Abstract
In Saccharomyces cerevisiae, telomeric DNA is protected by a nonnucleosomal protein complex, tethered by the protein Rap1. Rif and Sir proteins, which interact with Rap1p, are thought to have further interactions with conventional nucleosomic chromatin to create a repressive structure that protects the chromosome end. We showed by microarray analysis that Rif1p association with the chromosome ends extends to subtelomeric regions many kilobases internal to the terminal telomeric repeats and correlates strongly with the previously determined genomic footprints of Rap1p and the Sir2-4 proteins in these regions. Although the end-protection function of telomeres is essential for genomic stability, telomeric DNA must also be copied by the conventional DNA replication machinery and replenished by telomerase, suggesting that transient remodeling of the telomeric chromatin might result in distinct protein complexes at different stages of the cell cycle. Using chromatin immunoprecipitation, we monitored the association of Rap1p, Rif1p, Rif2p, and the protein component of telomerase, Est2p, with telomeric DNA through the cell cycle. We provide evidence for dynamic remodeling of these components at telomeres.
INTRODUCTION
Telomeres are nonnucleosomal protein–DNA complexes that prevent uncontrolled fusion, degradation, recombination, and elongation of chromosome ends (Muller, 1938; McClintock, 1941; Wright et al., 1992; Sandell and Zakian, 1993; Hande et al., 1999; Smith and Blackburn, 1999; Hackett et al., 2001). In Saccharomyces cerevisiae, terminal telomeric DNA is composed of ∼350 base pairs of short, degenerate TG1–3 repeats. One telomeric strand is polymerized by telomerase (Cohn and Blackburn, 1995) and forms an S-phase–specific TG1–3 overhang (Wellinger et al., 1993, 1996). The conventional DNA polymerase machinery is thought to synthesize the complementary C1–3 A strand. Duplex telomeric DNA repeats are bound by the sequence-specific binding protein Rap1p, which recruits proteins Rif1p and Rif2p as well as Sir3p and Sir4p via its C-terminal domain (Moretti et al., 1994; Moazed and Johnson, 1996; Moretti and Shore, 2001).
Telomeric regions in yeast also contain subtelomeric X-elements, which have only moderate homology to each other and are present at all chromosome ends, and the highly homologous Y′ elements located distal to the X elements (Chan and Tye, 1983) on about half of all chromosome ends. Y′ elements fall into 5.2- and 6.7-kb size classes (Louis and Haber, 1990; Louis and Haber, 1992), encode a helicase (Yamada et al., 1998), and are bounded by short (<150-base pair) tracts of internal telomeric TG1–3 sequence DNA. Because Y′ elements often occur in tandem arrays of two to four repeats, these TG1–3 tracts are found internal to the chromosome ends at distances depending upon the size class and number of Y′ elements present.
The Rap1p, Ku, and Sir2-4 proteins are cross-linkable to DNA as far in as 3–15 kb from the chromosome end, consistent with their simultaneous binding to TG1–3 repeat DNA both at chromosome ends and adjacent to the Y′ repeats (Hecht et al., 1996; Strahl-Bolsinger et al., 1997; Martin et al., 1999; Lieb et al., 2001). Recent evidence that Sir3p may simultaneously associate via different subdomains with Rap1p, Sir4p, and histones H3 and H4 suggests that Rap1p may spread over this large region both through direct sequence-specific binding to the TG1–3 DNA repeat blocks and through protein–protein interactions with Sir3p or Sir4p, which are spread into the Y′ elements (Moretti et al., 1994; Cockell et al., 1995; Moretti and Shore, 2001). One model of telomeric chromatin posits that the Rap1p and Sir proteins bound to the terminal telomeric TG1–3 tracts “fold back” to interact with internal histones, creating a higher order protective complex at the chromosome end (Grunstein, 1997).
Like the Sir proteins, the Rif1 and Rif2 proteins are also tethered to telomeric DNA through interactions with the Rap1p C terminus and one another. However, although SIR3 or SIR4 deletion only shortens telomeres slightly (Palladino et al., 1993), deletion of the Rif proteins causes dramatic telomere lengthening. Hence, the Rif1 and Rif2 proteins negatively regulate telomere length (Hardy et al., 1992; Wotton and Shore, 1997).
It has been suggested that the Rif proteins interact with the most distal Rap1p molecules on the terminal telomeric TG1–3 tracts, whereas the Sir2-4 proteins interact with more internal Rap1p molecules (Wotton and Shore, 1997). Rif1- and Rif2-fusion proteins joined to a transcriptional transactivator were experimentally capable of associating with internal tracts of TG1–3 repeat DNA linked to a HIS3 reporter gene (Bourns et al., 1998), but it was not determined whether Rif proteins are present on the internal Y′ telomeric tracts of native telomeres. Herein, we report that Rif1p binding extends many kilobases in from the terminal telomere TG1–3 tract and closely correlates with the Rap1p and Sir protein binding in these regions.
Although protection of chromosome ends through repressive chromatin is an important function of telomeres, telomeric DNA must also be replicated and replenished by telomerase. In vivo polymerization by telomerase has been observed in the late S phase and G2/M phases of the cell cycle (Diede and Gottschling, 1999; Marcand et al., 2000), whereas replication of chromosome end regions from late-activating origins is thought to occur starting from mid/late S phase in the cell cycle (Raghuraman et al., 2001). Telomerase action in vivo minimally requires the telomerase components Est2p, TLC1 RNA, Est1p (Evans and Lundblad, 1999; Pennock et al., 2001) and Est3p (Hughes et al., 2000), as well as Cdc13p (Nugent et al., 1998), Stn1p (Grandin et al., 2001; Pennock et al., 2001), and DNA polymerases Polα and Pol∂ (Diede and Gottschling, 1999) and is promoted by the Mre11p-, Rad50p-, and Xrs2p-containing (MRX) complex (Lendvay et al., 1996; Nugent et al., 1998; Diede and Gottschling, 1999, 2001; Ritchie and Petes, 2000; Tsukamoto et al., 2001) and the Ku proteins (Peterson et al., 2001). Thus, it is probable that repressive telomeric chromatin needs to be transiently disrupted to allow telomere replication to occur. In the simplest model, the negative regulators of telomere length (i.e., Rap1p, Rif1p, and Rif2p) would be present at chromosome ends at times when the positive regulators, such as telomerase, were not. Accordingly, we investigated the ability of Rap1p, Rif1p, Rif2p, and Est2p to immunoprecipitate telomeric DNA through the cell cycle. We report that all four proteins are cross-linkable to telomeric DNA and that the association, as measured in this way, changes significantly through the cell cycle. Chromatin spread analyses also indicate that the distributions and associations of Rap1p, Rif1p, and Rif2p are cell cycle dependent. These data provide new evidence for the active remodeling of telomeric chromatin through the cell cycle, in a manner that may involve long-range interactions over many kilobases of the chromosomal end region.
MATERIALS AND METHODS
Supplementary Data
All of the supplementary data referred to in the text can be obtained at http://biochemistry.ucsf.edu/∼blackburn/MBOC1.
Strain Construction
Strain genotypes are listed in the supplementary data Table A. The S288C yeast strain BY4736 was obtained from American Type Culture Collection (Manassas, VA) (Brachmann et al., 1998). Isogenic derivatives of BY4736 were used for all chromatin immunoprecipitation and microarray experiments. Epitope-tagged and deletion strains were generated using homologous polymerase chain reaction (PCR) recombination (Longtine et al., 1998). All epitope-tagged genes were constructed to retain their endogenous promoters and to be present at their normal chromosomal locations. EST2 was 13xMYC epitope tagged using existing constructs (Longtine et al., 1998), whereas RIF1 and RIF2 were “-proA” epitope tagged with two Z-domains (Nilsson et al., 1987) from protein A (gift of Dennis Wykoff). Briefly, the Fc-binding Z-domain of protein A was duplicated in tandem and cloned into pFA6a in frame to yield a C-terminal–tagging vector. Transformants were screened by PCR and Western blot analyses. The expression of epitope-tagged genes was confirmed by Western blotting analyses after immunoprecipitation (IP). All strains showed the single expected band upon Western blotting except for Rif2-proA, which exhibited a doublet band after IP (our unpublished data).
Plasmids containing tlc1-476A mutation (Chan et al., 2001) were transformed into epitope-tagged BY4736 strains. All strains containing the tlc1-476A mutation used in this study were heteroallelic for the TLC1 locus and also contained a wild-type (WT) copy of the TLC1 gene. The W303-1a and RIF1–9xMYC strains used in this study were obtained from David Shore (Mishra and Shore, 1999). Mating, sporulation, and tetrad dissection were done according to standard methods (Fink, 1991).
Southern Blotting and Hybridization Conditions
Genomic DNAs for Southern blots were prepared as previously described (Chan et al., 2001). Blots were UV cross-linked with 12 mjoules (Stratagene, La Jolla, CA) and hybridized with a γ-32P-labeled telomeric (TG3TG)4 oligonucleotide at 55°C for at least 6 h. Blots were washed twice and exposed to either phosphoscreens (Molecular Dynamics, Sunnyvale, CA) or Biomax film (Eastman Kodak, Rochester, NY). Exposures were taken in the linear range and analyzed with ImageQuant software (Molecular Dynamics).
