Abstract
The vertebrate nuclear pore is an enormous structure that spans the double membrane of the nuclear envelope. In yeast, most nucleoporins are found symmetrically on both the nuclear and cytoplasmic sides of the structure. However, in vertebrates most nucleoporins have been localized exclusively to one side of the nuclear pore. Herein, we show, by immunofluorescence and immunoelectron microscopy, that Nup98 is found on both sides of the pore complex. Additionally, we find that the pore-targeting domain of Nup98 interacts directly with the cytoplasmic nucleoporin Nup88, a component of the Nup214, Nup88, Nup62 subcomplex. Nup98 was previously described to interact with the nuclear-oriented Nup160, 133, 107, 96 complex through direct binding to Nup96. Interestingly, the same site within Nup98 is involved in binding to both Nup88 and Nup96. Autoproteolytic cleavage of the Nup98 C terminus is required for both of these binding interactions. When cleavage is blocked by a point mutation, a minimal eight amino acids downstream of the cleavage site is sufficient to prevent most binding to either Nup96 or Nup88. Thus, Nup98 interacts with both faces of the nuclear pore, a localization in keeping with its previously described nucleocytoplasmic shuttling activity.
INTRODUCTION
The nuclear pore complex (NPC) is composed of a central, eightfold symmetrical ring and spoke assembly, cytoplasmic fibers, and a filamentous nuclear basket (reviewed in Stoffler et al., 1999; Allen et al., 2000; Ryan and Wente, 2000; Vasu and Forbes, 2001). The total size of this immense structure is estimated at 120 MDa in vertebrates and approximately half that size in yeast (Yang et al., 1998). In the yeast, Saccharaomyces cerevisae, all of the nuclear pore components or nucleoporins (Nups) have now been identified (Rout et al., 2000). Considerable effort in the field continues to go toward determining where an individual protein is localized within the massive structure of the pore and with which other nucleoporins it interacts. Yeast genetics made possible a systematic and elegant approach of creating a series of strains in each of which a single nucleoporin was tagged with protein A. Using IgG-coated gold, an electron microscopy map was produced in which each nucleoporin was positioned within the pore (Rout et al., 2000). One of the surprising results from this map was the finding that most nucleoporins are distributed symmetrically on both faces of the pore complex. Of the ∼30 yeast nucleoporins, only five are restricted to a single face of the pore (Nup 159, Nup42, and Nup82 on the cytoplasmic side, and Nup60 and Nup1 on the nuclear side). An additional four nucleoporins seem to be biased to one side in their distribution (Nup116, Nup100, Nup145N, and Gle1).
In vertebrates, the full protein composition of the pore was very recently identified (Cronshaw et al., 2002). Unexpectedly, despite a mass twice that of the yeast structure, the vertebrate nuclear pore is also comprised of 30 nucleoporins. Of these 30 proteins, 22 have homologues or orthologues in yeast, and two others have possible functional equivalents. Because genetic approaches are more difficult in vertebrate systems, a complete map of nucleoporins in the vertebrate pore is not yet available. Determination of localization in vertebrate systems has generally required preparation of an antibody to a nucleoporin of interest and use of this antibody for immunofluorescence and electron microscopy studies. Not all antibodies have proven equally useful for electron microscopy studies, and thus a number of vertebrate nucleoporins have not been precisely localized or have conflicting localizations reported by different groups.
Although incomplete, the existing localization data on the vertebrate nuclear pore suggest that one difference from the yeast pore is the bias of nucleoporins to one side or the other of the NPC; most vertebrate nucleoporins have been ascribed a single localization site. Nup214, Nup88 (also known as Nup84), and Nup358, are reported to be exclusively on the cytoplasmic side (Kraemer et al., 1994; Wu et al., 1995; Bastos et al., 1997; Fornerod et al., 1997). Nup153, Nup98, Nup93, Nup188, Nup205 and Tpr have all been reported to be on the nuclear side of the pore (Radu et al., 1995; Bastos et al., 1996; Grandi et al., 1997; Miller et al., 2000; Frosst et al., 2002). The Nup62, Nup58, Nup54, Nup45 complex, as well as Nup155, are more centrally located within the “transporter” region of the nuclear pore (Radu et al., 1993; Hu et al., 1996b). Only one nucleoporin subcomplex, the Nup133 complex (Nups 160, 133, 107, 96 and sec13) has been reported to exist on both the nuclear and cytoplasmic faces of the vertebrate pore, although an alternate and exclusively nuclear localization was reported by a different laboratory (Belgareh et al., 2001; Vasu et al., 2001). Thus, given our current knowledge, the vertebrate nuclear pore complex seems to be both larger and potentially more asymmetric than the yeast pore.
The distribution of the GLFG family of nucleoporins within the pore is particularly interesting. In higher eukaryotes, only one GLFG repeat nucleoporin, Nup98, has been identified. However, there are three related proteins in S. cerevisae, Nup145, Nup116, and Nup100, and each of these nucleoporins possesses a subset of the features of Nup98. Both Nup98 and Nup145 undergo autocatalyzed proteolysis; Nup145 is cleaved to produce Nup145N and Nup145C, whereas Nup98 is cleaved from a Nup98/Nup96 polyprotein precursor (Emtage et al., 1997; Teixeira et al., 1997; Rosenblum and Blobel, 1999). Nup98 and Nup116 each contain a binding site for Gle2, a protein implicated in mRNA export (Murphy et al., 1996; Bailer et al., 1998; Pritchard et al., 1999). Strikingly, the GLFG nucleoporins comprise three of the four proteins whose distribution within the yeast pore is biased toward, but not restricted to, a single face of the pore complex. Nup145N, the portion of Nup145 homologous to Nup98, is enriched on the nuclear side, whereas Nup116 and Nup100 are biased toward the cytoplasmic face of the pore (Rout et al., 2000). Considering these localization data from the yeast GLFG nucleoporins, it is somewhat curious that Nup98, the only GLFG nucleoporin in vertebrates, has been found strictly on the nuclear side of the pore complex (Radu et al., 1995; Vasu et al., 2001; Frosst et al., 2002).
Here, we have continued to investigate the interaction between Nup98 and the nuclear pore complex. Recently, we reported that Nup98 is dynamically associated with the nuclear pore and shuttles between the nucleus and the cytoplasm (Griffis et al., 2002). This observed shuttling behavior suggested to us that Nup98 might have sites of association on both faces of the nuclear pore. Indeed, we show here that Nup98 can be found on both the nuclear and the cytoplasmic faces of the pore. Nup98 binds directly to Nup96, the C-terminal half of the proteolytically processed Nup98/Nup96 polyprotein and a component of the Nup133 subcomplex (Fontoura et al., 1999; Vasu et al., 2001). We now find that Nup98 also binds directly to another component of the nuclear pore, Nup88. Together with Nup214 and p62, Nup88 forms a subcomplex that is found only on the cytoplasmic face of the pore (Bastos et al., 1997; Fornerod et al., 1997). This same cytoplasmic orientation is also true for the yeast homolog of Nup88, Nup82 (Rout et al., 2000). Thus, Nup98 has two potential binding partners on the cytoplasmic face (Nup96 and Nup88) and one binding partner on the nuclear face of the pore (Nup96). We have mapped the sequences required for binding to each partner and find that the same region of Nup98 is similarly used for both interactions. Thus, it is unlikely that Nup98 acts as a bridge between the Nup133 complex and the Nup88/Nup214/Nup62 complex. Our results define a new interaction between a dynamic nucleoporin and the NPC, and suggest that the vertebrate nuclear pore may be more symmetrical than currently envisioned.
