM ATP and 20 μCi of γ-32P-ATP (4500 Ci/mmol, ICN Biochemicals, Costa Mesa, CA). Reactions were incubated for 30 min at 30°C and stopped by the addition of 30 μl 2× SDS sample buffer. Thirty microliters of each reaction was separated on 8% SDS-PAGE, and radiolabeled Apg1 was visualized using a Bio-Rad (Richmond, CA) personal molecular imager FX phosphorimager with Quantity One software.
Affinity Purification on IgG Sepharose
Yeast cells containing the desired prA-tagged gene were grown to midlog phase in YPD, and 40 A600 equivalents were spheroplasted with 100 μg/ml oxalyticase (Zymogenetics, Corvallis, OR) for 20 min in spheroplasting medium (YPD, 20 mM HEPES, pH 7.0, and 1 M sorbitol) at 30°C with shaking. The cells were then treated with or without 0.2 ng/ml rapamycin and incubated a further 15 min. Cells were then lysed in ice-cold lysis buffer (20 mM HEPES, pH 7.0, 0.5% Triton X-100, 5 mM EDTA, 1 mM MgCl2, 100 mM KCl, 5 mM potassium phosphate) supplemented with Complete EDTA-free protease inhibitor tablets and 1 μg/ml each of leupeptin and pepstatin A. Extracts were clarified by centrifugation at 100,000 × g for 15 min at 4°C in a Beckman Optima MAX-E ultracentrifuge (Berkeley, CA) and then incubated for 1.5 h at 4°C with 40 μl of a 50% slurry (vol/vol) of IgG sepharose beads (Amersham Biosciences, Piscataway, NJ). The beads were then washed once with lysis buffer, three times with wash buffer (20 mM HEPES, pH 7.0, 0.5% Triton X-100, 5 mM EDTA, 1 mM MgCl2, 200 mM KCl, 5 mM potassium phosphate) and once with preelution buffer (20 mM HEPES, pH 7.0, 5 mM EDTA, 1 mM MgCl2, 5 mM potassium phosphate) and eluted with 200 mM ammonium acetate (pH 3.5). Eluates were dried under vacuum and solubilized in SDS sample buffer.
Velocity Gradient Sedimentation
Yeast cells were grown as above and lysed in 20 mM HEPES, pH 7.0, 100 mM KCl, 5 mM potassium phosphate, 1 mM MgCl2, 5 mM EDTA, 0.2% Triton X-100, supplemented with Complete EDTA-free protease inhibitor tablets and 1 μg/ml each of leupeptin and pepstatin A. Extracts were clarified by a 15-min centrifugation at 100,000 × g in a Beckman Optima MAX-E ultracentrifuge and 100 μl (8 A600 equivalents) were loaded on a 5–20% sucrose gradient. The gradients were centrifuged at 259,000 × g in a TLS55 rotor in a Beckman Optima MAX-E ultracentrifuge for 7 h. Thirteen 100-μl fractions were collected, and samples were precipitated with 10% TCA, washed twice with cold acetone, and resuspended in SDS-PAGE sample buffer. Fractionation profiles were obtained by quantitative Western blotting using chemiluminescent detection with a Bio-Rad Fluor-S Max imager and software. Sedimentation constants for peak fractions were derived from published tables (McEwen, 1967). Molecular mass markers (Pharmacia, Piscataway, NJ) were run in parallel gradients under identical conditions: Aldolase (158 kDa), ferritin (440 kDa), and thyroglobulin (669 kDa).
In Vivo Phosphate Labeling and Immunoprecipitation
Yeast cells were grown to midlog in SD, washed, and resuspended in SD-phosphate. The cells were incubated a further 6 h in SD-phosphate, and 5 A600 equivalents were labeled with 150 μCi 32P (carrier free, ICN) for 1 h. Rapamycin or drug vehicle was added, and incubation was continued a further 20 min before the cells were harvested, treated with 10% TCA, and washed three times with acetone. The cell pellet was dried, and extracts were prepared by grinding in a TOMY mixer with 200 μl glass beads and 100 μl phosphate cracking buffer (100 mM potassium phosphate, pH 7.4, 6 M urea, 1% SDS, 5 mM EDTA) for 5 min. The extracts were diluted in phosphate-IP buffer (50 mM potassium phosphate, pH 7.4, 150 mM KCl, 5 mM EDTA, 0.5% Tween-20) and equivalent amounts of TCA-precipitable counts were immunoprecipitated with anti-Apg1 antibody and protein A sepharose. Immunoprecipitates were washed three times with urea wash buffer (50 mM Tris, pH 7.5, 2 M urea, 200 mM NaCl, 0.5% Tween-20) twice with TB (50 mM Tris, pH 7.5, 1% β-mercaptoethanol) and once with 50 mM Tris, pH 7.5. Beads were eluted with SDS loading buffer, and the eluate was electrophoresed on gels, dried, and exposed to phosphorimager plates.
Preparation of Whole Cell Extracts for Western Blot Analysis
Cells (10 A600 units) were grown to an A600 of 0.5–0.8 in SMD or YPD, treated with 10% TCA, and washed twice with acetone. The dry cell pellet was then resuspended in 100 ml cracking buffer (50 mM Tris, pH 6.8, 3.6 M urea, 1 mM EDTA, 1% SDS) and vortexed in a TOMY MT-360 mixer (Palo Alto, CA) at maximum speed, with an equal volume of acid-washed glass beads, for 15 min. Unlysed cells were removed by centrifugation, an equal volume of 2× SDS loading buffer was added, and the samples were incubated at 70°C for 10 min.
Alkaline Phosphatase Assay
Approximately four A600 equivalents of yeast cells were harvested, washed in ice-cold distilled water containing 2 mM PMSF, and resuspended in 100 μl lysis buffer (20 mM PIPES, pH 7.0, 0.5% Triton X-100, 50 mM KCl, 100 mM potassium acetate, 10 mM MgSO4, 10 μM ZnSO4, and 1 mM PMSF). An equal volume of acid-washed glass beads was added, the cells were lysed by vortexing for 4 min, and the lysate was diluted by adding 200 μl of lysis buffer. To start the assay, 20 μl of extract was added to 480 μl reaction buffer (250 mM Tris-HCl, pH 8.5, 0.4% Triton X-100, 10 mM MgSO4, 1.25 mM nitrophenyl phosphate), and samples were incubated 15 min at 37°C before terminating the reaction by adding 500 μl of stop buffer (2 M glycine, pH 11). Evolution of nitrophenol was monitored by measuring absorbance at 405 nM using a Beckman DU-640B spectrophotometer, and the time 0 blank was subtracted from each sample. Nitrophenol concentration was calculated using Beer's law with ε405 = 18,000 M−1 cm−1. Protein concentration in the extracts was measure with the Pierce BCA assay (Pierce Chemical Co., Rockford, IL), and one activity unit was defined as nmol nitrophenol/min/mg protein.
Yeast Two-Hybrid Assays
Two-hybrid assays were done as described by James et al. (1996). Briefly, PCR products of full-length APG1 and the kinase domain truncated variant (amino acids 326–897), carrying 5′ BamHI and 3′ SalI sites in their respective primers, were cloned into pGAD-C2 and pGBDU-C2. Plasmids were transformed into strain PJ69–4A, and interactions were assayed by streaking colonies on SD plates lacking either histidine or adenine and testing for growth.