Chromatin Immunoprecipitation and Analysis
Chromatin immunoprecipitations (ChIPs) were performed essentially as described previously (Hecht et al., 1996; Strahl-Bolsinger et al., 1997; Lieb et al., 2001). For each time course, three or more independent experiments were performed with independent yeast cultures. Yeast cultures (1.7 liters) were grown to 0.3–0.4 A600 and arrested for 3.5 h with 1 μg/ml alpha factor (Biosynthesis, Lewisville, TX). Arrests were confirmed by light microscopy and cultures were then washed twice in an equal culture volume of fresh YPD and released into the cell cycle. Cells were then sampled at 20-min time points for 2 h and fixed immediately with 1% formaldehyde (Sigma-Aldrich, St. Louis, MO). Cell budding index analyses and fluorescence-activated cell sorting (FACS) analyses were performed for each time course to validate the α-factor arrest and subsequent cell cycle staging. Cell pellets from 200 ml of culture were resuspended in 1 ml of lysis buffer plus protease inhibitors (Roche Diagnostics, Indianapolis, IN) and lysed using a bead beater (Biospec Products, Bartlesville, OK) three times for 1 min at 4°C. Lysates were sonicated (Branson Ultrasonics, Danbury, CT) three times for 15 s (constant output, 1.5 duty cycle) to a mean DNA length of 300 base pairs-1 kb. Lengths of both bulk and telomeric DNA were confirmed by agarose electrophoresis, ethidium staining, and Southern blot analysis. Clarified lysates were immunoprecipitated at 4°C with 50–75 μl of IgG-Sepharose (Pharmacia, Peapack, NJ) for -proA–tagged components, ∼30 μg of anti-MYC 9E10 (Covace, Princeton, NJ) with 50–75 μl of protein A-Sepharose (Sigma-Aldrich), or 1:25 dilution of α-RAP1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) with 50–75 μl of protein G-Sepharose (Sigma-Aldrich). Rap1p IPs were also done using a 1:150 dilution of a rabbit polyclonal antibody to Rap1p (Enomoto et al., 1997) (generous gift of Judith Berman, University of Minnesota, St. Paul, MN) with 50–75 μl of protein A-Sepharose. Typically, 90% of a given cell lysate was used for ChIPs, whereas 10% was set aside and used for the designated “input” samples. IPs were washed as described previously (Strahl-Bolsinger et al., 1997) and decross-linked at 65°C for at least 6 h. Decross-linked DNA was QiaQuick (QIAGEN, Valencia, CA) purified and eluted into 100 μl of buffer.
ChIP samples and their matched input dilutions were denatured in 1.5 M NaCl, 0.5 N NaOH for 15 min at room temperature and applied to a MiniFold-I dot blotter (Schleicher & Schuell, Dassel, Germany). Typically, 75% of ChIP elutions were loaded per time point, whereas 2.5–10% of input samples were loaded. Wells were rinsed alternately with 2 volumes of 3× SSC and denaturing solution. Blots were cross-linked, hybridized, washed, and exposed as described (see above).
For analyses, we wanted to calculate the percentage of telomeric DNA precipitated from the total telomeric DNA in a cell lysate. The “raw” amount of telomeric DNA precipitated for each protein and time point was determined by integrating the radioactive hybridization signals from dot blots with ImageQuant. The raw ChIP signals determined for each protein and time point were compared with the raw signal of matched, serially diluted input samples. Raw ChIP and input signals were then divided by the percentage of lysate used to obtain them (i.e., typically, 90% for ChIPs, 10% for inputs) and the percentage of the elution that was applied to the dot blot (i.e., 75% for ChIPs, 2.5–10% for inputs). This generated each “raw percentage of input” value. For example, raw percentage of input = (raw ChIP signal/(90% × 75%))/(raw input signal/(10% × 2.5%)). Over a given time course, the raw percentage of input values for each of the time points were normalized to the average value for all of the time points in the time course to give unbiased “fold change” information over the cell cycle. Est2p and Rif2p time course data was also normalized to the raw percentage of input values from mock ChIPs (i.e., without antibody) or from untagged control strains in data not shown. The statistical significance of differences between time points within a time course was determined using Student's t tests.
PCR Conditions
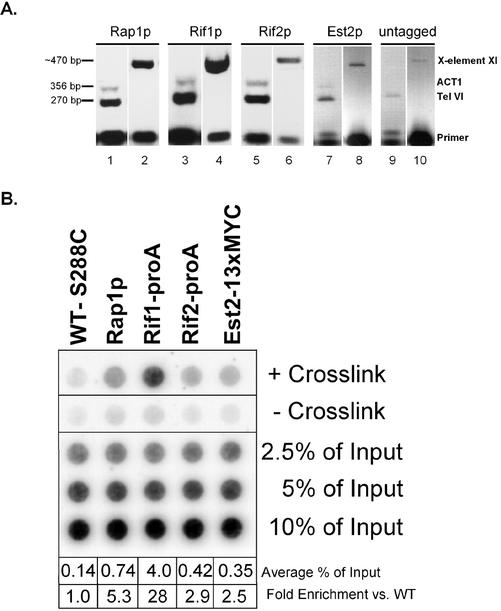
Eluted DNA from asynchronous chromatin immunoprecipitations (see above) was diluted and used as the template for PCR analyses. Three separate loci were assayed, two telomeric and one negative control locus: a sequence located ∼600 base pairs in from the end of the right telomere on chromosome VI (TEL-VI) (Strahl-Bolsinger et al., 1997) (5′-CAGGCAGTCCTTTCTATTTC, 5′-GCTTCTTAACTCTCCGACAG), the X-core element of chromosome XI located ∼200 base pairs in from the telomeric TG1–3 tract on chromosome XI (Fourel et al., 1999) (5′-TCCTGGATCCTTTGTTAACG, 5′-TCCTAGATCTACACCCACTACTCTAACCC), and the actin gene locus (ACT1) (Strahl-Bolsinger et al., 1997) (5′-CCAATTGCTCGAGAGATTTC, 5′-CATGATACCTTGGTGTCTTG) as a negative control. The TEL-VI and ACT products were generally amplified together in multiplex PCR reactions. The following conditions were generally used for PCR: 95°C 2 min following by 95°C for 15 s, 53–5°C for 1 min, followed by 72°C for 1 min for 25–30 cycles. PCR reactions were then loaded onto 2.5–3% agarose gels, resolved by electroporesis, ethidium stained, and imaged on a gel documentation system. The charge-coupled device (CCD) image color was inversed and recorded in the linear range of the CCD. Computer scans of thermal printouts were used for Figure 2A.
Figure 2.
Chromatin immunoprecipitation controls. (A) PCR of ChIP DNA is enriched for telomeric loci compared with the nontelomeric, nonspecific gene ACT1. Varying dilutions of ChIP DNA were PCR amplified with primers specific for the subtelomeric X-element core of chromosome XI L (lanes 2, 4, 6, 8, and 10) or for the subtelomere of the right arm of chromosome VI (TEL VI) and the Actin gene (ACT1) (lanes 1, 3, 5, 7, and 9). TEL VI and ACT1 were amplified in multiplex PCR reactions. The size of the PCR generated DNA fragments is shown at the left. (B) Chromatin immunoprecipitation of telomeric proteins is dependent upon the presence of cross-linking agent. A representative dot blot of ChIP samples is shown. Asynchronous yeast cultures were chromatin immunoprecipitated both in the presence (row 1) and absence (row 2) of 1% formaldehyde for 15 min at room temperature. The average percentage of total telomeric DNA immunoprecipitated is shown at the bottom as well as the fold-enrichment over control strains that were either mock immunoprecipitated without primary antibody, or contained no epitope tag. The numbers shown for the averages represent the amount of telomeric DNA immunoprecipitated over the entire timecourse for all replicates and do not reflect the representative blot shown above.
Microarray Production and Analysis
Microarrays were prepared as described previously (Gerton et al., 2000; Iyer et al., 2001; Lieb et al., 2001). Protocols for microarray preparation, hybridizations, ChIP amplification, and fluorescent dye coupling were those described and available from http://www.microarrays.org. Briefly, microarrays containing fragments homologous to all yeast open reading frames (ORFs) and intergenic regions were hybridized to ChIP samples from three independent, asynchronous Rif1-proA cultures. A common reference sample of amplified BY4736 genomic DNA was used as a control hybridization for all experiments. All genomic features (i.e., ORFs and intergenic fragments) were subjected to median rank analysis (Iyer et al., 2001; Lieb et al., 2001). Genomic features whose median percentile rank was in the top 10, 8, 5, and 3% of the immunoprecipitated fragments were compared with existing RAP1 and SIR data (Lieb et al., 2001). Features ranking in the top 5% or better were considered good Rif1p telomeric targets because they had a high correlation to Rap1p and the Sir2-4 proteins at telomeres. Top-ranking features were compared with the entire yeast genome by using BLAST to assess the extent of potential cross-hybridization. Fragments with >70% identity were considered “redundant” for analysis purposes, whereas those with <70% identity were considered “unique.” The distance of DNA association from the chromosome ends for Rif1-proA was expressed as the centromeric coordinate of all the top-ranking positive features that were located within 15 kb of the chromosome end. The innermost distance from the end measurement (IDE) was determined for each end and compared with the lengths determined for Rap1p and the Sir2-4 proteins by using the same analysis method (Lieb et al., 2001). The statistical significance of differences between IDE measurements was determined using t tests. DNA association maps were plotted using Promoter version 3.25. The supplemental data for the top-ranking RIF1 targets and their IDE measurements for individual chromosome ends is available at http://biochemistry.ucsf.edu/∼blackburn/MBOC1.