MATERIALS AND METHODS
DNA Constructs
Green fluorescent protein (GFP)-Nup98 and GFP-Nup98 single domain constructs were described previously (Griffis et al., 2002). The GFP-GLFG-C terminus construct (amino acids 224–920) was made by digesting the Nup98 cDNA with KpnI and XhoI and then ligating the fragment into pEGFP-C3 (BD Biosciences Clontech, Palo Alto, CA). To create the GST-C-term Nup98 construct (amino acids 506–920), the domain was amplified by polymerase chain reaction (PCR) and ligated into pGEX-6p (Amersham Biosciences, Piscataway, NJ). To make uncleavable Nup98 mutants, serine 864 was mutated to alanine (S864A) in the indicated plasmid templates as detailed under RESULTS. To produce the Nup98 C-terminal truncation mutants, a stop codon was added in place of serine 864 for the truncated mutant, in place of glutamic acid 873 for the 872-stop mutant, and in place of serine 883 for the 882-stop mutant. The hemagglutinin (HA)-tagged Nup84 (referred to herein as Nup88) plasmid was a gift of Dr. Brian Burke (Bastos et al., 1997). The HA-Nup88 truncation mutant was made by inserting two stop codons after amino acid 584 in the full-length Nup88 coding sequence. The huNup98/96 polyprotein plasmid was a gift of Dr. Beatriz Fontoura (Fontoura et al., 1999). To make the full-length Nup96 construct, the coding sequence of Nup96 was amplified by PCR by using the Nup98–96 precursor as template and ligated into pCDNA3. All mutations were created using the QuickChange PCR mutagenesis kit (Stratagene, La Jolla, CA) and were confirmed by DNA sequencing.
Cell Culture and Immunofluorescence
HeLa, COS1, and XL177 cells were maintained as described previously (Griffis et al., 2002) and transfections were performed using FuGENE 6 (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions. Immunofluorescence localization experiments were carried out as described previously (Griffis et al., 2002). For the digitonin permeabilization experiments, the following changes were made: after fixation in 4% paraformaldehyde, cells were permeabilized in digitonin diluted to 40 μg/ml in phosphate-buffered saline, and in subsequent steps Triton X-100 was omitted from the primary antibody block and wash solutions. To better visualize Nup98 at the nuclear envelope, cells were simultaneously fixed and permeabilized in 2% paraformaldehyde with 0.2% Triton X-100 for 10 min on ice. All subsequent steps were as described previously (Griffis et al., 2002). The following antibodies were used: anti-xNup98 (1:50; Powers et al., 1995), anti-hNup98 (1:3000; Griffis et al., 2002), anti-xLamins II and III (1:1000; Developmental Studies Hybridoma Bank, Iowa City, IA), anti-hLaminB (1:300; Santa Cruz Biotechnology, La Jolla, CA), anti-GFP (monoclonal antibody [mAb] 3E6, 1:200; Molecular Probes, Eugene, OR), anti-HA (mAb 3F10, 1:1000; Roche Diagnostics), mAb 414 (1:1000; Calbiochem, La Jolla, CA), goat anti-rabbit rhodamine isothiocyanate (1:800; Jackson Immunoresearch Laboratories, West Grove, PA), goat anti-rabbit Oregon Green (1:700; Molecular Probes), goat anti-mouse rhodamine isothiocyanate (1:800; Jackson Immunoresearch Laboratories), goat anti-mouse fluorescein isothiocyanate (1:800; Jackson Immunoresearch Laboratories), donkey anti-goat fluorescein isothiocyanate (1:400; Santa Cruz Biotechnology) and goat anti-rat Texas Red (1:1200; Jackson Immunoresearch Laboratories).
Images were captured using either a BX60 microscope (Olympus, Tokyo, Japan) with an 8-bit camera (Dage-MTI, Michigan City, IN) and IP Lab software (Scanalytics, Fairfax, VA) or an LSM 510 confocal microscope (Carl Zeiss, Thornwood, NY).
In Vitro Binding Assays and Coimmunoprecipitations
In vitro binding assays were performed as described previously (Hodel et al., 2002). For coimmunoprecipitation experiments, COS1 cells were released from a dish 40 h posttransfection by using 25 mM EDTA. Cells were pelleted and washed twice in 2 ml of buffer A (150 mM NaCl, 10 mM HEPES pH 7.45, 5 mM sodium pyrophosphate, 5 mM NaF, 2 mM sodium orthovanadate, 10 μg/ml aprotinin and leupeptin, 1 mM phenylmethylsulfonyl fluoride and 1× Complete protease inhibitors; Roche Diagnostics). After washing, cells were lysed in 1.2 ml of buffer B (150 mM NaCl, 10 mM HEPES pH 7.45, 5 mM sodium pyrophosphate, 5 mM NaF, 2 mM sodium orthovanadate, 10 μg/ml aprotinin leupeptin, 1 mM phenylmethylsulfonyl fluoride, 1× Complete protease inhibitors [Roche Diagnostics] and 0.2% NP-40) at 4°C for 30 min with gentle agitation. The lysate was cleared of aggregates by centrifugation for 10 min at 12,000 rpm in a refrigerated centrifuge (Tomy Seiko, Tokyo, Japan). Protein A-Sepharose beads (Sigma-Aldrich, St. Louis, MO) previously blocked in buffer B with 5 mg/ml bovine serum albumin were added and incubated for 1 h at 4°C. The samples were centrifuged for 10 min to remove the beads, and the supernatant was then divided into separate tubes. To these tubes either 5 μg of the appropriate antibody or 20 μl of anti-HA beads (Roche Diagnostics) was added to each tube, and the samples were rotated for 1 h at 4°C. Blocked protein A-Sepharose beads (20 μl) were added to non-HA samples and the tubes were rotated overnight. The samples were then washed three times in buffer B and twice in buffer A before being eluted from the beads with gel sample buffer. Western blots were performed as described previoulsy (Hodel et al., 2002). For Western blotting, the following antibodies were used: anti-HA (mAb 3F10, 1:1000; Roche Diagnostics), anti-hNup98 (1:3000), goat anti-rabbit horseradish peroxidase (HRP) (1:2000; Zymed Laboratories, South San Francisco, CA), and sheep anti-rat HRP (1:2000; Amersham Biosciences).
Immunoelectron Microscopy
Immunoelectron microscopy was performed as described previously (Griffis et al., 2002). However, to visualize Nup98 on the cytoplasmic side of the pore in HeLa cells, the permeabilization step using Triton X-100 was omitted. For immunogold electron microscopy, confluent HeLa cells were released from 10-cm dishes with 25 mM EDTA and pelleted. The cell pellet was washed twice with phosphate-buffered saline and then subjected to freeze-thaw permeabilization and preembedding immunogold labeling as described previously (Guan et al., 2000).
RESULTS
Nup98 Is Found on Both Faces of the Pore
We recently showed that Nup98 shuttles between the nucleus and the cytoplasm in a transcription-dependent manner (Griffis et al., 2002). The established localization for this nucleoporin has been on the nuclear face of the nuclear pore complex, at a distance from the membrane that is consistent with a position in the basket structure of the pore (Radu et al., 1995; Frosst et al., 2002). However, our observation that Nup98 crosses the pore led us to ask whether this nucleoporin could perhaps interact at more than one site within the nuclear pore complex. In particular, we asked whether a binding site might be found on the cytoplasmic face of the pore as a counterpart to the well-characterized localization on the nuclear face.