Microscopy
Cells were grown to midlog phase in SD medium lacking the appropriate selective nutrients and stained with 0.8 μM FM 4–64 (Molecular Probes, Eugene, OR) for 20 min. Cells were then washed in SD and resuspended in SD for a further 1 h before viewing or transfer into nitrogen starvation medium for 3 h. The cells were viewed using a Nikon E-800 microscope (Garden City, NY) fitted with DIC optics and a Hamamatsu Orca 2 digital camera (Bridgewater, NJ) with Openlab 3 software. Electron microscopy was as previously described (Abeliovich et al., 2000).
RESULTS
Inhibition of Apg1 Protein Kinase Activity Abrogates Precursor Ape1 Trafficking in Rich Medium but Not in Response to Nitrogen Starvation
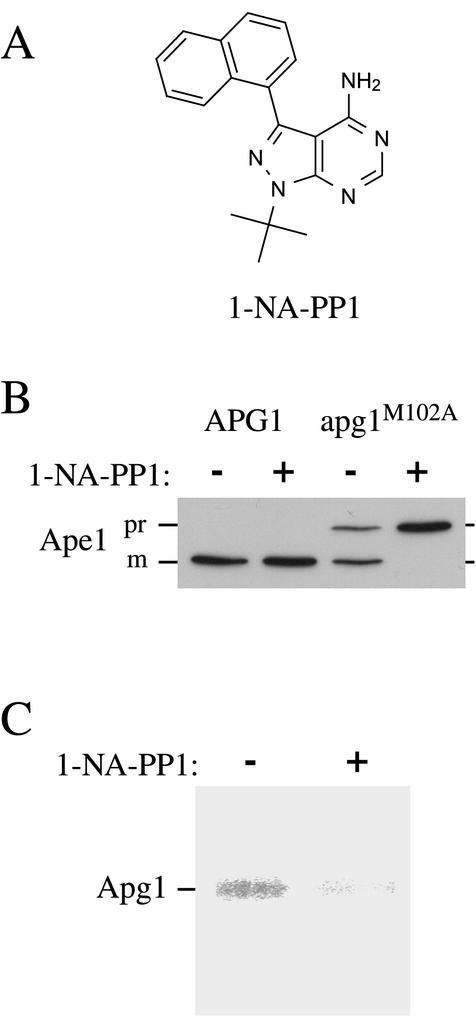
To address the requirement for Apg1 protein kinase activity in the induction of autophagy and in the Cvt pathway, we decided to generate a conditional inhibitor-sensitive allele of APG1. Accordingly, we introduced a mutation at position 102 of the Apg1 open reading frame, replacing a methionine residue with an alanine (Apg1M102A). This point mutation was designed based on previous work with Src and Cdc28, which showed that a mutation at this conserved position in the ATP binding pocket allowed binding of bulky analogs of the general Src-family kinase inhibitor, PP1 (structure shown in Figure 1A), that were not able to bind to the wild-type kinase (Liu et al., 1998; Bishop et al., 2000). Cells expressing Apg1M102A instead of the wild-type protein were able to mature prApe1 at close to normal levels (Figure 1B). In contrast, maturation of prApe1 in these cells was completely blocked by 20 μM 1-NA-PP1, an inhibitor that is unable to displace ATP in the active site of wild-type kinases (Figure 1B); this concentration of 1-NA-PP1 had no effect on prApe1 maturation in wild-type cells. As expected, protein kinase assays revealed that 1-NA-PP1 directly inhibited Apg1 autophosphorylation activity in immunoprecipitates prepared from cells expressing the apg1M102A mutant gene (Figure 1C).
Figure 1.
Precursor Ape1 maturation in the apg1M102A mutant is selectively inhibited by 1-NA-PP1. (A) Structure of 1-NA-PP1. (B) apg1Δ (HAY395) yeast cells carrying a wild-type APG1 or a mutant apg1M102A gene on pRS415 were grown to an A600 of 0.5 and treated with or without 20 μM 1-NA-PP1. The cells were incubated an additional 3 h and then harvested and processed for Western blotting as described in MATERIALS AND METHODS. (C) Kinase activity of Apg1M102A is inhibited by 20 μM 1-NA-PP1. Cell extract (2 mg per reaction) was immunoprecipitated with anti-Apg1 antiserum and assayed for autophosphorylation activity in the presence or absence of 20 μM 1-NA-PP1 as described in MATERIALS AND METHODS.
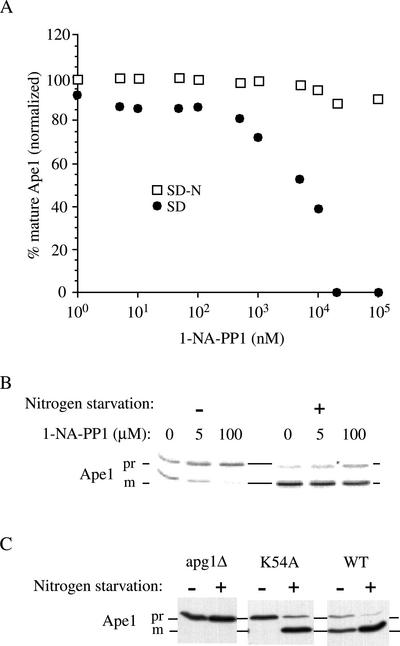
With this system, we were able to determine the effect of selectively inhibiting Apg1 kinase activity in vivo. An inhibition dose-response curve showed that half-maximal inhibition of prApe1 maturation occurred at ∼5 μM, in cells that were growing in standard synthetic medium (SD; Figure 2A). However, if cells were challenged with 1-NA-PP1 for 1 h and then transferred to nitrogen starvation medium in the presence of the same concentration of 1-NA-PP1, no inhibition was observed up to 100 μM (Figure 2, A and B).
Figure 2.
Inhibition of Apg1 kinase by 1-NA-PP1 blocks the Cvt pathway but allows autophagic maturation of prApe1. (A) Dose response for 1-NA-PP1 under standard growth conditions and under starvation. apg1Δ (HAY395) yeast cells expressing Apg1M102A from a plasmid were grown to midlog (A600 of 0.5), collected and resuspended in SD medium with the indicated concentrations of 1-NA-PP1. After 1 h incubation, one half of the cell culture was collected, washed with distilled water, and resuspended in nitrogen starvation medium (SD-N) in the presence of the same concentration of 1-NA-PP1. After a further incubation of 3 h, the cells were harvested and cells extracts were prepared. 0.5 A600 equivalents of each extract were resolved by SDS-PAGE and Ape1 was identified by Western blotting. The relative amount of prApe1 and mApe1 was quantified using a Bio-Rad Fluor-S MAX as described in MATERIALS AND METHODS. (B) Western blot of a subset of the samples described in A. (C) The kinase inactive apg1K54A mutant is blocked for prApe1 maturation in rich medium but not under starvation conditions. apg1Δ (HAY395) cells or these cells harboring a plasmid borne wild-type APG1 gene or apg1K54A allele were grown to midlog and either harvested immediately or transferred to nitrogen starvation medium for 3 h before extract preparation. Ape1 was detected by SDS-PAGE and Western blotting.