Chromatin Spreads
Duplicate yeast cultures were arrested in α-factor and released as described above. Time points were kept on ice for the duration of the time course and prepared simultaneously. Chromosome spreads were prepared as described previously (Loidl et al., 1991; Michaelis et al., 1997; Biggins et al., 2001). Samples were blocked with phosphate-buffered saline (PBS) + 1% bovine serum albumin and incubated overnight with precleared primary antibodies at room temperature. The mouse α-MYC 9E10 (Covance, Princeton, NJ) and rabbit α-RAP1 (Enomoto et al., 1997) primary antibodies were used at 1:1000 dilutions. Samples were washed twice for 5 min in PBS and overlaid with 1:1000 goat α-mouse-fluorescein isothiocyanate (FITC) or mouse α-rabbit-Cy3 conjugated secondary antibodies (Jackson Immunoresearch Laboratories, West Grove, PA) for 1 h at room temperature. Samples were washed twice in PBS, stained with 1 μg/ml DAPI for 5 min, and mounted with 90% glycerol, 1 mg/ml phenylenediamine pH 9 (Sigma-Aldrich).
Slides were visualized on an E600 microscope (Nikon, Tokyo, Japan) at 100× magnification and at least 10 fields were captured using a Coolsnap FX CCD (Roper Scientific, Tucson, AZ) for each time point. Approximately 75 DAPI-staining, spread nuclei were counted per time point. Fields were psuedo-colored blue for DAPI, red for Cy3, and green for FITC in Adobe Photoshop. The total numbers of discernible Cy3- and FITC-staining foci per spread nucleus were counted for both time courses. The numbers of colocalized Cy3- and FITC-staining foci per spread nucleus were also determined. For time-course analyses, data were processed similarly to ChIP data (see above).
RESULTS
Characterization of Epitope-tagged Strains and Chromatin Immunoprecipitation Controls
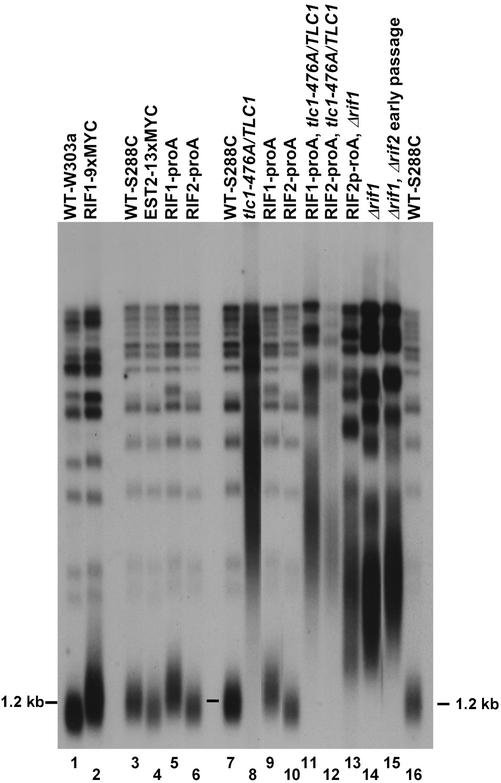
We epitope-tagged the RIF1, RIF2, and EST2 genes in strains by using PCR-based recombination. Each tagged gene was expressed from its normal chromosomal location under the control of its endogenous promoter. Only epitope-tagged strains with telomere lengths and distributions that were stably wild type or near wild type over repeated passaging (indicative of normal functioning of the tagged protein) were used for further studies. These included the Rif1-proA, Rif1–9xMYC, Rif2-proA, and Est2–13xMYC strains (Figure 1, lanes 1–6). After α-factor arrest and release, the cell cycle progression of each epitope-tagged strain was monitored by budding indices and FACS analyses and compared with WT. In all cases, the WT and tagged strains progressed through the cell cycle with similar budding and FACS profile kinetics after release from α-factor arrest (our unpublished data). These data suggested that these epitope-tagged strains had WT-like telomeric chromatin complexes.
Figure 1.
Characterization of telomeric length in strains. Telomere length phenotypes of epitope-tagged and tlc1-476A/TLC1 heteroallelic strains. Genomic DNAs were purified from haploid strains and heteroallelic tlc1-476A/TLC1 strains. Epitope-tagged strains do not show significant telomere lengthening or shortening (lanes 1–6). Strains containing tlc1-476A/TLC1 show elongated, deregulated telomeres. WT strains of both the W303a and S288C strain backgrounds are shown (lanes 1, 3, 7, and 16). RIF1 and RIF1, RIF2 deletion strains are also shown for reference (lanes 14 and 15). The Δrif1, Δrif2 strain used in this study (lane 15) was not extensively passaged and did not have fully elongated telomeres. The majority of Y′ telomeres migrate with the 1.2 kb marker shown.
We experimentally validated that the IP of telomeric chromatin from our yeast strains was telomere specific and dependent upon both the presence of the appropriate epitope and the treatment with cross-linking agent, by multiple controls. For background controls of Rap1p and Est2–13xMYC samples, the strains were “mock immunoprecipitated” without a primary antibody, but with protein A- or G-Sepharose beads. Control lysates for Rif1-proA and Rif2-proA IPs were made from the isogenic untagged (i.e., wild-type) cultures immunoprecipitated in the presence of IgG-Sepharose. We determined the specificity of our IP for telomeric loci compared with the nonspecific locus ACT1, by PCR (Figure 2A). Two separate telomeric loci were tested, the right subtelomere of chromosome VI (TEL-VI) and the X-core element of the left arm of chromosome XI (X-core). We tested the ability of Rap1p, Rif1p, Rif2p, Est2p, and untagged control strains to IP these three loci by PCR analyses. The TEL-VI region ∼600 base pairs in from the end of the chromosome VI right telomere was compared directly to the amount of ACT1 chromatin PCR amplified after IP. All of the epitope strains specifically immunoprecipitated significant amounts of the telomeric locus compared with both the untagged control strain and with the internal negative control loci, ACT1 (Figure 2A). Although ACT1 was undetectable in untagged strains (Figure 2A), there was an insignificant background at this locus in all of the tagged strains. Notably, although Rap1p and Rif1p IPs had very strong signals at both telomeric loci, Est2p and Rif2p had a weaker enrichment for these regions. These results reflect the trends in the relative telomeric association of these proteins in the dot blot assays described below (Figure 2B), and further suggest that these proteins bind with predominant specificity to telomeric DNA.
Determination of Genome-Wide RIF1 Targets by Microarray Analysis
To assess the association of the Rif1 protein with subtelomeric regions, we investigated association of Rif1-Pro1 with chromatin by using microarrays that contained all S. cerevisiae ORFs and intergenic regions (Gerton et al., 2000; Iyer et al., 2001; Lieb et al., 2001). We determined which telomeres were bound by Rif1-proA, how far in from the chromosome ends it associated, and whether Rif1-proA has nontelomeric binding sites. We used median rank analysis to determine which genome features or targets (i.e., ORFs and intergenic regions) were consistently enriched in at least two of three independent array experiments by using DNA precipitated from asynchronous Rif1-proA cultures in mid-log phase (Iyer et al., 2001; Lieb et al., 2001). Briefly, in this ranking method, those DNA targets are sorted in order of their red to green ratios (i.e., in order of decreasing binding). Features that are consistently enriched by the ChIP procedure show a higher median percentile rank. In previous experiments examining the genome-wide DNA association of chromatin factors, two general trends in the data have been observed: proteins that bind DNA directly seem to have a bimodal distribution of targets, with the highest-ranking targets forming a small peak at the edge of the main, normal distribution. An example of this is seen with Rap1p, which shows that the top 8% of the distribution is enriched for Rap1p binding (Lieb et al., 2001). In contrast, for factors that do not bind DNA directly, the distribution of features is seen as a roughly normal, rather than a clear bimodal, distribution. In these cases, it is not possible to unambiguously assign a threshold above which enriched features are deemed “significant binding targets.” In some cases, as with the G1/S-phase transcription factor MBF (MBP/Swi6 heterodimer) (Iyer et al., 2001), it is possible to correlate enriched ChIP features with mRNA expression.
Consistent with the previously reported results of Lieb et al. (2001), we observed the expected bimodal distribution for Rap1p. For Rif1-proA, its cross-linkability to genomic DNA formed a more normal distribution, which may be a consequence of the fact that its known association with DNA is indirect, occurring through Rap1p (Hardy et al., 1992). (We attempted similar experiments with Rif21-proA, but the signal-to-noise ratios obtained in the Rif2-proA coimmunoprecipitations were too low for reliable analysis, so results are presented herein only for Rif1-proA.) We analyzed the top 10, 8, 5, and 3% of enriched Rif1-proA ChIP targets and empirically determined that the top 5% of features from the distribution were consistently enriched for telomeric targets. This is consistent with the selective enrichment for telomeric DNA seen in Figure 2A.
Interestingly, Rif1-proA binding sites in telomeric regions correlated well with regions we found were bound with Rap1p, and with regions reported previously to be bound by Rap1p, Sir2p, Sir3p, and Sir4p (Lieb et al., 2001). Average values and values for two telomeres are shown in Figure 3 and the complete data set in supplementary data D and B. A total of 325 genomic features ranked in the top 5% of Rif1-proA immunoprecipitated fragments in at least two of three experiments (supplemental data C and E). A total of 84 of these, all located within 15 kb of the chromosome ends, were generally highly enriched in the Rif1-proA ChIPs. Thus, although the last 15 kb of all 32 telomeric regions represents only 4% of the total genomic sequence, 26% of the enriched genomic features were in these regions. We next compared the 325 Rif1-proA targets with the top-ranking 8% of Rap1p targets (Lieb et al., 2001). Rap1p and Rif1-proA shared only 85 targets. Strikingly, 75 of these, or 88%, were within 15 kb of the chromosome ends. This result suggests that Rap1p and Rif1-proA are highly correlated specifically at chromosome ends. Of the remaining 10 nontelomeric sites in common for these proteins, four were at the HMLa, HMRα, MATa, and MATα loci. In summary, although Rap1p and Rif1-proA are highly correlated at chromosome ends, they do not seem to have generally similar internal genomic targets other than HMR. This is consistent with previous observations that Rif1p, unlike Rap1p, is nonessential, and apparently is not required for any critical transcriptional activation functions mediated by Rap1p (Hardy et al., 1992).