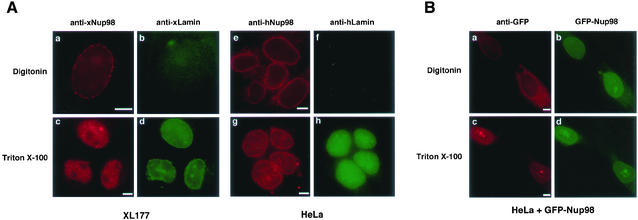
To address this question, we used multiple antibodies and cell types to localize Nup98 in both Triton X-100–permeabilized cells and digitonin-permeabilized cells. When cells are treated with Triton, both faces of the nuclear membrane are accessible; however, digitonin selectively permeabilizes the plasma membrane, allowing antibodies to access only the cytoplasmic face of the nuclear envelope. Immunofluorescence staining of digitonin-treated Xenopus XL177 cells with an antibody to Xenopus Nup98 (Powers et al., 1995) resulted in a punctate staining of the nuclear envelope characteristic of a nuclear pore complex localization (Figure 1A, a). Simultaneous staining with antibody to Xenopus lamins did not produce a signal, indicating that the nuclear lamina is not accessible and thus the nuclear envelope remains intact (Figure 1A, b). In contrast, permeabilization of XL177 cells with Triton resulted in a strong nuclear lamina stain and the characteristic intranuclear signal observed for xNup98 (Figure 1A, c and d). When antibodies to human Nup98 (Griffis et al., 2002) and human lamin B were used in the same experiment on HeLa cells, similar results were obtained (Figure 1A, e–h). Thus, in both human and Xenopus cells, a fraction of Nup98 is accessible on the cytoplasmic face of the nuclear pore.
Figure 1.
Nup98 localizes to the cytoplasmic side of the nuclear pore in digitonin-permeabilized cells. (A) XL177 (a–d) and HeLa cells (e–h) were fixed and then permeabilized using either digitonin or Triton X-100 before detecting Nup98 and lamins by immunofluorescence. Nup98 is visible at the nuclear rim in cells permeabilized with digitonin (a and e), whereas the intranuclear lamins are detected only when nuclei are permeabilized using Triton X-100 (d and h). (B) HeLa cells expressing GFP-Nup98 were fixed and permeabilized as described above and then GFP was detected directly (b and d) or indirectly using an anti-GFP mAb and rhodamine-isothiocyanate-labeled secondary (a and c). The GFP antibody clearly recognizes GFP-Nup98 on the nuclear pore in a digitonin permeabilized cell (a) and does not detect the intranuclear GFP-fusion protein unless the nucleus is permeabilized with Triton X-100 (c). Bars, 5 μm.
The Xenopus Nup98 antibody was raised and affinity-purified against the full-length endogenous protein, whereas the human Nup98 antibody was produced and purified using the bacterially expressed C-terminal domain of the protein. Thus, it was unlikely that both antibodies would result in the same nonspecific cross-reaction with the cytoplasmic face of the pore. However, to control for such a possibility, we transfected GFP-tagged huNup98 into HeLa cells and localized transfected Nup98 with a GFP-specific antibody after permeabilization with either digitonin or Triton (Figure 1B). Transfected cells were identified by the characteristic GFP-Nup98 fluorescence within the nucleus as well as at the nuclear envelope (Griffis et al., 2002). When cells were permeabilized with Triton, anti-GFP and rhodamine-labeled secondary antibody produced a fluorescence pattern identical to that observed with direct GFP fluorescence (Figure 1B, c and d). However, when cells were permeabilized with digitonin, staining with anti-GFP was seen only in the cytoplasm and on the nuclear rim; the intranuclear GFP-Nup98 was not detected by the antibody, demonstrating the integrity of the nuclear envelope (Figure 1B, a and b). Thus, the localization of the transfected protein confirms the results seen with the endogenous protein; a fraction of Nup98 is present on the cytoplasmic face of the nuclear pore.
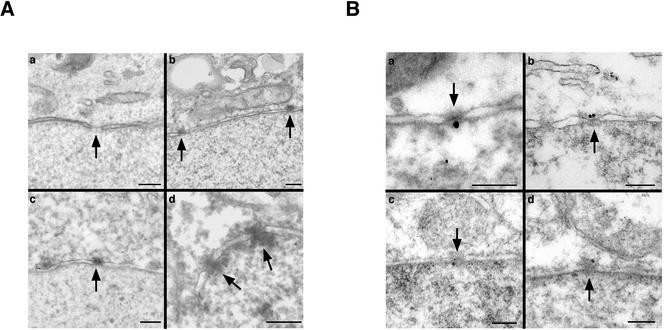
This novel localization of Nup98 within the pore was confirmed by immunoelectron microscopy. Either untransfected or GFP-Nup98–transfected HeLa cells were fixed, but their nuclei were not permeabilized with Triton. The cells were then stained with anti-huNup98, HRP-conjugated secondary antibody, and diamino-benzidine (DAB), which results in deposition of electron dense material at the site of antibody binding. In either untransfected (Figure 2A, b) or GFP-Nup98 transfected (Figure 2A, c and d) cells, DAB was found associated with the cytoplasmic face of the nuclear pore complex. Transfected cells showed an increased intensity of labeling, suggesting that it is possible for somewhat elevated amounts of Nup98 to accumulate at the pore when excess protein is expressed. In the absence of anti-hNup98, no deposition of DAB was observed at nuclear pores (Figure 2A, a). In a rare nucleus that did become permeabilized in the transfected sample, there was substantial deposition of DAB on both sides of the nuclear pore, in keeping with the previously established localization of Nup98 on the nuclear basket (Figure 2A, d).
Figure 2.
Nup98 localizes to both sides of the pore by immunoelectron microscopy. (A) Either untransfected (a and b) or GFP-Nup98 expressing cells (c and d) were treated for immunoelectron microscopy under conditions that maintained an intact nuclear envelope, and bound antibody was detected by the deposition of DAB. When primary antibody was omitted, no staining was seen at nuclear pores (a). Nup98 antibodies associated with the cytoplasmic side of the nuclear pore in both wild-type and transfected cells (b and c). In a rare nucleus that was permeabilized by the procedure, Nup98 was detected on both sides of the NPC (d). Nuclear pores are indicated by arrowheads. (B) Immunogold electron microscopy was used to detect Nup98 on both sides of the pore in HeLa cells after freeze-thaw permeabilization. Endogenous Nup98 was localized in HeLa cells by using an affinity-purified anti-hNup98 (a and b). GFP-Nup98 was also detected on both sides of the pore by using an anti-GFP monoclonal in a cell line stably expressing the protein (c and d). In all panels, orientation is with cytoplasm up, nucleus down. Bars, 250 nm.