To rule out the possibility that a nitrogen starvation–induced change in the ATP binding cleft rendered 1-NA-PP1 unable to inhibit the enzymatic activity, we tested the ability of a previously characterized kinase dead mutant (Kamada et al., 2000), a lysine-to-alanine substitution at position 54 (K54A), to mature prApe1 under nitrogen starvation conditions. Although cells expressing Apg1K54A were blocked for prApe1 maturation in rich medium, a 3-h incubation in nitrogen-starvation medium resulted in mostly mature Ape1 (Figure 2C). Thus, using two independent approaches, we have demonstrated that inhibition of Apg1 kinase activity does not significantly affect prApe1 maturation under conditions where autophagy is induced.
Inhibition of Apg1 Kinase Activity Allows Normal Levels of Autophagy
The maturation of prApe1 during nitrogen starvation or in the presence of rapamycin is a good assay for the induction of autophagic trafficking in mutants that are specifically blocked in the Cvt pathway. However, maturation of prApe1 through an autophagic mode does not automatically imply that normal autophagosomes are being formed. For example, we have previously demonstrated that abnormal autophagosomes form when autophagy is induced in aut7Δ cells (Abeliovich et al., 2000). Although these cells are able to partially mature prApe1 in SD-N, they are not normal for autophagy. In other words, prApe1 maturation under starvation conditions is not a reliable indicator for fully functional autophagy because small abnormal autophagosomes and possibly reduced numbers of normal autophagosomes may be able to import prApe1 but cannot sustain a sufficient volume of autophagy for physiological function. This is possible because prApe1 is a specific cargo of both autophagosomes and Cvt vesicles, and there is a redundant capacity for prApe1 trafficking such that maturation can occur despite a partially defective autophagic response.
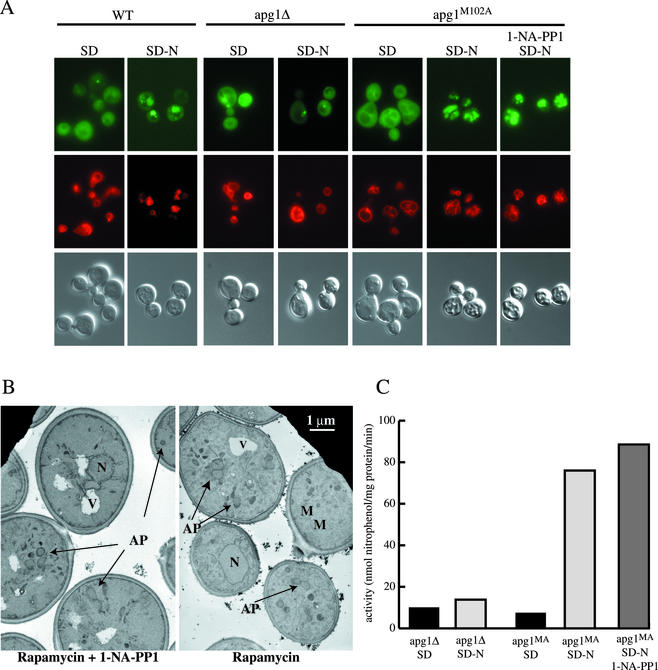
To analyze these possibilities more directly we first utilized an in vivo microscopy assay for autophagy. Aut7 is a component of Cvt vesicles and autophagosomes and is carried by these trafficking intermediates into the lumen of the vacuole (Kirisako et al., 1999; Huang et al., 2000). Cells that express GFP-tagged Aut7 accumulate the majority of the protein in the cytosol in nitrogen-rich medium. When these cells are undergoing autophagy, the bulk of this GFP fluorescence is transported into the vacuolar lumen (Kirisako et al., 1999; Huang et al., 2000). This allowed us to assay the ability of apg1M102A cells to accumulate vacuolar GFP in response to nitrogen starvation, in the presence and absence of 1-NA-PP1. When wild-type cells expressing GFP-Aut7 were grown in SD medium, GFP fluorescence was largely cytosolic, whereas a 4-h shift into nitrogen starvation medium resulted in primarily vacuolar staining (Figure 3A). This transport into the vacuolar lumen was dependent on Apg1, because it did not occur in apg1Δ cells. When cells expressing the Apg1M102A mutant were starved for nitrogen, GFP-Aut7 was transported into the vacuole regardless of whether they were treated with 30 μM 1-NA-PP1 (Figure 3A). Identical conclusions were reached by assaying the appearance of autophagic bodies in a protease-deficient strain (unpublished data; Takeshige et al., 1992). To assay the morphology of autophagosomes induced in the presence of 1-NA-PP1, we used a previously established morphological assay that relies on the accumulation of cytosolic autophagosomes in vam3ts cells when challenged with rapamycin at the nonpermissive temperature (Abeliovich et al., 1999, 2000). We found that vam3ts apg1Δ cells carrying the apg1M102A mutant gene on a centromeric plasmid induced autophagosomes in response to rapamycin, at the nonpermissive temperature, and the morphology of these autophagosomes was not altered when 30 μM 1-NA-PP1 was present (Figure 3B). Hence, inhibition of kinase activity does not affect autophagosome morphology.
Figure 3.
Inhibition of Apg1 kinase activity in the apg1M102A mutant does not block autophagy. (A) Wild-type (HAY75, WT), apg1Δ (HAY395) and apg1Δ cells harboring apg1M102A on pRS415 were transformed with a centromeric plasmid (pRS414) expressing Aut7-GFP under the control of the AUT7 promoter. Cells were grown to midlog and stained with FM 4–64 (0.8 μM) for 20 min, washed and incubated with or without 30 μM 1-NA-PP1 for 1 h in SD medium. Cells were then either viewed directly or washed with distilled water and transferred to nitrogen starvation medium for 4 h before viewing by light and fluorescence microscopy. Top panels: GFP fluorescence. Middle panels: FM 4–64 fluorescence. Bottom panels: DIC, differential interference contrast. (B) The vam3ts apg1Δ strain (HAY571) harboring apg1M102A on pRS415 was grown to midlog at 26°C, transferred for 1 h into medium containing 30 μM 1-NA-PP1 or mock-treated, and then transferred for 20 min to 37.5°C. Rapamycin was added to a final concentration of 0.2 μg/ml and the cells were incubated for a further 90 min before permanganate fixation and electron microscopy as described in MATERIALS AND METHODS. AP, autophagosome; M, mitochondrion; N, nucleus; V, vacuole. (C) The pho13Δ pho8Δ60 apg1Δ strain (HAY572) or HAY572 cells harboring apg1M102A on a plasmid were subjected to the same treatments as in (A) and alkaline phosphatase activity was measured in extracts as described in MATERIALS AND METHODS. Results represent duplicate measurements from two independent experiments.