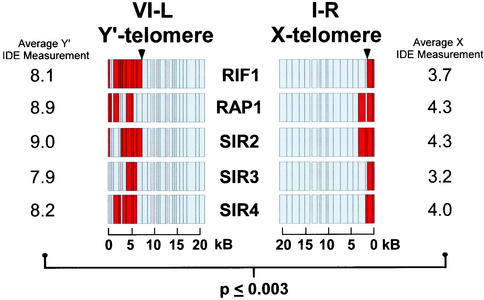
Figure 3.
RIF1, RAP1, and SIRs exhibit a broader region of association to Y′ telomeres than X telomeres. Rif1-proA IDE measurements. Multiple intergenic microarrays were hybridized with Rif1-proA ChIP samples from asynchronous cultures. The IDE was measured for targets in the top 5% of immunoprecipitated Rif1-proA fragments. RAP1, SIR2–4 data are reproduced from Lieb et al. (2001) and shown for comparison purposes. Physical maps of representative chromosome ends are show for X-element (I-R) and Y′ element (VI-L) telomeres for RIF1, RAP1, and the SIRs. Red indicates association detected on the microarray, whereas light gray indicates regions where no association was detected. Scale bars are shown at bottom. The Rif1-proA, Rap1p, and Sir2-4 protein average IDE measurements in kilobases are shown for chromosomes with X-elements or Y′ elements. A representative innermost associated DNA fragment is indicated by black arrowhead. A statistically significant length difference for Rif1-proA IDE measurements of X and Y′ element chromosomes is indicated. Rap1p and the Sir2-4 proteins were similarly significant.
The extent of Rif1-proA association to individual chromosome end regions was analyzed similarly to Rap1p and the Sir2-4 proteins described previously (Lieb et al., 2001). We chose 15 kb as the furthest distance from the end where we would begin to consider positive RIF1 targets, because this was the interval where Rap1p and the Sir2-4 proteins were the most colocalized (Lieb et al., 2001). We then took the centromere-promixal coordinate for those Rif1-proA targets and defined this value as the IDE, to which Rif1-proA bound. The IDE measurement was repeated for all 32 chromosome ends (supplemental data B). Using data from all detected chromosome ends, we determined that the IDE measurement for Rif1-proA averaged 6.4 kb in from the ends, with a minimum value of 0.47 kb and a maximum of 13 kb. It is important to note that the IDE-measurement does not imply that Rif1-proA is continuously associated from this point to the chromosome end. However, like Rap1p and the Sir2-4 proteins, Rif1-proA was generally found to be associated with a number of targets at each chromosome end (Figure 3).
We asked whether IDEs differed between chromosome ends that do or do not contain Y′ elements. Using the information gathered from comparing the identity of chromosome ends to one another and existing annotations (http://www.le.ac.uk/genetics/ejl12/EndsData.html), we separated telomeres into X-element (i.e., no Y′ elements) and Y′ element classes. We then compared the IDE measurements for Rif1-proA, Rap1p, and the Sir2-4 proteins for these two classes of chromosome ends (Figure 3). Strikingly, there was a highly significant (p ≤ 0.003) length difference based on the type of end. For X-element ends, the average Rif1-proA IDE-measurement was 3.7 kb by using data from all ends, 3.6 kb by using unique target data (see below), and 3.3 kb by using only the data from redundantly detected targets (see below) (Figure 3; supplemental data C). For telomeres with Y′ elements, the Rif1-proA IDE-measurement was 8.1 kb by using all end data, 7.5 kb for unique target data, and 8.9 kb for redundantly detected targets (Figure 3; supplemental data). There was not a significant difference in the IDE-measurements for chromosomes with Y′-long (6.7 kb) vs. Y′-short (5.2 kb) elements (our unpublished data). The difference between IDE measurements between X-element and Y′ element chromosome ends for Rap1p and the Sir2-4 proteins was highly correlated with Rif1-proA and (r > 0.98) and similarly significant (p ≤ 0.003). Thus, on average, telomeric proteins associate twice as far in from the chromosome ends on telomeres with Y′-elements than on those with only X-elements. Y′ elements are bounded at both ends by short, ∼150 base pairs, tracts of internal telomeric repeats. These internal tracts are shorter than the terminal telomeric tracts, but because they flank each of the 5.2- and 6.7-kb Y′ elements, they can be many kilobases in from the chromosome ends. These data suggest that the internal TG1–3 tracts that flank Y′ elements are capable of recruiting telomeric components and may contribute to overall telomere chromatin structure.
Many chromosome ends in S. cerevisiae are highly homologous to one another; specifically, 17 of the 32 chromosome ends contain the repetitive, highly homologous Y′ elements. This redundancy of subtelomeric DNA may lead to overestimation in the microarray data of the extent of Rif1-proA binding from the chromosome end. To address this issue we examined the Rif1-proA IDE measurements by using only targets with a homology to the rest of the genome of <70%. Using this unique target data set of 15/84 top-ranking features within the last 15 kb, we determined that the IDE for Rif1-proA was 5.3 kb (supplemental data E). Conversely, when we used only redundant targets, whose homology was greater than 70%, to estimate the Rif1-proA IDE, the result was 6.6 kb (supplemental data B). This range of Rif1-proA values did not differ significantly from those of Rap1p or the Sir2, Sir3, and Sir4 proteins, which were 6.8, 7.0, 6.3, and 6.6 kb, respectively (supplemental data B). These analyses indicate that for each chromosome end, the IDE to which Rif1-proA and the Sir2-4 proteins can associate is similar. Thus, the Rif1 and Sir2-4 proteins may not be strictly partitioned between the outer region of the telomere and the subtelomere as previously suggested (Wotton and Shore 1997), but may instead simultaneously occupy these regions.
Association of Rap1p with Telomeric Chromatin in the Cell Cycle
To examine the ability of telomeric components to IP telomeric DNA through the cell cycle, we performed time-course experiments. All IP time courses were repeated at least three times. Wild-type S288C strains were synchronized by α-factor arrest, released, and allowed to proceed through a complete cell cycle into the subsequent G1 phase. Lysates were made at each time point. Half of each lysate was immunoprecipitated with α-Rap1p antibodies, whereas the other half was mock immunoprecipitated without primary antibodies as described above, as a background control. The cell cycle stages after release from arrest were validated by FACS analysis and by budding indices. FACS analysis and budding indices were consistent with these cells being in G1 phase at the 20-min time point. However, because the nuclear morphology has been reported to be altered after α-factor arrest, for the analyses of the telomeric structural proteins Rap1p, Rif1p and Rif2p, we did not use the 20-min time points after release from arrest.
The average amount of telomeric DNA immunoprecipitated from mock IPs and untagged controls was comparable (0.14% of input; Figure 2B). For statistical analyses, the direct values obtained using both types of background controls were averaged. On average, telomeric DNA immunoprecipitated from wild-type strains by using α-Rap1p antibodies was enriched approximately fivefold over mock IPs (0.74% of input; Figure 2B), whereas Rif1-proA IPs enriched telomeric DNA ∼28-fold over IPs from untagged control strains (4% of input; Figure 2B). Rif2-proA IPs enriched telomeric DNA approximately threefold over IPs from untagged control strains (0.42% of input; Figure 2B), and Est2–13xMYC IPs enriched telomeric DNA ∼2.5-fold over mock IPs from wild-type strains (0.35% of input; Figure 2B). The detection of telomeric DNA in ChIPs assays was dependent on cross-linking agent in all cases, although in the Rap1p and Rif1p samples a detectable level of telomeric DNA was immunoprecipitated in its absence (Figure 2B). Because both Rif1p and Rif2p were -proA tagged, the amount of immunoprecipitated telomeric signal could be directly compared between the two proteins: in our assays Rif1-proA was, on average, 10-fold more cross-linkable to telomeric DNA than was Rif2-proA. Such direct comparison was not possible for the other proteins, because different antibodies were used to IP each component.
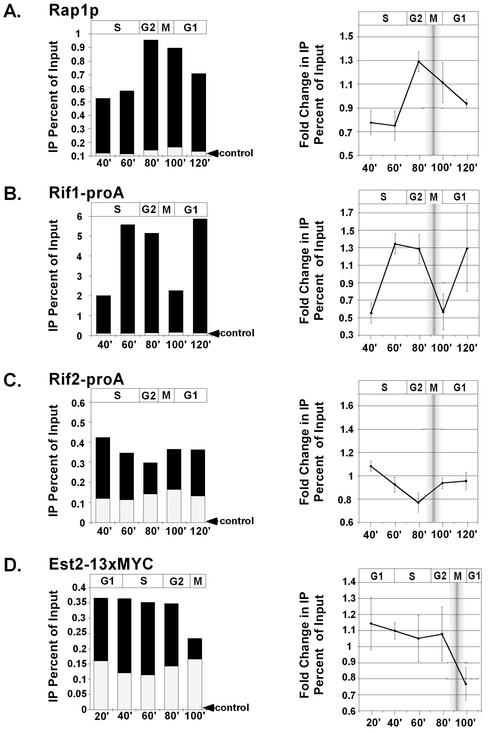
First, the cross-linkability of Rap1p to telomeric DNA was monitored throughout the cell cycle. Data are shown as both the raw immunoprecipitated telomeric signals (Figure 4A, left, black bars) and signals normalized to the average of all time points (Figure 4A, right). In Figure 4, left, the total height of each histogram bar represents the total amount of immunoprecipitated telomeric signal. The level of background signal from control mock IPs is shown on the same scale, overlaid as the white bar. The amount of telomeric DNA immunoprecipitated by Rap1p changed significantly over the cell cycle, whereas control mock IPs did not. The changes in Rap1p signal were apparent both as the raw percentage of input DNA (Figure 4A, left, black bars) and when individual time-point signals were normalized to the average immunoprecipitated signal to determine the fold change through the cell cycle (Figure 4A, right).