We also used immunogold electron microscopy on both HeLa cells and a HeLa-derived cell line that stably expresses GFP-Nup98. The cells were subjected to a freeze-thaw cycle to partially disrupt the nuclear envelope and permit antibody access to the nuclear interior (Guan et al., 2000), followed by fixation and immunostaining. In the absence of primary antibody the majority of nuclear pores had no associated gold particles. However, when the endogenous protein was detected with anti-huNup98, gold particles were found associated with both the cytoplasmic and nuclear faces of the pore (Figure 2B, a and b). Similar results were obtained when the GFP-Nup98 cell line was stained with anti-GFP (Figure 2B, c and d). Because the freeze-thaw protocol only partially disrupts the nuclear envelope, the concentration of antibody within the nucleus is most likely far lower than in the cytoplasm. Consequently, we did not determine a relative distribution of Nup98 by scoring gold particles on each face of the nuclear pore because this seemed likely to result in a highly inaccurate ratio. Taken together, the combination of immunofluorescence and immunoelectron microscopy strongly indicate that Nup98 is found on both surfaces of the nuclear pore complex.
C Terminus of Nup98 Mediates Interaction with the Pore
Before investigating how Nup98 associates with the cytoplasmic face of the pore, we first wanted to assess which sequences within Nup98 were responsible for its targeting to the NPC. Previously, we had localized the N-terminal (including the Gle2-binding site), GLFG repeat, and C-terminal domains as individual GFP fusion proteins. Unexpectedly, we found that none of the individual domains generated a significant nuclear pore-staining pattern in live cells (Griffis et al., 2002). However, the C-terminal domain of Nup98 has been shown to bind to a nuclear pore subcomplex containing Nups 160, 133, 107, 96 and sec13 (Vasu et al., 2001), and mutations that prevent proteolytic processing within the C-terminal domain also prevent association with the pore (Fontoura et al., 1999; Hodel et al., 2002). Thus, the C-terminal domain contains sequences that should be responsible, at least in part, for nuclear pore complex targeting.
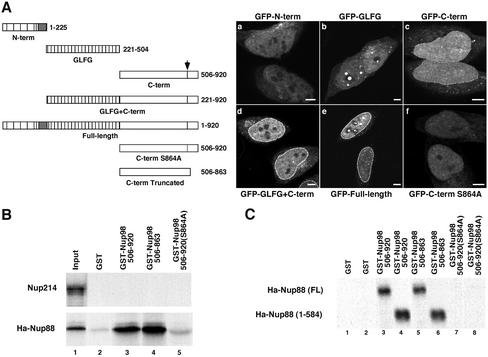
To determine whether a subfraction of any of the individual domains might have been localized to the nuclear pore, we observed the localization of GFP-fusion proteins following a modified fixation protocol that allows simultaneous fixation and permeabilization of the cell (2% paraformaldehyde plus 0.2% Triton X-100). This procedure allows some of the free protein to leak out of the cell during fixation, leaving behind an increased percentage of protein that is associated with structures. This treatment results in a stronger nuclear rim staining for the full-length Nup98 (Figure 3A, e) and uncovers a low but significant pore association of the C-terminal domain (Figure 3A, c). In contrast, the N-terminal and GLFG repeat domains of Nup98 still do not show any significant association with the nuclear pore (Figure 3A, a and b). Strikingly, when the GLFG repeat domain and the C terminus are combined, the fluorescence intensity is equivalent to that of full-length protein (Figure 3A, compare d and e). When the C-terminal domain carried a mutation that blocks autocatalytic cleavage, S864A, association of this domain with the nuclear pore is lost (Figure 3A, f). Similarly, full-length Nup98 could not be found at the NPC when the S864A mutation was present (Hodel et al., 2002). The C terminus could be weakly detected at the pore after digitonin permeabilization, indicating that at least some of this domain is present on the cytoplasmic face (Griffis and Powers, unpublished data). Thus, the C terminus of Nup98 most likely contains the only sequences for direct binding to the pore, and the GLFG repeat domain acts synergistically to enhance targeting.
Figure 3.
The C terminus of Nup98 binds to the cytoplasmic nucleoporin Nup88 in vitro. (A) N-terminal (a), GLFG (b), and C-terminal (c) domains of Nup98 were expressed as GFP-fusion proteins in HeLa cells, which were then fixed and permeabilized with 2% paraformaldehyde and 0.2% Triton X-100 before imaging on an LSM 510 confocal microscope. Only the C-terminal domain produces a nuclear rim stain. A construct containing the C-terminal and GLFG domains (d) results in a rim stain as intense as the full-length protein (e). Mutation of serine 864 to alanine in the C-terminal GFP fusion completely abolishes pore targeting (f). (B) Nup214 and Ha-tagged Nup88 were in vitro transcribed and translated in the presence of [35S]methionine and then mixed with glutathione-Sepharose beads bound to GST (lane 2), GST-Nup98 C terminus (amino acids 506–920, lane 3), truncated GST-Nup98 C terminus (amino acids 506–863, lane 4), and uncleavable GST-Nup98 C terminus (amino acids 506–920 S864A, lane 5). Lane 1 represents the input translation (Ha-Nup88) or the input translation pulled down with GST-TAP (Nup214). (C) To map the domain of Nup88 required to bind to Nup98, a stop codon was inserted after amino acid 584 of Nup88 to eliminate the C-terminal coiled-coil domain. Full-length and truncated Nup88 constructs were in vitro transcribed and translated and then mixed with beads carrying GST (lanes 1 and 2), GST-Nup98 C terminus (lanes 3 and 4), the truncated GST-Nup98 C terminus (lanes 5 and 6), and uncleavable GST-Nup98 C terminus (S864A; lanes 7 and 8). Bars, 5 μm.
Nup98 Binds Nup88 In Vitro
If the C-terminal domain of Nup98 contains the sequences involved in binding at both the nuclear and cytoplasmic faces of the pore, there are several candidates for binding partners. The Nup133 complex is known to interact with the C terminus of Nup98 (Vasu et al., 2001). Within this complex, Nup98 binds directly to Nup96 (Hodel et al., 2002). The localization of the Nup133 complex within the pore is debated; it is consistently found on the nuclear side of the pore, and one group has additionally localized components of this complex to the cytoplasmic side of the pore as well, although others have not seen this localization (Belgareh et al., 2001; Vasu et al., 2001). Thus, Nup96, in addition to its role in localizing Nup98 to the nuclear face of the pore, could perhaps provide a cytoplasmic side-binding partner for Nup98. Surprisingly, in Nup98 knockout cells, proteins thought to be strictly on the cytoplasmic face of the pore, including Nup214 and Nup88, were displaced from the pore (Wu et al., 2001). This unexpected finding might be explained if one of these proteins were in fact a binding partner for Nup98 on the cytoplasmic face. To investigate this possibility, we used an in vitro binding assay to test for direct binding between the C-terminal domain of Nup98 (amino acids 505–920) and Nups 214 and 88. Figure 3B demonstrates that Nup88, but not Nup214, binds to the C-terminal domain in a GST pull-down assay. Our result is in agreement with previous reports that the yeast homologue of Nup88, ScNup82, interacts with Nup116, one of the three yeast Nup98 homologues (Bailer et al., 2000; Ho et al., 2000). This C-terminal fragment of Nup98 is active in autoproteolysis and cleaves after F863 to allow release of the peptide tail. Binding between Nup98 and Nup88 is completely inhibited by the S864A point mutation that prevents proteolytic processing of the Nup98 C terminus (Figure 3B, lane 4; Hodel et al., 2002) and blocks nuclear pore targeting in vivo (Figure 3A, f). In contrast, truncation of the GST-C terminus at the Nup98 proteolytic cleavage site (Figure 3B, lane 3) has no effect on binding to either Nup88 or to Nup96 (Hodel et al., 2002).