A more quantitative assay for autophagy measures nonspecific uptake of a cytosolic protein, Pho8Δ60, into the lumen of the vacuole (Noda et al., 1995). Pho8Δ60 is a truncated version of yeast vacuolar alkaline phosphatase (encoded by the PHO8 gene) lacking the N-terminal transmembrane domain that functions as an internal uncleaved signal sequence. As a result, Pho8Δ60 cannot enter the ER and is only delivered to the vacuole through autophagy. On vacuolar delivery, the C-terminal propeptide is proteolytically removed. The resulting activation of the zymogen can be measured by a simple colorimetric assay. Because this protein is not specifically taken up by autophagosomes or Cvt vesicles and because only a small proportion of Pho8Δ60 is delivered to the vacuole, the amount matured is proportional to the “autophagic volume” that is delivered into the vacuolar lumen, giving a quantitative output. This allows us to distinguish whether kinase activity is altogether redundant for induction of autophagy or plays some facilitative role either in the enlargement of the autophagosome or in determining the number of autophagosomes. We constructed a Pho8Δ60 strain that expresses Apg1M102A instead of the wild-type protein and measured induction of alkaline phosphatase activity in response to starvation in the presence and absence of 1-NA-PP1. As seen in Figure 3C, induction of alkaline phosphatase activity in cell extracts of apg1M102A cells was not reduced by 1-NA-PP1 at a concentration of 30 μM. Rather, it was slightly increased in two independent experiments. Thus, by three different criteria, Apg1 kinase activity does not seem to be required for induction of autophagosomes.
The C Terminus of Apg1 Is Subject to Reversible Sequestration
Apg1 is proposed to be a component of a multiprotein complex (Kamada et al., 2000; Scott et al., 2000; Kim et al., 2001), although the precise nature of this putative complex has not been determined. To better understand the physical interactions of Apg1 under various physiological conditions, we chromosomally tagged the C terminus of Apg1 with a tandem repeat of the IgG binding domain from S. aureus prA. This tag did not interfere with normal function of Apg1 because prApe1 was normally matured and the cells were not sensitive to nitrogen starvation (Figure 4). We reasoned that a chromosomal tag expressed from its endogenous promoter would allow a more faithful analysis of protein complexes and their dynamics under different physiological conditions. Analyses of Apg1 protein–protein interactions were previously conducted with overexpressed pairs of proteins (Kamada et al., 2000; Scott et al., 2000; Kim et al., 2001). Although this approach allows sensitive detection of pairwise interactions, it may not yield information on higher order complexes (because of competitive effects) or regulatory mechanisms (because of saturation). Unlike the studies with pairwise overexpression of proteins, we were unable to identify specific stoichiometrically associating proteins that copurified with Apg1, either when examining silver-stained gels or when immunoblotting for specific proteins that interact with Apg1 when overexpressed or in yeast two-hybrid systems. This result is similar to that seen in a genome wide survey of protein–protein interactions in S. cerevisiae, which included Apg1; that is, no interacting partners for prA-tagged Apg1 were identified (Ho et al., 2002).
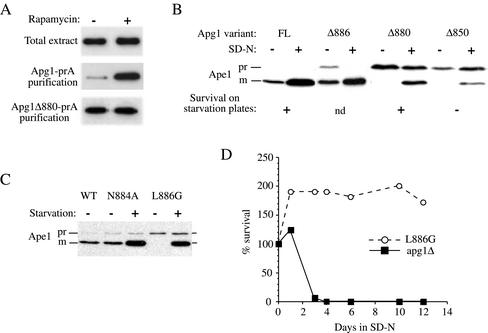
Figure 4.
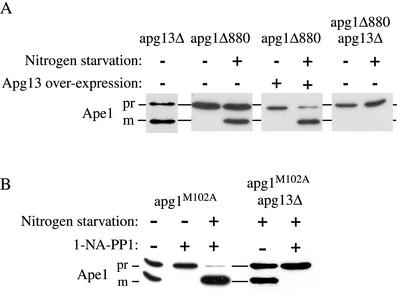
The C terminus of Apg1 is engaged in a Cvt pathway-specific interaction. (A) The pep4Δ Apg1-prA strain (HAY437) expressing a full-length Apg1-prA fusion, or the pep4Δ apg1Δ880-prA strain (HAY478) expressing an Apg1Δ880 truncation fused to prA were grown to midlog in YPD medium and converted to spheroplasts. The spheroplasts were treated with or without 0.2 μg/ml rapamycin for 15 min before cell lysis, and the Apg1-prA fusions were purified from the extracts using IgG sepharose as described in MATERIALS AND METHODS. Apg1-prA was identified and quantified in the extracts by SDS-PAGE and immunodetection with anti-prA antibodies. (B) Truncation of the last 18 amino acids of Apg1 results in a Cvt pathway-specific phenotype. The apg1Δ850-prA (HAY454), apg1Δ880-prA (HAY455), apg1Δ886-prA (HAY518) and full-length (FL) Apg1-prA (HAY370) strains were grown to midlog. One-half of each culture was transferred to nitrogen starvation medium for 3 h before extract preparation and the other half was harvested directly. Precursor Ape1 maturation and the levels of the fusion proteins were assessed by Western blotting (0.5 A600 equivalents per lane). Survival of yeast in nitrogen starvation conditions was determined by patching cells on phloxine B plates (Tsukada and Ohsumi, 1993); nd, not determined. (C) A leucine to glycine mutation at position 886 results in a Cvt pathway-specific phenotype. WT (HAY75), apg1N884A, and apg1L886G yeast cells were grown to midlog in SD and either harvested directly or first transferred to nitrogen starvation medium for 3 h before extract preparation. Precursor Ape1 maturation was assayed by Western blotting. (D) apg1L886G cells do not lose viability in nitrogen starvation. The apg1L886G (HAY602) and apg1Δ (HAY395) cells were grown to midlog and transferred to nitrogen starvation medium. At indicated time points, viable cell counts were performed by removing an aliquot and plating an appropriate dilution on YPD.
Surprisingly, we found that the efficiency of purification of Apg1-prA on IgG sepharose depended on the physiological condition of the cells before lysis. If spheroplasted cells were treated with rapamycin for 15 min before detergent lysis, we observed a much higher yield of the purified fusion protein, both by immunoblotting (Figure 4A) as well as by silver staining (unpublished data). This increase did not reflect an increase in the amount of Apg1-prA in the extract because the total amount of the protein in the extract was constant, consistent with previous data (Matsuura et al., 1997). Thus, the protein-A tag was excluded from interacting with IgG sepharose in the absence of rapamycin, suggesting that a protein modification was occurring, which allowed the interaction between Apg1-prA and IgG sepharose after rapamycin treatment. The “Coils” program (Lupas et al., 1991), which predicts the propensity of an amino acid sequence to form coiled-coils, gives a reasonable probability that the last 20 amino acids of Apg1 engage in a coiled-coils interaction (unpublished data).