Figure 4.
Telomeric proteins change their cross-linkability to telomeric DNA during the cell cycle. α-Factor synchronized cultures were released into the cell cycle and 1% formaldehyde fixed at 20-min intervals. Rap1p (A), Rif1-proA (B), Rif2-proA (C), and Est2–13xMYC (D) samples were subjected to ChIP and associated DNA was probed on dot blots with a telomeric oligo. Raw telomeric signal as a percentage of total input DNA is shown in left-hand panel (black bars), with control mock IPs or untagged controls superimposed (white bars, indicated by black arrowheads). The approximate stage of the cell cycle, as determined by budding indices, is shown above graphs. Right-hand panels show fold change in telomeric signals that have been normalized to the average signal for all time points in the time course. SD of the means are shown. Shaded bar represents the approximate range of time of mitosis for the cell population. In the Est2p-13xMYC experiments, the 120-min time point samples did not yield reproducible values, possibly because of loss of synchrony by this time, so data are shown up to 100 min.
Cross-linkability of Rap1p to telomeric DNA was minimal 40–60 min after α-factor release, corresponding to early and mid S phase. Cross-linkability then increased rapidly from 60–80 min and then decreased during mitosis and through G1 of the next cell cycle. Both the timing and extents of decrease in Rap1p cross-linkability to telomeric DNA are consistent with previous microscopic studies that indicated that half the Rap1p is displaced from telomeres as cells pass through mitosis (Laroche et al., 2000).
RIF1 Association with Telomeric Chromatin in the Cell Cycle
The ability of Rif1-proA to IP telomeric DNA was examined throughout the cell cycle (Figure 4B). Rif1-proA–associated telomeric signal was minimal at 40 and 100 min after α-factor release, corresponding to the beginnings of S phase and G1, respectively, and was maximal in mid/late S phase and G2 in the cell cycle (60 and 80 min time points). This increase in the chromatin association of Rif1-proA in late S phase and G2 is consistent with the increase in RIF1 gene transcription observed during S phase (Cho et al., 1998). Notably, 60 min after α-factor release, Rif1-proA became relatively more cross-linkable to telomeric DNA than Rap1p (Figure 4, A and B, right). This may reflect a tighter association of Rif1-proA with the remaining telomere-bound fraction of Rap1p at this time point, possibly occurring through a binding partner other than Rap1p, or higher accessibility of Rif1-proA on telomeres than Rap1p. Like Rap1p, Rif1-proA was displaced at some point between 80 and 100 min, which respectively correspond to entry into mitosis and entry into the next cell cycle.
RIF2 Association with Telomeric Chromatin in the Cell Cycle
The amount of telomeric DNA immunoprecipitated by Rif2-proA, relative to untagged controls, changed significantly through the cell cycle (Figure 4C). Notably, the trend over the time course was distinct from those of both Rap1p and Rif1-proA. The amount of telomeric DNA immunoprecipitated by Rif2-proA progressively decreased from 40 to 80 min after α-factor release, corresponding to progression from S to G2. Although the absolute enrichment of Rif2-proA–cross-linked telomeric DNA over untagged controls was less than that of Rap1p or Rif1-proA, this decrease through the cell cycle was reproducible and statistically significant. Rif2-proA slightly increased its association with telomeric DNA through mitosis and into the next cell cycle. Similar changes were observed whether the numbers were analyzed as raw signals (Figure 4C, left) or as signals normalized to the average signal for the time course (Figure 4C, right). When raw Rif2-proA telomeric signal was further normalized to the signal of untagged controls, a similar decreasing trend through the cell cycle was seen; however, this decrease began 40 min after α-factor release instead of immediately (supplementary data G). We conclude that, regardless of the signal correction or analysis method used, there is a robust decrease in Rif2-proA telomere cross-linkability through S and the G2 phases of the cell cycle. This is inverse to the general trend of increased telomeric cross-linkability for Rif1-proA and Rap1p through S phase and G2 (compare Figure 4, A with B, to C). Similarly, through mitosis and the subsequent G1 phase, the amount of cross-linkable telomeric DNA seemed to increase for Rif2-proA, whereas it decreased for Rif1-proA and Rap1p.
EST2 Association with Telomeric Chromatin in the Cell Cycle
The known time of telomere elongation by telomerase is in late S phase through G2 (Diede and Gottschling, 1999; Marcand et al., 2000). Recent studies of replication timing have shown that origins within 35 kb of the telomere begin replicating in early/mid S phase, and replication of the telomeric regions is largely complete by mid S phase. Strikingly, however, our data indicate that Est2–13xMYC is associated with telomeric DNA significantly before either telomere replication occurs or telomerase acts. Cross-linkability of Est2–13xMYC to telomeric DNA changed during the cell cycle (Figure 4D). There was a statistically significant decrease (p < 0.03; Student's t test) in immunoprecipitated telomeric signal at 100 min after α-factor release, corresponding to the end of mitosis (Figure 4D, right) vs. all of the earlier time points. Together, these data suggest that Est2–13xMYC may remain associated with the telomere throughout much of the cell cycle and became displaced or inaccessible to ChIP as cells pass through mitosis.
Association of Rif1p and Rif2p Is Relatively Increased on Partially Uncapped Telomeres
To gain more insight into the regulation of the chromatin association of Rap1p, Rif1-proA, and Rif2-proA, we investigated two different situations in which cells are healthy but the ability to strictly regulate telomere length homeostasis is compromised. First, we examined heteroallelic tlc1-476A/TLC1 strains. The tlc1-476A allele contains a C-to-A transversion in the template region that is copied into the core binding site for Rap1p, disrupting this binding site (Chan et al., 2001). This mutation preserves both in vitro and in vivo telomerase activity, and in the homozygous state leads to elongated, deregulated, extensively degraded telomeres containing aberrantly long (TG)n tracts of up to 60 base pairs, and to significant defects in growth and chromosome segregation (Chan et al., 2001) (Smith and Blackburn, unpublished data). On the other hand, heteroallelic tlc1-476A/TLC1 strains have normal chromosome segregation and no detectable cell growth defects. This indicates that their telomeres, which are still elongated (Figure 1, lane 12), are largely functional.
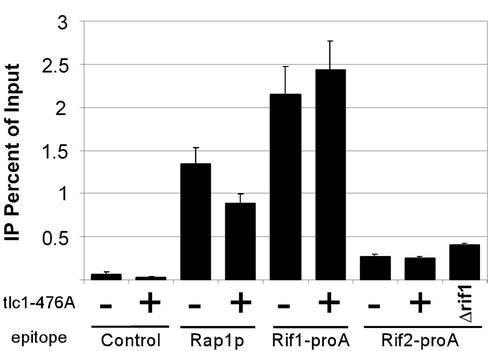
We generated heteroallelic tlc1-476A/TLC1 strains by integrating the tlc1-476A template mutant allele next to the WT telomerase RNA gene, TLC1. We confirmed that their terminal telomeric tract lengths were significantly longer than WT (Figure 1, lanes 8, 11, and 12). Correspondingly, the telomeric DNA hybridization, as detected by Southern blot and dot-blotting analyses by using a WT telomeric probe, was two- to fivefold greater in tlc1-476A/TLC1 strains than in WT (our unpublished data). We assessed the quantitative ability of Rap1p, Rif1-proA, and Rif2-proA to IP telomeric DNA in these cells. Interestingly, the percentage of input telomeric DNA that could be immunoprecipitated by α-Rap1p antibodies in tlc1-476A/TLC1 strains was only 66% of that in the control WT strain (Figure 5). Thus, despite the increase in total telomeric DNA in tlc1-476A/TLC1 cells, the absolute amount of associated Rap1p decreased. This decrease could reflect the presence of (GT)n tracts lacking Rap1p consensus binding sequences, because Rap1p was found to be unable to bind such disrupted site-sequence telomeric repeat DNA in vitro (Prescott and Blackburn, 2000). Surprisingly however, in contrast to Rap1p, the absolute amounts of telomeric DNA immunoprecipitated by both Rif1- and Rif2-proA increased, with the percentage of input telomeric DNA that could be immunoprecipitated by Rif1- and Rif2-proA not being significantly decreased in the tlc1-476A/TLC1 cells compared with WT cells (Figure 5).
Figure 5.
RIF proteins increase their association to deregulated telomeres, whereas RAP1 does not. ChIP assays were performed on asynchronous haploid cultures that had either TLC1 (−) or tlc1-476A/TLC1 (+) at the TLC1 locus, or contained a deletion at the RIF1 locus. Epitopes detected are indicated. Antibodies used for each strain are described under MATERIALS AND METHODS. The percentage of immunoprecipitated telomeric DNA as a function of total telomeric DNA input is shown.
We also examined another situation in which telomere length control is compromised, a Δrif1 strain. As shown in Figure 1 (lanes 12 and 13), telomeres in these cells and tlc1-476A/TLC1 strains were lengthened to similar extents. Again, in the Δrif1 cells, as in the tlc1-476A/TLC1 heteroallelic strains, the relative cross-linkability of Rif2-proA with telomeric DNA was significantly higher than in the WT control (Figure 5).