Nup88 can be divided into two structural domains; the N-terminal two-thirds of the protein (amino acids 1–584) has no obvious structural motifs, whereas the C-terminal one-third (amino acids 585–742) is predicted to be largely coiled-coil in structure (Fornerod et al., 1997). When the N-terminal domain of Nup88 was tested independently in the GST pull-down assay, we found that it was fully functional for binding to Nup98. Furthermore, we observed the same interaction pattern seen with the full-length Nup88 protein; the Nup88 N terminus bound only those forms of the Nup98 C-terminal domain that are functionally targeted to the nuclear pore (Figure 3C). From these data, we conclude that the N-terminal, noncoiled coil domain of Nup88 mediates interaction with Nup98.
Nup98 Interacts with Nup88 In Vivo
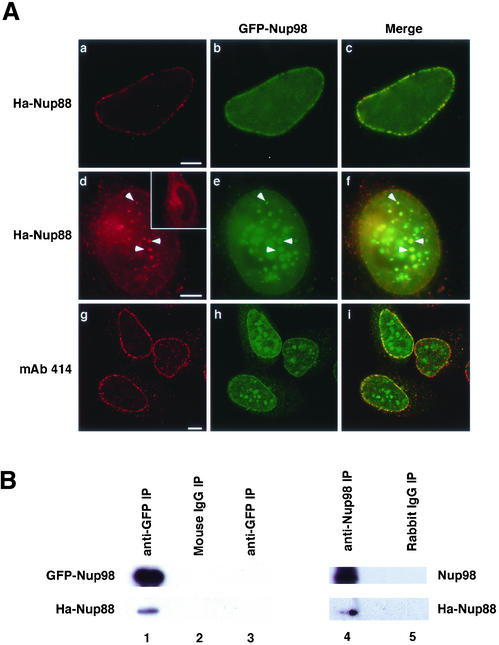
The biochemical assay described above clearly demonstrated that Nup98 and Nup88 interact in vitro. To determine whether interaction between these proteins also occurs in the cell, we investigated the localization of HA-Nup88 and GFP-Nup98 after cotransfection. At low expression levels, Nup88 and Nup98 were colocalized at the nuclear pore complex (Figure 4A, a–c). When Nup88 is overexpressed individually, even at very high levels, it is never found within the nucleoplasm (Bastos et al., 1997; Figure 4d, inset). However, in cells with high levels of Nup98 expression, HA-Nup88 could also be found in intranuclear foci together with Nup98 (Figure 4A, d–f). This was not a nonspecific effect of overexpression causing generalized mislocalization of pore proteins because immunostaining with mAb 414, which recognizes multiple nucleoporins, did not show any colocalization with Nup98 foci (Figure 4A, g–i).
Figure 4.
Nup98 and Nup88 interact in vivo. (A) HeLa cells coexpressing GFP-Nup98 and Ha-Nup88 were processed for immunofluorescence by using appropriate antibodies as detailed under MATERIALS AND METHODS. When GFP-Nup98 (b) and Ha-Nup88 (a) were expressed at low levels, they colocalized only at the nuclear rim (c). When coexpressed at high levels, (d–f) Ha-Nup88 (d) was also observed in the intranuclear foci along with GFP-Nup98 (f, arrowheads). In the absence of Nup98 overexpression, Nup88 never accumulated in the nucleus even when highly overexpressed (d, inset). Monoclonal 414 (g) did not detect any colocalization of FXFG nucleoporins in the intranuclear foci even with extremely high levels of GFP-Nup98 expression (i). Bars, 5 μm. (B) Cos1 cells expressing GFP-Nup98 and Ha-Nup88 were lysed and the lysate was immunoprecipated with anti-GFP (lane 1) or nonspecific mouse IgG (lane 2). When cells expressed only Ha-Nup88, anti-GFP antibodies did not coprecipitate any Ha-Nup88 (lane 3). Cells expressing Ha-Nup88 alone were lysed and the lysate was immunoprecipated with anti-hNup98 (lane 4) or a nonspecific rabbit IgG (lane 5).
To further confirm these results, we cotransfected plasmids encoding tagged versions of each of the two partners and tested the ability of Nup98 and Nup88 to coimmunoprecipitate (Figure 4B). Antibody to the GFP tag on Nup98 also precipitated some HA-Nup88 (Figure 4B, lane 1), an interaction that was not observed when GFP-Nup98 was not present (Figure 4B, lane 3). Similarly, when HA-Nup88 alone was transfected into cells and antibody to endogenous Nup98 was used for immunoprecipitation, HA-Nup88 was observed to interact with the Nup98 protein (Figure 4B, lane 4).
Nup96 and Nup88 Have Equivalent Requirements for Binding Nup98
Our observation that Nup88 could be mislocalized to the nucleoplasm when coexpressed with Nup98, suggested that Nup98 can actively relocate Nup88 into the nucleus. Nup98 has been previously described to be responsible for the import of Nup96 into the nucleus, after which Nup96 is incorporated into the nucleoplasmic side of the nuclear pore complex (Fontoura et al., 1999). Because Nup98 seemed to similarly escort Nup88, we asked whether the binding interaction between Nup88 and Nup98 might be analogous to the interaction between Nup96 and Nup98.
Nup98 can be produced in two distinct forms that derive from alternative splicing. The larger transcript encodes the Nup98/Nup96 polyprotein that is autocatalytically cleaved to generate the two nucleoporins. The smaller transcript encodes only Nup98, which then undergoes the same autocatalytic cleavage, resulting in the 90-kDa protein referred to as Nup98 and an 8-kDa C-terminal tail. In vitro, the tail peptide remains noncovalently associated with the 90-kDa body of Nup98 (Fontoura et al., 1999). In vivo, release of the tail peptide from the body of the protein is essential for targeting to the nuclear pore complex; uncleavable Nup98 mutants cannot bind in trans to Nup96 and do not associate with the pore (Fontoura et al., 1999; Hodel et al., 2002; Figure 5). The C-terminal tail peptide most likely acts as a competitive inhibitor of Nup96 binding because all but the last five amino acids of the 57 amino acid tail peptide are identical to the N terminus of Nup96. Intriguingly, we had observed the same pattern of interaction with respect to binding to Nup88; both the complete, proteolytically processed Nup98 C terminus and the truncated C terminus bind to Nup88, but the uncleavable mutant does not (Figure 3, B and C). This result suggested to us that the peptide might also be a competitive inhibitor of Nup88 binding and that the same binding site might be used for interaction with both Nup96 and Nup88.
Figure 5.