The desequestration of the Apg1-prA fusion protein in response to rapamycin treatment could be explained if a Cvt pathway-specific interaction was localized to the C terminus of the protein. If this is the case, one would expect that truncation of the C-terminal amino acids that encompass the ultimate coiled-coil domain would result in a Cvt pathway-specific defective phenotype. Indeed, a truncated fusion protein, Apg1Δ880-prA that lacks the C-terminal 18 amino acids, was completely blocked for prApe1 maturation in standard medium but displayed ∼50% maturation of prApe1 under starvation conditions (Figure 4B). In contrast, a truncation of 12 amino acids, Apg1Δ886-prA, showed little effect on Cvt trafficking of prApe1, whereas further truncating the protein to amino acid 850 reduced the degree of bypass in nitrogen starvation and also abrogated survival on SD-N plates (Figure 4B). Thus, residues between amino acids 880 and 886 may have a specific role in the Cvt pathway function of Apg1. Consistent with this, we find that a leucine to glycine mutation at amino acid 886 (L886G) resulted in a Cvt pathway-specific block (Figure 4C). Furthermore, the apg1L886G mutant was not defective in survival under nitrogen starvation conditions, confirming that autophagy is functional in this mutant (Figure 4D). Although the truncation mutation at position 880 appeared to have some effect on the autophagic bypass, the L886G mutant showed essentially complete bypass and maturation of prApe1 upon starvation. In contrast with glycine substitutions, alanine substitutions in the 880–886 region of Apg1 had little effect on prApe1 maturation (Figure 4C, unpublished data). This is consistent with a requirement for an alpha-helical conformation in this region and fits with the Coils program prediction that the extreme C terminus of Apg1 forms a coiled-coil structure.
Apg13 Has an Autophagy-specific Function and Is Required for the Autophagic Bypass of 1-NA-PP1–mediated Inhibition of Apg1M102A
Because the apg1Δ880 mutant is blocked in the Cvt pathway and this block is largely bypassed by starvation, we used this phenotype to test genetic interactions with Apg13, a protein that preferentially associates with Apg1 under autophagic conditions (Funakoshi et al., 1997; Kamada et al., 2000) and is thought to be required for Apg1 function. Overexpression of Apg13 in the apg1Δ880 truncation mutant strain had no effect on prApe1 maturation in SD medium but drastically increased the efficiency of bypass under nitrogen starvation conditions (Figure 5A). This strongly suggests that the effect of Apg13 on Apg1 function is exerted under autophagic conditions and not in nitrogen-rich medium and supports previous findings that demonstrate an increased interaction between the two proteins upon induction of autophagy (Kamada et al., 2000). Corroborating this, an apg1Δ 880 apg13Δ double mutant was unable to mature Ape1 under all circumstances. The apg13Δ single mutant was only marginally impaired in prApe1 maturation in SD medium, indicating that the inability of the apg1Δ880 apg13Δ double mutant to bypass the prApe1 block is due to a defect in autophagy and not the Cvt pathway.
Figure 5.
Apg13 has an autophagy-specific function. (A) Logarithmically growing apg1Δ880 (HAY455), apg13Δ (ResGen) or apg13Δ apg1Δ880 (HAY487) cells either containing or not containing the APG13 gene on a multicopy plasmid were either directly harvested or first transferred to nitrogen starvation medium and then harvested. Protein extracts were prepared and Ape1 was detected by Western blotting. (B) Starvation-dependent maturation of prApe1 in the presence of 20 μM 1-NA-PP1 requires Apg13. apg1Δ (HAY395) or apg1Δ apg13Δ (HAY603) cells carrying the apg1M102A gene on pRS415 were grown to midlog and treated with or without 20 μM 1-NA-PP1 for 1 h, and either starved for nitrogen in the presence of 1-NA-PP1 or allowed to remain in SD medium, for a further 3 h before preparation of protein extracts and SDS-PAGE. Ape1 was detected by Western blotting.
To further test whether Apg13 functions by altering Apg1 kinase activity, we constructed a double apg1M102A apg13Δ mutant strain. In the absence of 1-NA-PP1, the apg1M102A apg13Δ strain behaves similar to the apg1M102A strain in that it was able to mature prApe1 under starvation conditions. In the double mutant strain, however, this maturation was completely blocked by treatment with 1-NA-PP1 (Figure 5B). Because our previous experiments show that induction of the Cvt pathway, but not autophagy, depends on Apg1 kinase activity, this result implies that maturation of prApe1 in the apg13Δ background under starvation conditions is carried out by the (Apg13-independent) Cvt pathway. In other words, Apg13 appears to be required for autophagy but is not essential for the Cvt pathway, similar to Apg17 (Kamada et al., 2000). Furthermore, the inability of apg1M102A apg13Δ cells to overcome the prApe1 defect when treated with 1-NA-PP1 under starvation conditions confirms that the starvation-induced bypass seen with apg1M102A cells (Figure 2) was Apg13-dependent and was due to autophagy. Finally, because the Apg13-dependent bypass in apg1M102A cells occurs in the presence of 1-NA-PP1, the effect of Apg13 on Apg1 function during induction of autophagy is largely independent of kinase activity. Hence, the kinase-independent function of Apg1 in autophagy requires intact Apg13.
Apg1 Sediments as a 33S Complex and Undergoes a Shift to 25S On Treatment with Rapamycin
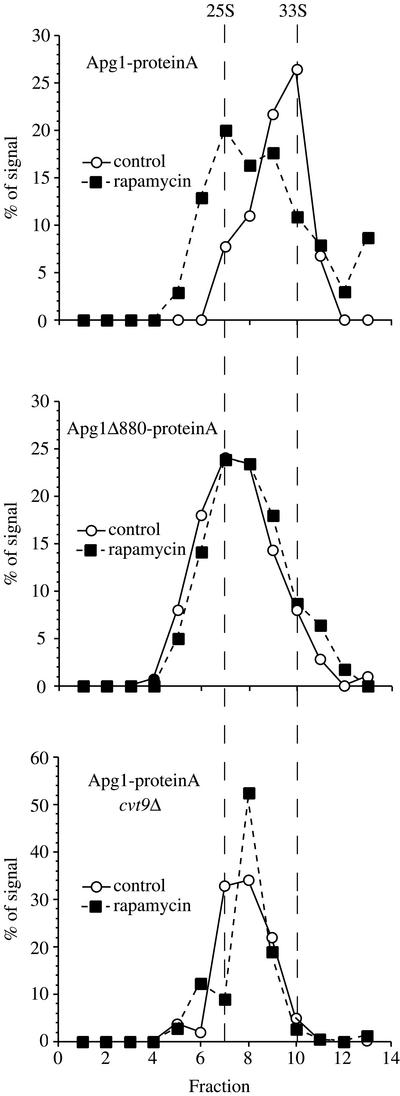
Apg1 is required for the induction of autophagy (Matsuura et al., 1997; Kamada et al., 2000). However, our data imply that Apg1 has a noncatalytic (or nonkinase) role in this process. One possible explanation is that the switch from Cvt to autophagic trafficking involves a structural change in Apg1 that does not require kinase activity. To determine if such a structural change occurs, we analyzed the hydrodynamic properties of the Apg1-prA fusion, in extracts that were pretreated with rapamycin or mock-treated. Yeast cells expressing a full-length chromosomally tagged Apg1-prA fusion protein were converted to spheroplasts and challenged with or without rapamycin for 15 min before detergent lysis. The lysates were precleared at 100,000 × g for 15 min to avoid aggregates and then separated by centrifugation through a 5–20% sucrose gradient (see MATERIALS AND METHODS). Apg1-prA from untreated cells sediments as a 33S particle. In contrast, in extracts from rapamycin-treated cells the protein was reproducibly shifted to a sedimentation profile with a peak at 25S, implying that a structural change had occurred (Figure 6).
Figure 6.