Colocalization of RAP1 with RIF1
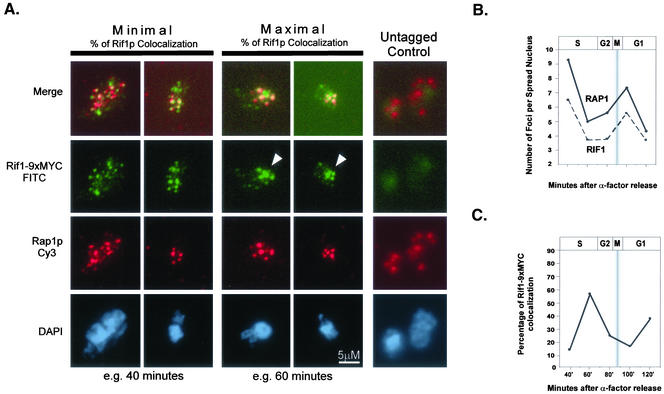
As an independent method to assess the interaction of Rap1p and Rif1–9xMYC with chromosomes and one another through the cell cycle, we localized these proteins by immunofluorescence in chromatin spreads (Klein et al., 1992; Michaelis et al., 1997). Previous cytological evidence has shown that Rap1p colocalizes with Y′ elements at the telomeres in relatively intact nuclei (Gotta et al., 1996) and that Rif1p and Rap1p generally colocalize in vivo (Mishra and Shore, 1999). In the chromatin spread method, spheroplasts are gently disrupted and their nuclear contents are spread locally on the slide. Associated proteins are then paraformaldehyde fixed to the chromatin, but are dispersed over a larger area than in intact nuclei, facilitating their visualization. Chromatin spreads were performed using cells of the W303-a strain background. Comparison of budding indices and FACS profiles from control α-factor arrest/releases showed that untagged and epitope tagged W303-a and S288C strains had a similar response to α-factor and similar kinetics of release and progression through the cell cycle (our unpublished data). For each cell cycle time point, the total number of Rap1p- and Rif1–9xMYC-staining foci per spread nucleus was counted for ∼75 DAPI staining areas (i.e., nuclei) (Figure 6A). By using this chromatin spread method, the overall DAPI-staining area per nucleus did not change significantly between the different time points analyzed (our unpublished data). The average number of staining foci per spread nucleus was then calculated for each protein and time point. We observed between four and nine (average of 6.4) Rap1p foci per spread nucleus through the cell cycle (Figure 6B, black line). This is similar to the reported number of Rap1p foci visible in intact cells (Laroche et al., 2000). We observed between four and six Rif1–9xMYC foci (average of 4.9) per spread nucleus through the cell cycle (Figure 6B, dashed line). The number of visible, discrete Rap1p and Rif1–9xMYC foci was highest 40 min after α-factor release, in early S phase, and lowest 60–80 min after α-factor release, corresponding to late S phase and G2 (Figure 6B). Overall, the trends in the number of foci for Rap1p and Rif1–9xMYC over the cell cycle were well correlated. As the number of Rif1–9xMYC spots decreased in number they also tended to increase in size. When the number of Rif1–9xMYC spots per spread nucleus was lowest (i.e., 60–80 min after α-factor release), the percentage of spread nuclei with a few, relatively large, Rif1–9xMYC spots was maximal (Figure 6A, white arrowheads). Although the number of Rap1p spots also decreased through the cell cycle, the spots did not seem to cluster in the same manner as Rif1–9xMYC.
Figure 6.
RAP1 and RIF1 colocalize and cluster through the cell cycle. α-Factor synchronized cultures were released into the cell cycle and collected at 20-min intervals. Spheroplasts were spread on glass slides and immunofluorescence performed for Rap1p (Cy3, red) and Rif1–9xMYC (FITC, green). Representative examples of maximally colocalized and minimally colocalized spots are shown as well as untagged control (A). White arrowheads indicate large, clustered Rif1–9xMYC foci (A). The average number of Rap1p and Rif1–9xMYC foci per nucleus was quantified for each time point (B). The percentage of Rif1–9xMYC colocalization to Rap1p through the cell cycle was also quantified (C). The approximate stage of the cell cycle, as determined by budding indices is shown above graphs. Shaded bar in B and C represents the approximate range of mitosis.
Next, we assessed the extent of colocalization of Rap1p and Rif1–9xMYC through the cell cycle. In G1 and early S phase, when more spots were present, there was a low percentage of colocalization (Figure 6, A and C), and as the number of spots decreased, the “coalescence” and the percentage of Rif1–9xMYC colocalization increased (Figure 6, A and C). Thus, when there were the fewest Rif1–9xMYC spots (i.e., 60 min after α-factor release) there was the highest percentage of the remaining Rif1–9xMYC spots colocalizing with Rap1p spots. Strikingly, from mid S phase to G2, when the number of Rap1p and Rif-9xMYC foci was lowest, their ability to IP telomeric DNA was greatest in the ChIP assays (Figure 4, A and B). This suggests that the few, clustered spots remaining were associated with telomeric DNA.
DISCUSSION
Herein, we have presented evidence that regulated changes in the association properties of telomeric proteins remodel chromatin at the chromosome ends over the cell cycle. We demonstrated that Rif1p in the telomeric regions associates with regions extending in from the chromosome ends that correlate well with such regions bound by the Rap1 and Sir2-3 proteins. Our results therefore suggest that Rif1p, like Rap1p and the Sir2-4 proteins, also associates with the telomeric DNA regions that include and/or flank Y′ elements. Furthermore, we found that Rap1p and Rif1p are maximally associated with telomeres in late S phase and G2, whereas Rif2p steadily decreases its telomeric association through S phase. Previous immunofluorescence data in yeast suggested that Rap1p, Sir3p, and Sir4p are partially displaced from telomeres in late G2/M (Laroche et al., 2000). S-phase–specific association of the human Xrs2p homolog NBS1 with telomeres has also been observed (Wu et al., 2000; Zhu et al., 2000). In yeast, although in vivo telomeric DNA addition occurs in late S phase and G2/M (Diede and Gottschling, 1999; Marcand et al., 2000), it had not been determined when in the cell cycle telomerase is associated with, or gains access to, its telomeric substrate. Our data suggest that the core telomerase protein Est2p is associated with the chromosome end through most of the cell cycle, only being displaced in mitosis.
Considered together with published work (Laroche et al., 2000), the ChIP data for Rap1p and the tagged Rif1, Rif2, and Est2 proteins suggest a finely tuned, dynamic interplay between the telomeric components at the chromosome ends. Notably, there was generally no greater than a 2.5-fold change in the cross-linkability of the telomeric factors investigated in this study, which is comparable with the changes in chromatin association seen with other factors (Guacci et al., 1994). This may reflect telomere homeostasis being achieved through a dynamic equilibrium of lengthening and shortening activities and that only small changes in the chromatin can tip the balance between telomere lengthening or end protection.
A Model for Cell Cycle Regulation of Telomeric Chromatin
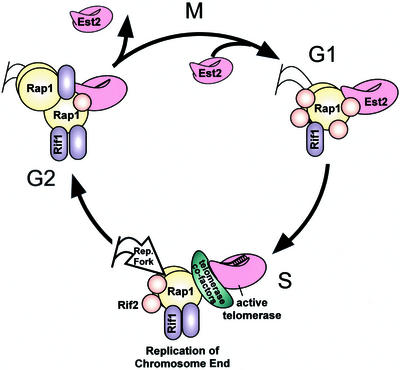
We propose a minimal model for the regulation of telomeric chromatin in the cell cycle to account for the available experimental data (Figure 7). This model assumes, for simplicity of discussion, that the ability of a telomeric protein to IP telomeric DNA primarily reflects association with the telomeric chromatin, rather than altered epitope ability. The repressive chromatin complex, composed minimally of Rap1p, Rif1p, and Rif2p, is dynamically remodeled over the course of the cell cycle. Our results suggest that core telomerase associates with the chromosome end through much of the cell cycle and is displaced at G2/M after the last telomeres are fully replicated. We suggest that the progressive Rif2p displacement from telomeres through S phase opens the telomeric chromatin structure to loading of telomerase cofactors, thus allowing telomerase action by late S phase. Such opening of telomeric chromatin may also involve disrupting interactions between those Rap1p, Rif1p, and Sir2-4 proteins spread along internal Y′ elements with those at the terminal telomeric TG1–3 tracts. We propose that as replication forks move through the end regions and telomeres are replicated, Rap1p and Rif1p quickly reassociate to protect the ends and inhibit overelongation by telomerase. Once telomere replication is complete by the end of G2, and as cells enter mitosis, Est2p, Rif1p, and Rif2p are strongly displaced from the chromatin. Reassociation of Rif2p by the next S phase inhibits premature telomerase action or recombination.
Figure 7.
A model for telomeric chromatin remodeling through the cell cycle. In this speculative model showing the dynamics of telomeric remodeling during the cell cycle, in G1, Rif1 (purple ovals), Rif2 (orange circles), EST2 (pink shape), and additional Rap1 (yellow circles) are loaded onto chromatin. Rif2 is maximally associated in G1. Its gradual dissociation from telomeric DNA through S phase releases inhibition of telomerase. Telomerase, its associated cofactors (green oblong), and the replication fork act on telomeres in mid/late S phase. Telomerase is displaced in G2 when Rap1p and Rif1p are maximally associated. As cells undergo mitosis, Rif1p strongly disassociates from telomeres, allowing restoration of binding in the subsequent G1 phase.
