Abstract
We have used GST pulldowns from A431 cell cytosol to identify three new binding partners for the γ-adaptin appendage: Snx9, ARF GAP1, and a novel ENTH domain-containing protein, epsinR. EpsinR is a highly conserved protein that colocalizes with AP-1 and is enriched in purified clathrin-coated vesicles. However, it does not require AP-1 to get onto membranes and remains membrane-associated in AP-1–deficient cells. Moreover, although epsinR binds AP-1 via its COOH-terminal domain, its NH2-terminal ENTH domain can be independently recruited onto membranes, both in vivo and in vitro. Brefeldin A causes epsinR to redistribute into the cytosol, and recruitment of the ENTH domain requires GTPγS, indicating that membrane association is ARF dependent. In protein-lipid overlay assays, the epsinR ENTH domain binds to PtdIns(4)P, suggesting a possible mechanism for ARF-dependent recruitment onto TGN membranes. When epsinR is depleted from cells by RNAi, cathepsin D is still correctly processed intracellularly to the mature form. This indicates that although epsinR is likely to be an important component of the AP-1 network, it is not necessary for the sorting of lysosomal enzymes.
INTRODUCTION
Epsin N-terminal homology (ENTH) domains have been found in a number of proteins implicated in membrane traffic. Epsin itself was originally discovered as a binding partner for Eps15, an EH domain-containing protein first identified as a substrate for the EGF receptor tyrosine kinase (Chen et al., 1998). Epsin and Eps15 bind not only to each other, they also both bind to the COOH-terminal appendage domain of the α-adaptin subunit of the AP-2 adaptor complex, which facilitates clathrin-mediated endocytosis. More recently two epsin homologues have been identified in mammals and named epsins 2 and 3 (Rosenthal et al., 1999; Spradling et al., 2001). All three epsins have a similar domain organization, consisting of an NH2-terminal ENTH domain, a ubiquitin interaction motif (UIM), and a COOH-terminal domain with little or no conventional secondary structure, but containing a series of motifs including DPW motifs for binding to the α appendage, NPF motifs for binding to Eps15, and clathrin-binding motifs. Yeast contains two homologues of epsin, Ent1p and Ent2p, as well as two other ENTH domain-containing proteins, Ent3p and Ent4p (Wendland et al., 1999). Like the mammalian epsins, Ent1p and Ent2p have not only ENTH domains, but also UIMs, NPFs, and clathrin-binding motifs. Studies in both mammalian cells and yeast indicate that the epsins and Ent1p/Ent2p are essential for clathrin-mediated endocytosis, although their precise role is less clear. Proposed functions include bringing different components of the coat together (Kalthoff et al., 2002), recruiting ubiquitinated cargo into coated pits (Riezman, 2002) and helping to change the curvature of the membrane during vesicle formation (Ford et al., 2002).
The ENTH domain of epsin 1 has been shown to bind to PtdIns(4,5)P2 (PIP2; Itoh et al., 2001). The AP-2 adaptor complex and several of the other AP-2–associated proteins, including AP-180/CALM and dynamin, can also bind PIP2, although structural studies show that each one does so in a different manner (Itoh et al., 2001; Collins et al., 2002; Ford et al., 2002). Because PIP2 is primarily generated on the plasma membrane, it has been proposed that these interactions help to target these proteins to the appropriate location. This hypothesis is supported by mutagenesis studies, which show that proteins with mutations in their PIP2 binding sites fail to localize properly (Gaidarov and Keen, 1999; Ford et al., 2002).
Epsin, AP180/CALM, and dynamin are just some of the many proteins involved in endocytosis that bind either directly or indirectly to the α appendage (Slepnev and De Camilli, 2000). Most of the direct binding partners have one or more DPF/DPW motifs, although in vitro studies indicate that any protein containing the consensus sequence DΦF can potentially bind to the α appendage (Owen et al., 2000). The β2 appendage is structurally very similar to the α appendage, and the two appendages have been shown to share some of the same binding partners, at least in vitro (Owen et al., 2000). Recently the structures of two other appendage domains have been solved: the AP-1 γ appendage and the GGA1 appendage (Kent et al., 2002; Lui et al., 2003; Nogi et al., 2002). The GGAs are monomeric adaptors that facilitate cargo selection at the TGN (Robinson and Bonifacino, 2001). AP-1 was also initially assumed to act at the TGN, although recent studies suggest that it may be more important in retrieving cargo from a post-TGN compartment (Meyer et al., 2000, 2001; Valdivia et al., 2002). The γ and GGA appendages are structurally nearly identical to each other, and they are also very similar to the NH2-terminal subdomains of the α and β2 appendages (Lui et al., 2003). However, the accessory protein binding sites on the α and β2 appendages, as mapped by mutagenesis, are not in their NH2-terminal subdomains but in their COOH-terminal subdomains (Owen et al., 1999, 2000). So far very few partners have been found for the γ and GGA appendages, and the consensus sequence(s) for binding has not yet been identified. The only protein that clearly binds to the γ appendage in vivo as well as in vitro is γ-synergin, an EH domain-containing protein of unknown function (Page et al., 1999). Several other proteins have been shown to bind to the γ appendage in vitro, including rabaptin-5 and Eps15, although the physiological relevance of these interactions is less clear. The GGAs also bind γ-synergin in vitro although apparently not in vivo, as well as a bona fide partner, p56, another protein of unknown function (Lui et al., 2002).