Structural changes occur in Apg1 upon induction of autophagy and depend on Cvt9 and the C terminus of Apg1. The Apg1-protein A pep4Δ (HAY437), Apg1Δ880-protein A pep4Δ (HAY478), and Apg1-protein A cvt9Δ pep4Δ (HAY591) strains were grown to midlog in YPD and converted to spheroplasts. Spheroplasts were treated with or without rapamycin (0.2 μg/ml) for 15 min before lysis. Lysates were precleared by differential centrifugation at 100,000 × g for 15 min and loaded onto a 5–20% sucrose gradient. The gradients were centrifuged at 259,000 × g for 7 h, fractions were precipitated with 10% TCA and Apg1-prA was identified by Western blotting with anti-prA antiserum and quantified using a Bio-Rad Fluor-S MAX. The migration of the two peaks observed in wild-type cells is denoted by the vertical dashed lines (calculated to be 25S and 33S for the slow and fast moving components, respectively).
Consistent with the desequestration data shown in Figure 4, the truncated fusion protein Apg1Δ880-prA sedimented at ∼25S in untreated extracts and did not show a shift in sedimentation profile upon rapamycin treatment (Figure 6), confirming that the C-terminal 18 amino acids are important for a Cvt pathway-specific interaction that is abrogated upon induction of autophagy. We also tested the hydrodynamic properties of Apg1-prA in cvt9Δ cells (Figure 6). Cvt9 is a protein required specifically in the Cvt pathway and has been shown to coprecipitate with Apg1, when both proteins are overexpressed (Kim et al., 2001). We found that the sedimentation profile of Apg1-prA was shifted to more slowly sedimenting fractions in cvt9Δ cells. Thus, Cvt9 seems to be involved in the structural changes that contribute to Apg1 function, and this protein appears to function upstream of Apg1. Because Cvt9 does not seem to be present as a stoichiometric member of the Apg1 complex, it is likely that it exerts this effect either catalytically or through a transient association.
Although pairwise interactions of Apg1 with Cvt9 (Kim et al., 2001) and of Apg1 with Apg13 (Kamada et al., 2000) have been reported when the proteins were overexpressed from multicopy plasmids, both we and others have been unable to identify stoichiometrically associating proteins that will purify with Apg1 in the absence of overexpression (Ho et al., 2002; unpublished data). Accordingly, we tested whether Apg1 self interacts. We found that the C-terminal, noncatalytic domain of Apg1 self-interacts in two-hybrid assays (unpublished data). However, because the full-length Apg1 construct auto-activates transcription, we were not able to further deduce the structure–function relationship of this self-interaction using the full-length protein. Because no other protein apart from Apg1 itself appears to be included stoichiometrically in the complex, we surmise that the changes in sedimentation behavior are either a result of conformational changes in the Apg1 molecule itself, a change in the number of molecules per complex, or both of these effects.
The Apg1Δ880 Protein Is Hypophosphorylated
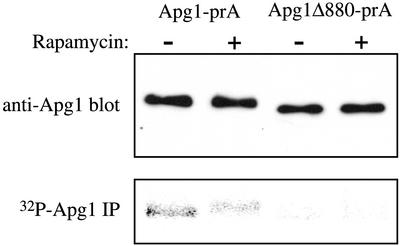
Apg1 has been shown to be a phosphoprotein in vivo and autophosphorylates in vitro. The phosphorylation level of Apg1 drops upon induction of autophagy, and this is reflected by a drop in autophosphorylation (Matsuura et al., 1997). One explanation for our data would be that autophosphorylation controls a structural change in Apg1 that is dependent on the C terminus. Thus, mutants lacking the C-terminal coiled-coil domain would be stuck in the “autophagic mode” and would be unable to mature prApe1 in the absence of starvation or rapamycin treatment. We therefore tested the phosphorylation state of Apg1Δ880 in the presence and absence of rapamycin. Although full-length Apg1-prA is phosphorylated and undergoes dephosphorylation in response to rapamycin (by ∼50%), as previously reported (Matsuura et al., 1997), the Apg1Δ880 mutant is strongly hypophosphorylated under all conditions (Figure 7). Because this mutant is also sedimenting as a 25S species in sucrose gradients (Figure 6), this is again consistent with autophosphorylation being important for the Cvt pathway-competent mode of Apg1 and not the autophagic mode. Although the region between amino acids 880 and 886 contains a serine residue, mutation to alanine did not affect prApe1 maturation (unpublished data), implying that the phenotypes of the apg1Δ880 mutant were not directly due to the lack of phosphorylation on this amino acid.
Figure 7.
The Apg1Δ880 mutant is hypophosphorylated. Cells expressing full-length prA-tagged Apg1 (HAY370) or the Δ880 truncation fused to prA (HAY455) were grown to midlog, and labeled with 150 μCi 32P for 1 h before treatment with 0.2 ng/ml rapamycin or mock solution. After 20 min exposure to rapamycin, protein extracts were prepared and Apg1 was immunoprecipitated as in MATERIALS AND METHODS. One half of the immunoprecipitate (IP) was separated by SDS-PAGE and exposed for phosphorimaging while the other half was separated, transferred to nitrocellulose, and immunoblotted for Apg1.
DISCUSSION
Apg1 Has a Catalytic Role in the Cvt Pathway and a Nonkinase Role in Autophagy
Autophagy is a catabolic membrane trafficking phenomenon that depends on signal transduction events. The Apg1 protein kinase is a central player in the induction of autophagy and is also essential for cytoplasm to vacuole targeting (Cvt), a related constitutive trafficking pathway. According to current models, induction of autophagy subverts the machinery of the Cvt pathway into the formation of autophagosomes. In this article we describe experimental evidence for molecular changes in Apg1 that occur upon induction of autophagy, confirming these hypotheses. We find that although the kinase activity of Apg1 is essential in the Cvt pathway, it is not necessary or significantly less so, in the induction of autophagosomes. On the other hand, sedimentation velocity studies show that induction of autophagosomes is accompanied by structural changes in Apg1 that correlate with dephosphorylation.
Protein kinases can be thought of as having different types of cellular functions. Classically, as exemplified by the glucagon receptor/cAMP-dependent phosphorylation cascade, kinases pass on a “signal” by changing their activity in response to an upstream stimulus and by phosphorylating different substrate proteins, in response to this signal. It is the substrates, then, that perform the regulated process, whereas the kinase serves as an intermediary that is capable of amplifying the signal. More recently, however, examples have arisen that paint a different picture. When the analog- sensitive approach was applied to the Cdc28 protein kinase in yeast, a cyclin-dependent kinase, it was found that kinase activity was required for the G2/M transition, in contrast to studies with temperature-sensitive alleles, which showed a block in G1 (Bishop et al., 2000). Similarly, analog-sensitive alleles of Cla4 did not show aberrations in actin polarization in response to kinase inhibition, whereas mutants depleted for Cla4 did show such aberrations (Weiss et al., 2000). It has been suggested that these discrepancies arise from kinase-independent functions of these proteins, most likely as structural scaffolds (Bishop et al., 2001).