Several lines of evidence support such a model for telomeric chromatin remodeling. We have shown that Est2–13xMYC specifically enriches telomeric loci over the negative control locus ACT1 and is capable of cross-linking to telomeric DNA through most of the cell cycle except mitosis, when its cross-linkability to telomeric DNA drops to levels only slightly above background (Figure 4D, right). Thus, during mitosis, telomerase is either displaced from telomeres or becomes inaccessible to antibodies. Telomeric DNA, like repetitive rDNA sequences (Guacci et al., 1994), may be highly condensed during mitosis. However, Est2–13xMYC cross-linkability to telomeric DNA was comparable at times in the cell cycle when association of the repressive chromatin factors Rap1p and Rif1-proA was both high and low (compare Figure 4D, 40–80 min, with Figure 4, A and B, 40–80 min), suggesting that the Est2–13xMYC ChIP signals reflect association with telomeric DNA, rather than masking of the 13xMYC epitope by these telomeric chromatin factors. Our results suggest that Est2p may load early in the cell cycle, in G1 or early S phase, even though in G1, telomerase neither extends de novo-cut ends (Diede and Gottschling, 1999), nor acts on intact, shortened telomeres in vivo (Marcand et al., 2000). This suggests that to begin to elongate telomeres in late S phase, telomerase cofactors such as Cdc13p, Est1p, or replication factors, all known to be required for telomerase action in vivo (Lundblad and Szostak, 1989; Lendvay et al., 1996; Diede and Gottschling, 1999), may be loaded. It is not known whether, for example, for telomeric DNA addition to occur, Cdc13p needs to interact with the replication protein Pol1p (Qi and Zakian, 2000), possibly at replication forks that have progressed toward telomerase at the chromosome ends in S phase. Transcription of the EST1 gene increases in mid G1 phase of the cell cycle (Spellman et al., 1998), raising the additional possibility that Est1p may be a limiting cofactor in the telomerase complex until late in S phase.
One intriguing possibility suggested by the association of Est2p with telomeres through G1 and S is that telomeric chromatin acts to sequester telomerase at the chromosome end through most of the cell cycle. Telomeric chromatin is thought to act as a “reservoir” of DNA damage response factors, as well as factors that silence rDNA (Kennedy et al., 1997; Smith et al., 1998) and silent mating type loci (Buck and Shore, 1995; Marcand et al., 1996). Telomeric sequestration of telomerase could serve to prevent accidental de novo telomeric DNA addition onto inappropriate DNA substrates such as double-strand breaks, preventing potentially promiscuous and detrimental chromosome “mishealing” events at nontelomeric breaks (Jager and Philippsen, 1989).
Rap1p and Rif1p coimmunoprecipitation with telomeric DNA was highest in G2 (Figure 4, A and B), when chromosome condensation and packaging is greatest (Guacci et al., 1994), and coinciding with when the fewest positive staining foci were seen in chromatin spread assays. Conversely, in G1/early S phase, when there were greater numbers of Rap1p foci, the need to package and cluster telomeres would be least. The genome is actively transcribed and replicated in G1 and S phases, when the requirement of Rap1p for the transcriptional control of several genes involved in translation and carbohydrate metabolism (Shore, 1994; Lieb et al., 2001) would be expected to be greatest. Consistent with our association data for Rap1p, the Y′-element helicase is transcriptionally up-regulated in the M/G1 phase of the cell cycle (Spellman et al., 1998), suggesting that these subtelomeric ORFs are accessible to transcription factors and not packaged in repressive telomeric chromatin at this time. In sum, these results suggest that fewer, clustered Rap1p foci are correlated with increased telomeric association and condensation, while more numerous foci are correlated with less condensed telomeric chromatin.
Rif1-proA had a pattern of telomeric chromatin association in ChIP analyses that was similar to, but distinct from, that of Rap1p. The number of Rif1–9xMYC foci observed through the cell cycle in chromatin spread assays also closely mirrored that of Rap1p and, as the Rif1-proA/Rap1p foci coalesced, the telomeric association of Rif1p in ChIP assays increased. However, a notable difference between Rif1p and Rap1p was the association of Rif1-proA to telomeric DNA earlier in the cell cycle than Rap1p. Whereas Rif1-proA chromatin association with telomeric DNA might be expected to depend on Rap1p DNA binding, it is unknown whether other protein–protein interactions can recruit Rif1-proA to DNA. Indeed, the microarray data for Rif-1proA identified a total of 239 targets associated with Rif1-proA that were not associated with Rap1p. Hence, Rif1p association to these nontelomeric genomic targets may be independent of Rap1p (supplementary data C and D). However, other than Rap1p and Rif2p, no other candidates have been identified that interact with Rif1p in high-throughput 2-hybrid screens (Uetz and Hughes, 2000; Ito et al., 2001).
Of particular interest were a number of nontelomeric Rif1p targets that were genes involved in various aspects of telomere maintenance. Specifically, the TEL1, Ku70, POL1, SME1, MLP1, and MEC1 genes were found among the top 3% of nontelomeric targets. The TEL1 and MEC1 genes are thought to be involved in sensing telomere length or signal transduction at the telomere (Craven and Petes, 1999). The SME1 gene is an SM-family protein and is involved in processing the telomerase RNA gene (TLC1) (Seto et al., 1999). The MLP1 gene encodes a nuclear pore component that interacts with MLP2, which also interacts with the Ku proteins and provides a possible link between the telomeric chromatin and nuclear periphery (Galy et al., 2000). The likelihood of randomly enriching six genes involved in telomere functions in ChIPs is very low (p < 8 × 10−13). It will be of interest to determine whether Rif1p regulates these genes.
Although the telomeric association of Rap1p and Rif1-proA increased dramatically through S phase until G2/M, Rif2-proA steadily decreased its association to telomeres through the cell cycle. Genetic evidence suggests that Rif1p and Rif2p act in distinct pathways to maintain telomeres, because deletion of both genes results in synergistic loss of length control (Wotton and Shore, 1997). Our model could explain this observed synergism. One possibility is that Rif1p plays a more structural role at the chromosome ends, whereas the primary role for Rif2p is to inhibit telomerase and recombination activities (Diede and Gottschling, 1999; Teng et al., 2000). Rif1p is considerably larger than Rif2p and associated 10 times more strongly with DNA than Rif2p in the ChIP assays (Figures 2B and 4B and C, left). We found that, like Rap1p and the Sir2-4 proteins (Strahl-Bolsinger et al., 1997; Lieb et al., 2001), Rif1p is spread over many kilobases of DNA at the end regions. In Δrif2 strains, telomeres elongate but remain well regulated. Previous studies have shown that Rif2p is particularly important in preventing the terminal telomeric tract from participating in the RAD52-dependent type II survivor pathway (Teng et al., 2000) and is also important in directly inhibiting telomerase addition to de novo cut telomeric ends (Diede and Gottschling, 1999). The known times in the cell cycle of in vivo telomeric DNA addition, late S phase and G2, coincide with when the ChIP association of Rif2p with telomeric DNA was lowest. Therefore, we propose that Rif2p directly inhibits telomerase at the telomeres, and that RIF2 deletion frees telomerase of this inhibition, so that it adds telomeric DNA earlier in the cell cycle, resulting in the observed longer telomeres. In this situation, the repressive structural effects of Rap1p and Rif1p are still present and capable of preventing runaway telomere lengthening or deregulation. In a Δrif1 strain, telomere lengthening is more substantial, and interaction between Rap1p and Rif2p is increased (Wotton and Shore, 1997). Similarly, in our experiments the association of Rif2p with telomeric DNA was modestly increased in a Δrif1 strain (Figure 5). The telomere lengthening observed in Δrif1 strains (Hardy et al., 1992; Wotton and Shore, 1997) (Figure 1, lanes 14 and 15) and our ChIP data suggest that this increased Rif2p association alone to telomeric DNA is not sufficient to reestablish a repressive chromatin structure and inhibit telomerase. Thus, the disruption of telomeric chromatin in cells might result in greater telomere elongation than in Δrif2 strains by both disrupting repressive Rap1p-Rif1p chromatin and by eliminating the Rif1p-Rif2p interaction that helps recruit more Rif2p to telomeres to inhibit telomerase.
Interestingly, the elongated, deregulated telomeres of tlc1-476A/TLC1 cells showed increased Rif1p and Rif2p association with telomeric DNA in ChIP analyses, even though Rap1p association decreased. Therefore, we propose that increased recruitment of Rif1p and Rif2p in tlc1-476A/TLC1 cells, and of Rif2p in Δrif1 strains, is a cellular response to the partial uncapping effects of these mutations, possibly as a mechanism to regain length control. Recent high-throughput two-hybrid studies of Rif2p suggest that it may have as many as 80 binding partners, including Sir2p (Ito et al., 2001), raising the possibility that other protein–protein interactions may also be important for Rif protein recruitment to telomeric chromatin.
This work, taken together with previous studies, provides evidence that telomere chromatin dynamically changes through the cell cycle. Although the fundamental signals for telomeric chromatin remodeling have yet to be elucidated, it is likely that cell cycle regulation of other telomeric and general chromatin factors such as the Kus, SIRs, nucleosomes, and condensins also occurs. It will be of great interest to understand how these as well as the damage and replication machineries modulate telomeric chromatin through the cell cycle to maximally protect the chromosome ends through cell growth, DNA replication, and nuclear division.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Shore for providing the W303–1a and RIF1–9xMYC strains and Judith Berman for providing polyclonal α-Rap1p antibody. We also thank Jason Leib for the permission to reprint RAP1 and SIR protein chromatin association data in Figure 3 and for advice with the analysis of our microarray data. We are grateful to Inna Botchkina for performing FACS analyses and David Smith for computer support and assistance with microarray data processing. We also thank Daniel Levy and Jue Lin for helpful comments on the manuscript. This work was supported by National Institutes of Health grant GM-26259 (to E.H.B.) and a National Science Foundation predoctoral fellowship (to C.D.S.).