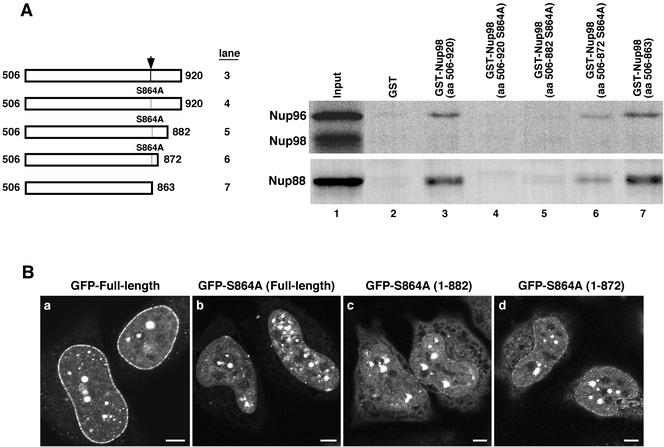
Nup88 and Nup96 bind to the same domain of Nup98. (A) Truncations were made to the uncleavable GST-Nup98 S864A mutant to remove portions of the C-terminal tail peptide. These truncations were tested in in vitro binding assays with both translated Nup96 and Ha-Nup88. The wild-type and truncated GST fusions bound both translated proteins at the same level (lanes 3 and 7, respectively). The full-length S864A mutant and the 882 truncation mutant did not bind to either Nup96 or Nup88 (lanes 4 and 5). The uncleavable mutant truncated at amino acid 872 bound some Nup88 and Nup96 (lane 6). (B) Truncation mutations were localized as GFP-Nup98 fusions in HeLa cells fixed with 2% paraformaldehyde, 0.2% Triton X-100. The wild-type protein gives the typical strong rim stain observed under these conditions (a), whereas the S864A mutant gives no appreciable rim staining (b). The 882 (c) and 872 (d) truncation mutants give progressively stronger nuclear rim stains. Bar, 5 μm.
We recently reported the crystal structure of the C-terminal domain of Nup98 (Hodel et al., 2002). In this structure, the very N terminus of the tail peptide (amino acids 864–870) could be seen to make multiple contacts with the body of Nup98. Presumably these same contacts are made between Nup98 and the N terminus of Nup96 and mediate interaction of the two proteins after cleavage of the polyprotein precursor. Indeed, when we either deleted the first nine amino acids of Nup96 or added an epitope tag to its N terminus, binding to Nup98 was abolished (Xu and Powers, unpublished data). The remainder of the tail peptide is disordered and not visible in the crystal structure; however, this region of the peptide contains a cluster of negatively charged residues (amino acids 872–877) that could potentially interact with a nearby positively charged loop in the body of the protein.
To test whether the interactions of Nup98 with Nup96 and Nup88 are truly equivalent, we truncated the 57 amino acid tail peptide after nine amino acids (506–872) or after 19 amino acids (506–882), in the context of the uncleavable S864A mutant to prevent release of the tail (Figure 5A). We then tested the ability of each of these forms of Nup98 to interact with both Nup96 and Nup88 (Figure 5A), as well as to bind to the nuclear pore in vivo (Figure 5B). An uncleavable tail of 19 amino acids was sufficient to fully inhibit binding to both Nup96 and Nup88 in vitro (Figure 5A, lane 5). Even nine amino acids, approximately the length of tail sequence that was ordered in the crystal structure, was sufficient to block most interaction between Nup98 and either of its binding partners (Figure 5A, lane 6). The same C-terminal truncations were then made in the context of the GFP-Nup98 S864A protein, and their association with the NPC was observed after transfection into cells. In vivo, the truncations of the tail peptide did allow some increased interaction of the uncleavable Nup98 with the nuclear pore complex, but this was much reduced from the stronger nuclear pore targeting observed with the wild-type protein (Figure 5B, compare c and d with a). Nuclear pore complex association was minimal with the 882 truncation (Figure 5B, c) and somewhat greater with the 872 truncation (Figure 5B, d), in keeping with the limited activity of this form in the binding assay. Taken together, both the in vitro binding data and in vivo localizations are most consistent with a model in which the same interaction site within Nup98 is used for interaction with both Nup96 and Nup88.
DISCUSSION
Understanding the spatial organization of individual proteins within the nuclear pore complex is essential for determining their specific roles in the mechanism and regulation of nuclear trafficking. Herein, we have used a combination of immunofluorescence and immunoelectron microscopy to demonstrate that Nup98 associates with both the nuclear and cytoplasmic faces of the nuclear pore complex. These distinct localizations result from interaction with two distinct binding partners, Nup96 on the nuclear side of the pore and Nup88 on the cytoplasmic side of the pore. Strikingly, the same site within the C terminus of Nup98 seems to mediate both interactions; binding to both Nup96 and Nup88 showed the same sensitivity to mutations that alter the C terminus of Nup98.
It is curious that Nup98 has previously been observed exclusively on the nuclear side of the pore complex, whereas we have repeatedly seen Nup98 on the cytoplasmic side of the pore in digitonin-permeabilized cells, and on both sides of the pore by electron microscopy (Radu et al., 1995; Vasu et al., 2001; Frosst et al., 2002). We obtained identical results in both human and Xenopus cells and with several different antibody preparations. The fact that we have not seen staining in digitonin-permeabilized cells by using one antibody to Tpr, three antibodies that recognize Nup153, and two antibodies to lamins confirms that our experimental conditions do not permeabilize the nuclear envelope (Griffis and Powers, unpublished data). We are uncertain why other groups have not detected Nup98 on the cytoplasmic face of the pore as well. Possibly the difference lies in epitope accessibility or epitope preferences of antibodies used by different investigators.
Previous studies reported that the C-terminal domain of Nup116 directs that protein to the yeast nuclear pore complex (Bailer et al., 2000; Ho et al., 2000) and indicated that the C terminus of Nup98 could function in yeast. However, it was also noted that nuclear pore targetting by the C-terminal domain of Nup116 was very inefficient compared with the full-length protein (Bailer et al., 2000). We similarly found that the C terminus of Nup98 contains the minimal pore-targeting sequence. We have further shown that the GLFG domain acts synergistically with the C-terminal domain to promote a strong nuclear pore complex interaction. The mechanism by which this synergism occurs is currently unclear. It is possible that the contribution of the GLFG domain results from interactions with nuclear transport factors and export substrates. If one role of a dynamic nucleoporin might be to aid in directing export complexes to the pore, then a mechanism that promotes the pore targeting of Nup98 when it is bound to receptor/cargo complexes would be beneficial. Alternatively, the synergy of these two domains in pore targeting could result from an interaction of the hydrophobic GLFG repeat domain with the proposed central hydrophobic phase of the nuclear pore (Ribbeck and Gorlich, 2002). In support of this, the GLFG domain alone has a very weak, but reproducible, interaction with the pore, which could only be detected by high-resolution confocal microscopy (Griffis and Powers, unpublished data). However, this minimal interaction would not seem to be sufficient to account for the very considerable difference in affinity of the C-terminal domain with or without the GLFG domain. Further experiments to address the potential role of receptors and cargo in the interaction between Nup98 and the nuclear pore will be essential.
Our results indicate that the same site within the C-terminal domain of Nup98 binds to both Nup96 and Nup88. We previously characterized the uncleavable S864A mutant by crystallography and showed that it does not differ in structure from the wild-type C-terminal domain; except for the severed peptide bond, when the tail peptide remains, it is associated in the same conformation observed in the uncleavable mutant. Therefore, the inhibitory effect of the truncated uncleavable mutants on nuclear pore binding is unlikely to occur through alteration of the overall structure. In vivo, however, the tail peptide seems to dissociate from the body of Nup98 (Hodel et al., 2002). It is most probable that in the uncleavable mutant, the nonremovable tail peptide acts as a competitive inhibitor of both Nup96 and Nup88 binding. Because the contacts between the tail peptide and Nup98 involve only a limited number of amino acids, these same residues most likely represent the binding site for both Nup96 and Nup88. Utilization of the same site for binding would seem to rule out the possibility that 98 forms a bridge or link between the two complexes. This conclusion fits with our model that 98 is a dynamic, rather than a key structural, element of the pore.