Apg1 is a protein kinase that is essential for two related membrane trafficking phenomena, autophagy, and the Cvt pathway (Harding et al., 1996; Matsuura et al., 1997). To gain a better understanding of the role of this protein kinase, we created an analog-sensitive allele of APG1 that was specifically sensitive to 1-NA-PP1. Strikingly, 1-NA-PP1 inhibited Cvt pathway-dependent maturation of prApe1 but did not inhibit autophagy or autophagic trafficking of prApe1, at concentrations in excess of 20-fold of the apparent IC50 for Cvt pathway inhibition. At these concentrations we would predict a saturation of the ATP binding site of the kinase. Accordingly, we should observe inhibition of autophagic trafficking if kinase activity is essential during the autophagic response. To further test this point we constructed a kinase dead mutant form of Apg1, with the idea that it would mimic a “titration end point” for kinase inhibition. Although the kinase dead mutation possibly has other effects on the function of Apg1, it largely recapitulates the findings with the analog-sensitive inhibition when assayed for prApe1 maturation. This leads us to suggest that Apg1 has kinase-independent and -dependent functions. Apg1 kinase activity is required in the Cvt pathway but is not absolutely essential in the induction of autophagy.
The kinase dead Apg1K54A mutant was previously shown to have a reduced autophagic response as measured by induction of Pho8Δ60 alkaline phosphatase activity (Kamada et al., 2000), even though it was completely inactive in phosphorylation assays. We can rationalize the apparent discrepancy with our inhibition studies (which show no such effect; Figure 3) because the Apg1K54A kinase mutant is constitutively inactive, allowing for secondary indirect effects to have an impact on its ability to sustain autophagy. Thus, in our assay that depends on maturation of prApe1 (Figure 2C), we do not see this decrease in Apg1K54A activity because prApe1 trafficking is less sensitive to quantitative changes in the amplitude of the autophagic response, due to the redundant capacity of autophagosomes to transport prApe1 (Abeliovich et al., 2000). In addition, the analogous mutation in protein kinase A was shown to increase the Km by at least fivefold and would therefore have an effect on the amount of nucleotide bound by the protein. If the amount of bound ATP/ADP in itself affects Apg1 function (as opposed to kinase activity), then we can also explain the slight increase in autophagic trafficking that is observed in the inhibitor-sensitive mutant upon addition of saturating amounts of 1-NA-PP1 (which is a nucleotide analog).
Previous studies of Apg1 and its function did not reach a consensus regarding the role of the kinase activity. Matsuura et al. (1997) reported that Apg1 is autophosphorylated and that autophosphorylation is strongly inhibited by induction of autophagy. This contrasted with the report by Kamada et al. (2000), which showed that induction of autophagy entailed an increased affinity of Apg13 with Apg1 and is correlated with an increase in kinase activity measured in vitro toward an exogenous substrate, myelin basic protein. Thus, one report implied a decrease in kinase activity (toward an endogenous substrate, Apg1), whereas the other reported an increase in kinase activity (toward myelin basic protein) under autophagic conditions. Our data are more consistent with the earlier report, in that inhibition of kinase activity had no effect on autophagy but completely blocked the Cvt pathway. Because our results also demonstrated a change in the tertiary/quaternary structure of Apg1, it is possible that the access of the exogenous substrate (myelin basic protein) is limited in extracts from cells in rich medium and that access is increased in extracts from starved cells, much like the C-terminal prA tag on Apg1 was not fully accessible in extracts from vegetative cells. Interestingly, the starvation-dependent prApe1 maturation that bypasses the 1-NA-PP1 block of the Cvt pathway was dependent on Apg13. This implies that the bypass-dependent maturation reflects autophagic trafficking because it requires a factor specific for autophagy. In addition, it also indicates that at least part of the function of Apg13 in autophagy does not involve the kinase activity of Apg1.
Conformational Changes in Apg1 Are Mediated Through the C Terminus
A second set of data that we present here suggests that Apg1 undergoes conformational changes in response to starvation stimuli and that the C terminus of the protein is important in mediating these changes. Thus, it is likely that these structural changes are important in the induction of autophagy. To further characterize these changes, we tried to see if we could copurify Apg1-associated factors by IgG sepharose chromatography, but these efforts were unsuccessful, and a recent study of whole genome purification of prA-tagged proteins from yeast also reached a similar conclusion for Apg1 (Ho et al., 2002). It is possible then, that Apg1 interacts primarily with itself, as suggested by the autophosphorylation data and that the interactions with other proteins that have been observed with yeast two-hybrid systems and overexpressed proteins reflect transient or nonstoichiometric interactions. Indeed, two-hybrid analysis showed that the noncatalytic region of Apg1 self-interacts (unpublished data). However, the full-length protein autoactivates transcription when fused to a DNA binding domain, and this precludes simple structure–function correlations regarding the role of specific subregions in the C terminus.
The region of amino acids 880–886 in Apg1 is essential for the Cvt pathway and appears to be part of a potential coiled-coil domain that is required for self-association. The fact that the Apg1Δ880 truncation mutant sediments at 25S suggests that the shift between the Cvt pathway mode and the autophagic function of Apg1 involves a change in the degree of Apg1 self-assembly. The conversion to autophagy is accompanied by reduced autophosphorylation, which would account for the decrease in Apg1 phosphorylation upon induction of autophagy, as shown in Figure 7. The fact that the Apg1Δ880 mutant is hypophosphorylated strongly suggests that it is the self-assembly that determines the autophosphorylation activity, which in turn contributes to determining the functional state of the protein.
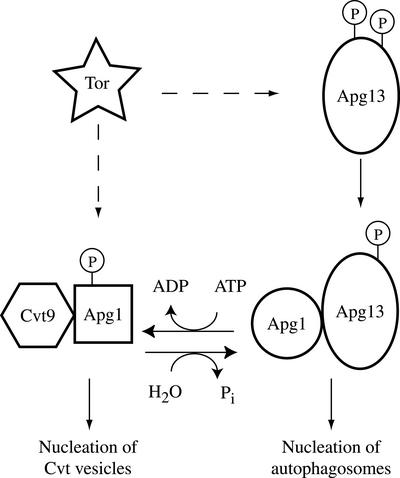
Our data, taken together with previous findings, are consistent with the following model: In response to starvation or stimulation with rapamycin, a signal transduction mechanism is induced that results in structural changes in Apg1 mediated through changes in the state of upstream factors such as Apg13. These changes then allow nucleation of autophagosomes, instead of nucleation of Cvt vesicles (Figure 8). How could these structural changes be linked to phosphorylation? We propose that the autophosphorylation activity may be required to “freeze” Apg1 in a Cvt pathway-competent state. In response to autophagic stimulation, the kinase activity is inhibited (or a phosphatase activity is stimulated) and Apg1 is dephosphorylated. This results in conversion to a second structural state that is compatible with autophagic trafficking, but not with Cvt trafficking. Cvt9 is a protein that is required specifically in the Cvt pathway, but not for autophagy, and we find that in cells lacking Cvt9, Apg1 does not undergo this structural conversion. These results, taken together, imply a role for Cvt9 upstream of Apg1 function, although Cvt9 does not seem to be required for the phosphorylation of Apg1 (unpublished data).
Figure 8.