Footnotes
This article contains supplementary data. The supplementary data is available at www.molbiolcell.org.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–08–0457. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–08–0457.
REFERENCES
- Biggins S, Bhalla N, Chang A, Smith DL, Murray AW. Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics. 2001;159:453–470. doi: 10.1093/genetics/159.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourns BD, Alexander MK, Smith AM, Zakian VA. Sir proteins, Rif proteins, and Cdc13p bind Saccharomyces telomeres in vivo. Mol Cell Biol. 1998;18:5600–5608. doi: 10.1128/mcb.18.9.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Buck SW, Shore D. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 1995;9:370–384. doi: 10.1101/gad.9.3.370. [DOI] [PubMed] [Google Scholar]
- Chan SW, Chang J, Prescott J, Blackburn EH. Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr Biol. 2001;11:1240–1250. doi: 10.1016/s0960-9822(01)00391-8. [DOI] [PubMed] [Google Scholar]
- Chan CS, Tye BK. Organization of DNA sequences and replication origins at yeast telomeres. Cell. 1983;33:563–573. doi: 10.1016/0092-8674(83)90437-3. [DOI] [PubMed] [Google Scholar]
- Cho RJ, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- Cockell M, Palladino F, Laroche T, Kyrion G, Liu C, Lustig AJ, Gasser SM. The carboxy termini of Sir4 and Rap1 affect Sir3 localization: evidence for a multicomponent complex required for yeast telomeric silencing. J Cell Biol. 1995;129:909–924. doi: 10.1083/jcb.129.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M, Blackburn EH. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- Craven RJ, Petes TD. Dependence of the regulation of telomere length on the type of subtelomeric repeat in the yeast Saccharomyces cerevisiae. Genetics. 1999;152:1531–1541. doi: 10.1093/genetics/152.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr Biol. 2001;11:1336–1340. doi: 10.1016/s0960-9822(01)00400-6. [DOI] [PubMed] [Google Scholar]
- Enomoto S, McCune-Zierath PD, Gerami-Nejad M, Sanders MA, Berman J. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- Fink CGaGR. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press; 1991. [Google Scholar]
- Fourel G, Revardel E, Koering CE, Gilson E. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 1999;18:2522–2537. doi: 10.1093/emboj/18.9.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Olivo-Marin JC, Scherthan H, Doye V, Rascalou N, Nehrbass U. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 2000;403:108–112. doi: 10.1038/47528. [DOI] [PubMed] [Google Scholar]
- Gerton JL, DeRisi J, Shroff R, Lichten M, Brown PO, Petes TD. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 2001;20:1173–1183. doi: 10.1093/emboj/20.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol. 1997;9:383–387. doi: 10.1016/s0955-0674(97)80011-7. [DOI] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Feldser DM, Greider CW. Telomere dysfunction increases mutation rate and genomic instability. Cell. 2001;106:275–286. doi: 10.1016/s0092-8674(01)00457-3. [DOI] [PubMed] [Google Scholar]
- Hande MP, Samper E, Lansdorp P, Blasco MA. Telomere length dynamics and chromosomal instability in cells derived from telomerase null mice. J Cell Biol. 1999;144:589–601. doi: 10.1083/jcb.144.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CF, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6:801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Evans SK, Weilbaecher RG, Lundblad V. The Est3 protein is a subunit of yeast telomerase. Curr Biol. 2000;10:809–812. doi: 10.1016/s0960-9822(00)00562-5. [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Jager D, Philippsen P. Stabilization of dicentric chromosomes in Saccharomyces cerevisiae by telomere addition to broken ends or by centromere deletion. EMBO J. 1989;8:247–254. doi: 10.1002/j.1460-2075.1989.tb03370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Gotta M, Sinclair DA, Mills K, McNabb DS, Murthy M, Pak SM, Laroche T, Gasser SM, Guarente L. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell. 1997;89:381–391. doi: 10.1016/s0092-8674(00)80219-6. [DOI] [PubMed] [Google Scholar]
- Klein F, Laroche T, Cardenas ME, Hofmann JF, Schweizer D, Gasser SM. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J Cell Biol. 1992;117:935–948. doi: 10.1083/jcb.117.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche T, Martin SG, Tsai-Pflugfelder M, Gasser SM. The dynamics of yeast telomeres and silencing proteins through the cell cycle. J Struct Biol. 2000;129:159–174. doi: 10.1006/jsbi.2000.4240. [DOI] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- Loidl J, Nairz K, Klein F. Meiotic chromosome synapsis in a haploid yeast. Chromosoma. 1991;100:221–228. doi: 10.1007/BF00344155. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Louis EJ, Haber JE. The subtelomeric Y′ repeat family in Saccharomyces cerevisiae: an experimental system for repeated sequence evolution. Genetics. 1990;124:533–545. doi: 10.1093/genetics/124.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis EJ, Haber JE. The structure and evolution of subtelomeric Y′ repeats in Saccharomyces cerevisiae. Genetics. 1992;131:559–574. doi: 10.1093/genetics/131.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- Marcand S, Brevet V, Mann C, Gilson E. Cell cycle restriction of telomere elongation. Curr Biol. 2000;10:487–490. doi: 10.1016/s0960-9822(00)00450-4. [DOI] [PubMed] [Google Scholar]
- Marcand S, Buck SW, Moretti P, Gilson E, Shore D. Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by Rap 1 protein. Genes Dev. 1996;10:1297–1309. doi: 10.1101/gad.10.11.1297. [DOI] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Mishra K, Shore D. Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by rif proteins. Curr Biol. 1999;9:1123–1126. doi: 10.1016/s0960-9822(99)80483-7. [DOI] [PubMed] [Google Scholar]
- Moazed D, Johnson D. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell. 1996;86:667–677. doi: 10.1016/s0092-8674(00)80139-7. [DOI] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- Moretti P, Shore D. Multiple interactions in sir protein recruitment by rap1p at silencers and telomeres in yeast. Mol Cell Biol. 2001;21:8082–8094. doi: 10.1128/MCB.21.23.8082-8094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. The remaking of chromosomes. Collecting Net. 1938;8:182–195. , 198. [Google Scholar]
- Nilsson B, Moks T, Jansson B, Abrahmsen L, Elmblad A, Holmgren E, Henrichson C, Jones TA, Uhlen M. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1987;1:107–113. doi: 10.1093/protein/1.2.107. [DOI] [PubMed] [Google Scholar]
- Nugent CI, Bosco G, Ross LO, Evans SK, Salinger AP, Moore JK, Haber JE, Lundblad V. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol. 1998;8:657–660. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- Palladino F, Laroche T, Gilson E, Axelrod A, Pillus L, Gasser SM. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, Tzoneva M, Gottschling DE. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet. 2001;27:64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- Prescott J, Blackburn EH. Telomerase RNA template mutations reveal sequence-specific requirements for the activation and repression of telomerase action at telomeres. Mol Cell Biol. 2000;20:2941–2948. doi: 10.1128/mcb.20.8.2941-2948.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- Ritchie KB, Petes TD. The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics. 2000;155:475–479. doi: 10.1093/genetics/155.1.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Zakian VA. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- Schwarz SE, Matuschewski K, Liakopoulos D, Scheffner M, Jentsch S. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc Natl Acad Sci USA. 1998;95:560–564. doi: 10.1073/pnas.95.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- Seufert W, Futcher B, Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Smith CD, Blackburn EH. Uncapping and deregulation of telomeres lead to detrimental cellular consequences in yeast. J Cell Biol. 1999;145:203–214. doi: 10.1083/jcb.145.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Pillus L, Boeke JD. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics. 1998;149:1205–1219. doi: 10.1093/genetics/149.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Teng SC, Chang J, McCowan B, Zakian VA. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol Cell. 2000;6:947–952. doi: 10.1016/s1097-2765(05)00094-8. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y, Taggart AK, Zakian VA. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr Biol. 2001;11:1328–1335. doi: 10.1016/s0960-9822(01)00372-4. [DOI] [PubMed] [Google Scholar]
- Uetz P, Hughes RE. Systematic and large-scale two-hybrid screens. Curr Opin Microbiol. 2000;3:303–308. doi: 10.1016/s1369-5274(00)00094-1. [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Ethier K, Labrecque P, Zakian VA. Evidence for a new step in telomere maintenance. Cell. 1996;85:423–33. doi: 10.1016/s0092-8674(00)81120-4. [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Wolf AJ, Zakian VA. Saccharomyces telomeres acquire single-strand TG1–3 tails late in S phase. Cell. 1993;72:51–60. doi: 10.1016/0092-8674(93)90049-v. [DOI] [PubMed] [Google Scholar]
- Wotton D, Shore D. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 1997;11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- Wright JH, Gottschling DE, Zakian VA. Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev. 1992;6:197–210. doi: 10.1101/gad.6.2.197. [DOI] [PubMed] [Google Scholar]
- Wu G, Lee WH, Chen PL. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres. J Biol Chem. 2000;275:30618–30622. doi: 10.1074/jbc.C000390200. [DOI] [PubMed] [Google Scholar]
- Yamada M, Hayatsu N, Matsuura A, Ishikawa F. Y'-Help1, a DNA helicase encoded by the yeast subtelomeric Y′ element, is induced in survivors defective for telomerase. J Biol Chem. 1998;273:33360–33366. doi: 10.1074/jbc.273.50.33360. [DOI] [PubMed] [Google Scholar]
- Zhu XD, Kuster B, Mann M, Petrini JH, Lange T. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.