There is some conservation between the tail peptide binding site of Nup98 and the equivalent site in the three yeast homologues Nup145, Nup116, and Nup100, although only Nup145 carries out autoproteolysis to produce Nup145N and Nup145C (Hodel et al., 2002). Despite this conservation, the yeast family members show strong preferences in their binding partners. In yeast, Nup116 has been shown to bind to Nup82, the homolog of vertebrate Nup88, and consequently to localize primarily, but not exclusively, to the cytoplasmic face of the pore (Bailer et al., 2000; Ho et al., 2000). Nup100 is also found primarily on the cytoplasmic face of the pore and binds to Nup82 in a two-hybrid assay. In contrast, Nup145N did not bind to Nup82, but instead binds to Nup145C, the orthologue of vertebrate Nup96 (Ho et al., 2000). Although Nup145C is symmetrically distributed across the pore, Nup145N seems to preferentially bind at the nucleoplasmic face (Rout et al., 2000). It is possible that the conserved tail-binding site within Nups116 and 100 allows for some recognition of Nup145C, but the higher affinity of these proteins for Nup82 results in their bias toward the cytoplasmic face of the pore. It is interesting to note that again, as for binding to Rae1/Gle2 and autoproteolytic cleavage, Nup98 represents an amalgam of the properties of the yeast GLFG family.
It is somewhat challenging to reconcile the structural asymmetry of the nuclear and cytoplasmic faces of the NPC with the mostly symmetrical distribution of Nups in yeast and what may be the increasing symmetry of Nups in vertebrates. A few proteins are restricted to a single side of the pore and these alone may be responsible for much of the asymmetry of the structure. For example, recent depletion experiments suggest that Nup358 may be the single essential component of the cytoplasmic fibers (Walther et al., 2002). Additionally, symmetrically distributed Nups can be found in different subcomplexes at different positions in the pore. For example, Nup62 is found in a complex with Nup214 and Nup88 on the cytoplasmic face, with Nups 58, 54, and 45 in the central region of the pore, and with Nup93, Nup205, and Nup188 on the nuclear face (Hu et al., 1996a; Fornerod et al., 1997; Miller et al., 2000). Similarly, we have shown that Nup98 can interact with distinct complexes at different positions within the pore. On the nuclear face of the pore, Nup98 interacts with the Nup133 complex; all evidence suggests that Nup88 is not present on the nuclear face of the pore. On the cytoplasmic face, Nup98 can bind to the Nup214 complex via Nup88. Our current data cannot rule out that Nup98 also interacts with the Nup133 complex on the cytoplasmic side of the pore.
Like Nup98, Nup153 has been reported to be dynamically associated with the nuclear pore (Nakielny et al., 1999; Daigle et al., 2001). Nup153 has a well established localization on the nuclear face of the pore complex where it binds to the same Nup133 subcomplex as does Nup98, although the direct binding partner of Nup153 remains to be identified (Vasu et al., 2001). It is not clear whether Nup153 is ever released into the cytoplasm or if it moves off the pore only into the nucleus. In contrast to Nup98, Nup153 is not accessible to antibodies after fixation and digitonin permeabilization of cells (Bastos et al., 1996; Nakielny et al., 1999). Thus, it seems that the dynamic roles played by Nup98 and Nup153 are distinct. Nup98 transits the pore, with sites of association on both faces and can exit the pore into either the nuclear or cytoplasmic compartments. In contrast, Nup153 may not associate with the cytoplasmic face of the pore or may have only a very transient exposure to the cytoplasm.
Interestingly, Nup153 seems to significantly contribute to the structural organization of the nuclear basket, a role that is somewhat difficult to reconcile with its dynamic association with the pore. When Nup153 was depleted from Xenopus extracts, the reconstituted nuclei were lacking several other nucleoporins, including Nup98, Nup93, and Tpr, and the structure of the nuclear face of the pore was substantially altered although cytoplasmically oriented nucleoporins were unaffected (Walther et al., 2001). One report has suggested an essential structural role in the pore for Nup98 as well. Disruption of the Nup98 gene in the mouse resulted in early embryonic lethality. However, on examination of cultured embryonic cells from this knockout, Wu et al. (2001) found that the absence of Nup98 led to loss of multiple proteins from the pore, primarily those with cytoplasmic orientation (Nup358, Nup214, Nup88, Nup62). This was unexpected as no previous evidence had connected Nup98 with the cytoplasmic face of the pore. Our results demonstrate that there is indeed a specific interaction between Nup98 and one or, possibly two, complexes on the cytoplasmic side of the pore. However, this does not yet provide a fully satisfying explanation for the phenotype observed in the cells from the Nup98 knockout mouse, and it is again puzzling to reconcile both a structural and a dynamic role. Possibly Nup98 plays a role in regulating dynamic organization of the cytoplasmic structures of the pore, as has been proposed for Nup153 on the nuclear side.
Intriguingly, the result observed in the Nup98 knockout cells might be related to a phenotype previously characterized for aberrant expression of Nup116 in yeast (Ho et al., 2000). Overexpression of the C-terminal domain of Nup116 in a wild-type background had no phenotype. In contrast, overexpression of this domain in a Nup116 null cell was lethal and led rapidly to displacement of Nup82 from the nuclear pore. In the Nup98 knockout mouse, the absence of Nup98 was demonstrated by immunoblot by using an antibody to the Rae1/Gle2 binding site near the N terminus. However, Nup96, the downstream half of the Nup98/Nup96 polyprotein was expressed in these cells and processed to the correct molecular weight (Wu et al., 2001). The autocatalytic cleavage that produces Nup96 requires >200 amino acids at the C-terminal domain of Nup98 to fold into the active autocatalytic domain (Rosenblum and Blobel, 1999; Hodel et al., 2002). Therefore, the presence of Nup96 suggests the, as yet untested, possibility that a portion of the C-terminal domain of Nup98 is expressed in the knockout mouse. Indeed, multiple alternatively spliced versions of Nup98/Nup96 have been reported, lending some support to this possibility (Fontoura et al., 1999; Enninga et al., 2002).
In summary, we have demonstrated that Nup98, which transits the nuclear pore complex, has sites of association on both the nuclear and cytoplasmic faces of the pore. Nup98 has two different binding partners, Nup96 on the nuclear side and Nup88 on the cytoplasmic side. Because the same site is used for both of these interactions, it is unlikely that Nup98 acts to form a bridge between the separate subcomplexes containing the two partner proteins. Our results provide further insight into the workings of a dynamic nucleoporin, but clearly there is much yet to be understood about the role of Nup98 in the organization and function of the nuclear pore.
ACKNOWLEDGMENTS
We thank Erica Phillips and Melanie Blevins for excellent technical assistance and advice, and Hong Yi (Emory Neuroscience Microscopy Facility) for expert assistance with electron microscopy. We are grateful to Dr. Alec Hodel for helpful discussions, and Drs. Victor Faundez, Katharine Ullman, Win Sale, and Barry Shur for helpful discussions and comments on the manuscript. The Xenopus lamins II and III mAb developed by Dr. Michael Klymkowsky were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa (Department of Biological Sciences). We thank Dr. Beatriz Fontoura for the Nup98/96 plasmid and Dr. Brian Burke for the Nup88 plasmid. This work was supported by grant GM-59975 from the National Institutes of Health (to M. A. P.). E.R.G. is a predoctoral trainee of the National Institutes of Health (T32 GM08367-13).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–09–0582. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–09–0582.