Model for mechanistic coupling between kinase activity and nucleation of autophagosomes. Under normal growing conditions Tor kinase activity induces Apg1 kinase autophosphorylation, either through modulation of Apg13 phosphorylation or through a direct effect on Apg1. Inhibition of Tor results in a partial dephosphorylation of Apg13, and a decrease of Apg1 kinase autophosphorylation, leading to dephosphorylation and a structural change in Apg1. This structural change is correlated with an increased interaction with Apg13, and involves Cvt9. The dephosphorylated form of Apg1 directs autophagosome formation when associated with Apg13, whereas the phosphorylated form directs nucleation of Cvt vesicles.
Apg13 is absolutely required for autophagy, as defined by the Pho8Δ60 assay and by measuring the accumulation of autophagic bodies in the absence of vacuolar protease activity (Funakoshi et al., 1997, and our unpublished observations). In this study we show that Apg13 is not absolutely required for prApe1 trafficking through the Cvt pathway but is required for switching Apg1 from the kinase-dependent mode to the kinase-independent autophagic mode (Figure 5). One explanation for these results is that Apg13 is part of the toggle mechanism that switches the system between a (kinase-dependent) Cvt mode and a (kinase-independent) autophagic mode. In the absence of Apg13, the system cannot be shunted to the autophagic mode and therefore remains sensitive to the Apg1 kinase inhibitor.
A basic question regarding yeast autophagy relates to its relationship to the Cvt pathway. Several possibilities have been raised. One hypothesis suggests that the difference between the two pathways is only in the size of Cvt vesicles vs. the size of autophagosomes. This view is inconsistent with the existence of Cvt pathway-specific genes, which are not required for autophagy, and cvt mutants in which autophagosomes but not Cvt vesicles are formed. In addition, there are genes such as APG17 and SEC12 that are required for autophagy, but not for the Cvt pathway, and APG13, which in our hands is only marginally required for prApe1 maturation in SD medium. Thus, there are basic genetic differences between the pathways, and these differences relate to early events that precede the sequestration of prApe1 into Cvt vesicles and autophagosomes. Along these lines, some elements of the SNARE machinery are required only in the Cvt pathway, and not in autophagy (Abeliovich et al., 1999), implying some basic differences in the membrane trafficking requirements of the two pathways. Are these pathways active simultaneously? Our results show that a single protein involved in both pathways, Apg1, undergoes a structural change upon induction of autophagy. These data imply a qualitative difference between nucleation of autophagosomes and Cvt vesicles and supports a toggle mechanism, whereby machinery common to both pathways is mobilized by Apg-specific factors such as Apg13 and shunted to the formation of autophagosomes.
ACKNOWLEDGMENTS
We thank Drs. Scott Emr, Takahiro Shintani, and Lois Weisman for reagents and helpful discussion; Dr. Yoshinori Ohsumi for the TN124 strain and the APG1 knockout cassette; Drs. Robert Fuller and Fulvio Reggiori for helpful comments; and members of the Klionsky lab for stimulating discussions. This work was supported by National Institutes of Health Public Health Service grants GM53396 to D.J.K and AI44009 to K.M.S. and grant NSF MCB-9817002 to W.A.D.
Abbreviations used:
- 1-NA-PP1
4-amino-1-tert-butyl-3-(1′-naphthyl)pyrazolo[3,4-d]pyrimidine
- Ape1
aminopeptidase I
- Cvt
cytoplasm-to-vacuole targeting
- PP1
4-amino-1-tert-butyl-3-phenylpyrazolo[3,4-d]pyrimidine
- prA
protein A
- prApe1
precursor aminopeptidase I
- SD
synthetic minimal medium with dextrose
- SD-N
synthetic minimal medium with dextrose but lacking nitrogen
- YPD
1% bacto-yeast extract, 2% bacto-peptone, 2% dextrose
Footnotes
DOI: 10.1091/mbc.E02–07–0413
REFERENCES
- Abeliovich H, Darsow T, Emr SD. A t-SNARE/Sec1p complex composed of Tlg2p and Vps45p is required for cytoplasm to vacuole targeting of aminopeptidase I. EMBO J. 1999;18:6005–6016. doi: 10.1093/emboj/18.21.6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeliovich H, Dunn WA, Jr, Kim J, Klionsky DJ. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol. 2000;151:1025–1033. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeliovich H, Grote E, Novick P, Ferro-Novick S. Tlg2p, a yeast syntaxin homolog that resides on the Golgi and Endocytic structures. J Biol Chem. 1998;273:11719–11727. doi: 10.1074/jbc.273.19.11719. [DOI] [PubMed] [Google Scholar]
- Abeliovich H, Klionsky DJ. Autophagy in yeast: mechanistic insights and physiological function. Microbiol Mol Biol Rev. 2001;65:463–479. doi: 10.1128/MMBR.65.3.463-479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AC, Buzko O, Shokat KM. Magic bullets for protein kinases. Trends Cell Biol. 2001;11:167–172. doi: 10.1016/s0962-8924(01)01928-6. [DOI] [PubMed] [Google Scholar]
- Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Bishop AC, Kung C-y, Shah K, Witucki L, Shokat KM, Liu Y. Generation of monospecific nanomolar tyrosine kinase inhibitors via a chemical genetic approach. J Am Chem Soc. 1999;121:627–631. [Google Scholar]
- Dunn WA., Jr Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol. 1990a;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn WA., Jr Studies on the mechanism of autophagy: maturation of the autophagic vacuole. J Cell Biol. 1990b;110:1935–1945. doi: 10.1083/jcb.110.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast Saccharomyces cerevisiae. Gene. 1997;192:207–213. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- Gerhardt B, Kordas TJ, Thompson CM, Patel P, Vida T. The vesicle transport protein Vps33p is an ATP-binding protein that localizes to the cytosol in an energy-dependent manner. J Biol Chem. 1998;273:15818–15829. doi: 10.1074/jbc.273.25.15818. [DOI] [PubMed] [Google Scholar]
- Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J Biol Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Huang W-P, Scott SV, Kim J, Klionsky DJ. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Klionsky DJ. Autophagy, cytoplasm-to vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annu Rev Biochem. 2000;69:303–342. doi: 10.1146/annurev.biochem.69.1.303. [DOI] [PubMed] [Google Scholar]
- Kim J, et al. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Shah K, Yang F, Witucki L, Shokat KM. Engineering Src family protein kinases with unnatural nucleotide specificity. Chem Biol. 1998;5:91–101. doi: 10.1016/s1074-5521(98)90143-0. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Phillipsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- McEwen CR. Tables for estimating sedimentation through linear concentration gradients of sucrose solution. Anal Biochem. 1967;20:114–149. doi: 10.1016/0003-2697(67)90271-0. [DOI] [PubMed] [Google Scholar]
- Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- Pogulis RJ, Vallejo A, Pease LR. In vitro recombination and mutagenesis by overlap extension PCR. Methods Mol Biol. 1996;57:167–176. doi: 10.1385/0-89603-332-5:167. [DOI] [PubMed] [Google Scholar]
- Scott SV, Baba M, Ohsumi Y, Klionsky DJ. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, et al. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- Sikorsky RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Weiss EL, Bishop AC, Shokat KM, Drubin DG. Chemical genetic analysis of the budding-yeast p21-activated kinase Cla4p. Nat Cell Biol. 2000;2:677–685. doi: 10.1038/35036300. [DOI] [PubMed] [Google Scholar]