REFERENCES
- Allen TD, Cronshaw JM, Bagley S, Kiseleva E, Goldberg MW. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J Cell Sci. 2000;113:1651–1659. doi: 10.1242/jcs.113.10.1651. [DOI] [PubMed] [Google Scholar]
- Bailer SM, Balduf C, Katahira J, Podtelejnikov A, Rollenhagen C, Mann M, Pante N, Hurt E. Nup116p associates with the Nup82p-Nsp1p-Nup159p nucleoporin complex. J Biol Chem. 2000;275:23540–23548. doi: 10.1074/jbc.M001963200. [DOI] [PubMed] [Google Scholar]
- Bailer SM, Siniossoglou S, Podtelejnikov A, Hellwig A, Mann M, Hurt E. Nup 116p and Nup 100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor Gle2p. EMBO J. 1998;17:1107–1119. doi: 10.1093/emboj/17.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R, Lin A, Enarson M, Burke B. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J Cell Biol. 1996;134:1141–1156. doi: 10.1083/jcb.134.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R, Ribas de Pouplana L, Enarson M, Bodoor K, Burke B. Nup84, a novel nucleoporin that is associated with CAN/Nup214 on the cytoplasmic face of the nuclear pore complex. J Cell Biol. 1997;137:989–1000. doi: 10.1083/jcb.137.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle N, Beaudouin J, Hartnell L, Imreh G, Hallberg E, Lippincott-Schwartz J, Ellenberg J. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J Cell Biol. 2001;154:71–84. doi: 10.1083/jcb.200101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emtage JL, Bucci M, Watkins JL, Wente SR. Defining the essential functional regions of the nucleoporin Nup145p. J Cell Sci. 1997;110:911–925. doi: 10.1242/jcs.110.7.911. [DOI] [PubMed] [Google Scholar]
- Enninga J, Levy DE, Blobel G, Fontoura BM. Role of nucleoporin induction in releasing an mRNA nuclear export block. Science. 2002;295:1523–1525. doi: 10.1126/science.1067861. [DOI] [PubMed] [Google Scholar]
- Fontoura BM, Blobel G, Matunis MJ. A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J Cell Biol. 1999;144:1097–1112. doi: 10.1083/jcb.144.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Deursen J v, Baal S v, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. The human homologue of yeast C.R.M1 is in a dynamic subcomplex with C.A.N/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosst P, Guan T, Subauste C, Hahn K, Gerace L. Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J Cell Biol. 2002;156:617–630. doi: 10.1083/jcb.200106046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Dang T, Pane N, Shevchenko A, Mann M, Forbes D, Hurt E. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol Biol Cell. 1997;8:2017–2038. doi: 10.1091/mbc.8.10.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. Nup98 Is a Mobile Nucleoporin with Transcription-dependent Dynamics. Mol Biol Cell. 2002;13:1282–1297. doi: 10.1091/mbc.01-11-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan T, Kehlenbach RH, Schirmer EC, Kehlenbach A, Fan F, Clurman BE, Arnheim N, Gerace L. Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol Cell Biol. 2000;20:5619–5630. doi: 10.1128/mcb.20.15.5619-5630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho AK, Shen TX, Ryan KJ, Kiseleva E, Levy MA, Allen TD, Wente SR. Assembly and preferential localization of Nup116p on the cytoplasmic face of the nuclear pore complex by interaction with Nup82p. Mol Cell Biol. 2000;20:5736–5748. doi: 10.1128/mcb.20.15.5736-5748.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel A, Hodel M, Griffis E, Hennig K, Ratner G, Xu S, Powers M. The three-dimensional structure of the autoproteolytic, nuclear pore-targeting domain of the human nucleoporin nup98. Mol Cell. 2002;10:347. doi: 10.1016/s1097-2765(02)00589-0. [DOI] [PubMed] [Google Scholar]
- Hu T, Guan T, Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J Cell Biol. 1996a;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T, Guan T, Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J Cell Biol. 1996b;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer D, Wozniak RW, Blobel G, Radu A. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc Nat Acad Sci USA. 1994;91:1519–1523. doi: 10.1073/pnas.91.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BR, Powers M, Park M, Fischer W, Forbes DJ. Identification of a new vertebrate nucleoporin, nup188, with the use of a novel organelle trap assay [In Process Citation] Mol Biol Cell. 2000;11:3381–3396. doi: 10.1091/mbc.11.10.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R, Watkins JL, Wente SR. GLE2, a Saccharaomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol Biol Cell. 1996;7:1921–1937. doi: 10.1091/mbc.7.12.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Shaikh S, Burke B, Dreyfuss G. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 1999;18:1982–1995. doi: 10.1093/emboj/18.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MA, Macaulay C, Masiarz FR, Forbes DJ. Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J Cell Biol. 1995;128:721–736. doi: 10.1083/jcb.128.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard CE, Fornerod M, Kasper LH, van Deursen JM. RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J Cell Biol. 1999;145:237–254. doi: 10.1083/jcb.145.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Blobel G, Wozniak RW. Nup155 is a novel nuclear pore complex protein that contains neither repetitive sequence motifs nor reacts with WGA. J Cell Biol. 1993;121:1–9. doi: 10.1083/jcb.121.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Ribbeck K, Gorlich D. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 2002;21:2664–2671. doi: 10.1093/emboj/21.11.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum JS, Blobel G. Autoproteolysis in nucleoporin biogenesis. Proc Natl Acad Sci USA. 1999;96:11370–11375. doi: 10.1073/pnas.96.20.11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KJ, Wente SR. The nuclear pore complex: a protein machine bridging the nucleus and cytoplasm. Curr Opin Cell Biol. 2000;12:361–371. doi: 10.1016/s0955-0674(00)00101-0. [DOI] [PubMed] [Google Scholar]
- Stoffler D, Fahrenkrog B, Aebi U. The nuclear pore complex: from molecular architecture to functional dynamics. Curr Opin Cell Biol. 1999;11:391–401. doi: 10.1016/S0955-0674(99)80055-6. [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Siniossoglou S, Podtelejnikov S, Benichou JC, Mann M, Dujon B, Hurt E, Fabre E. Two functionally distinct domains generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins. EMBO J. 1997;16:5086–5097. doi: 10.1093/emboj/16.16.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu S, Shah S, Orjalo A, Park M, Fischer WH, Forbes DJ. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J Cell Biol. 2001;155:339–354. doi: 10.1083/jcb.200108007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu SK, Forbes DJ. Nuclear pores and nuclear assembly. Curr Opin Cell Biol. 2001;13:363–375. doi: 10.1016/s0955-0674(00)00221-0. [DOI] [PubMed] [Google Scholar]
- Walther TC, Fornerod M, Pickersgill H, Goldberg M, Allen TD, Mattaj IW. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 2001;20:5703–5714. doi: 10.1093/emboj/20.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Pickersgill HS, Cordes VC, Goldberg MW, Allen TD, Mattaj IW, Fornerod M. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J Cell Biol. 2002;158:63–77. doi: 10.1083/jcb.200202088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Kasper LH, Mantcheva RT, Mantchev GT, Springett MJ, van Deursen JM. Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proc Natl Acad Sci USA. 2001;98:3191–3196. doi: 10.1073/pnas.051631598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]