The approach that we have used in the past to search for binding partners for the γ and GGA appendages has been to carry out GST pulldowns using pig brain cytosol, followed by MALDI-TOF or nanoelectrospray mass spectrometry (Hirst et al., 2000; Lui et al., 2003). In this way we have been able to identify all of the major bands in the pulldowns, but only γ-synergin and p56 have been shown definitively to bind to the γ and GGA appendages under more physiological conditions. However, brain may not be the best source of γ appendage binding partners, because it is highly enriched in proteins involved in synaptic vesicle recycling and relatively depleted in proteins involved in other trafficking pathways. Therefore, we have now carried out GST pulldowns using human tissue culture cell cytosol and rat liver clathrin-coated vesicle extracts to try to identify additional γ appendage binding partners. Among other proteins, we find a novel ENTH domain–containing protein. Here we present an initial characterization of this protein, which we propose should be called epsinR. We show that it colocalizes with AP-1 by immunofluorescence, and we investigate how it interacts with the γ appendage. We also examine how epsinR is recruited onto membranes, both by expressing and localizing truncated versions of the protein and by manipulating other proteins and lipids in the cell. Finally, we investigate the function of epsinR using RNA interference.
MATERIALS AND METHODS
Plasmid Construction and GST Pulldowns
Standard molecular biology techniques were used throughout this study (Sambrook and Russell, 2001). For pulldown experiments, GST-appendage domain fusion proteins were made and pulldowns were carried out essentially as previously described (Hirst et al., 2000), using A431 cytosol prepared in PBS containing 0.1% NP-40 and a protease inhibitor cocktail (Complete Mini), at a protein concentration of 1.5 mg/ml. An epsinR construct was also made, using PCR to amplify a fragment containing amino acids 324–428, which was then ligated into pGEX-4T-1 (Pharmacia, Piscataway, NJ). Bound proteins were eluted with sample buffer and subjected to SDS-PAGE. Gels were either stained with Coomassie blue for matrix-assisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometry or transferred onto nitrocellulose for Western blotting.
MALDI-TOF Mass Spectrometry
Coomassie blue–stained gel bands were excised, washed, in-gel digested with trypsin, and subjected to MALDI-TOF (Jensen et al., 1997). All samples were analyzed with a Voyager-DE STR mass spectometer (Perseptive Biosystems, League City, TX). Database searches using peptide masses were performed with the Mascot program (http://www.matrixscience.com). Eleven tryptic peptides were matched to human ARF GAP1 (sequence coverage, 33%) from the 40-kDa band. In the case of the 75-kDa band, 17 peptides matched Snx9 (sequence coverage, 35%), and tryptic peptides matched KIAA0171 (epsinR; sequence coverage, 13%). From the clathrin-coated vesicle sample, 11 peptides from the 75-kDa band matched mouse epsinR (gi:13278582) (sequence coverage, 22%), and 9 peptides from the 50-kDa band matched mouse epsinR (sequence coverage, 20%). From the pulldown using the epsinR construct, 13 peptides from the 170-kDa band matched human clathrin heavy chain (sequence coverage, 7%), 21 peptides from the 110-kDa band matched β1 (sequence coverage, 17%), and 15 peptides from the 95-kDa band matched γ (sequence coverage, 17%).
Antibodies and Blotting
Antibodies against Snx9, rat ARF GAP1, and Eps15 were kind gifts from Carl Blobel (Howard et al., 1999), Dan Cassel (Cukierman et al., 1995), and Paolo Di Fiore (Chen et al., 1998), respectively. Antibodies against the Xpress epitope (Invitrogen, Carlsbad, CA), GM130 (BD Transduction Labs, Lexington, KY), γ-adaptin (mAb 100/3; Sigma Chemical, St. Louis, MO), and GFP (Qbiogene, Carlsbad, CA) were purchased from the manufacturers. Rabbit polyclonal antibodies against γ-synergin, γ-adaptin, and brain-specific α-adaptin were raised in-house and have already been described (Seaman et al., 1993; Page et al., 1999). To raise antibodies against epsinR, the KIAA0171 cDNA clone was obtained from the Kazusa Institute, and amino acids 165–625 were amplified by PCR and ligated into pGEX-4T-1 (Pharmacia Biotechnology, Piscataway, NJ). We also raised an antibody against Snx9 by amplifying the full-length coding sequence from a plasmid kindly provided by Carl Blobel and ligating it into pTrcHisA (Invitrogen). Constructs were expressed in MC1061 cells and purified according to the manufacturers' instructions. The immunization protocol and affinity purification of the resulting antisera were performed as previously described (Hirst et al., 2000).
Western blots were probed with various antibodies, followed by 125I-protein A as previously described (Hirst et al., 2000). Samples included GST pulldowns, whole cell extracts, clathrin-coated vesicles purified from rat liver (Page et al., 1999), and samples from in vitro recruitment experiments (see below). Ligand blotting was also carried out, using various GST fusion proteins (described above) followed by anti-GST and 125I-protein A (Hirst et al., 2000). Samples analyzed by ligand blotting included purified clathrin-coated vesicles, the His/Xpress-tagged full-length Snx9 described above, and epsinR NH2-terminal (amino acids 1–165) and COOH-terminal (amino acids 165–625) domains and the GAP1 COOH-terminal domain (124–406), all prepared in pTrcHisA. For protein-lipid overlay assays, phosphoinositide arrays were purchased from Echelon (Echelon Biosciences, Salt lake City, Utah) and probed according to the manufacturer's instructions, using a GST-epsinR ENTH domain construct (amino acids 1–165) amplified by PCR, and ligated into pGEX-4T-1.
Transfection, Recruitment, and Immunolocalization
GFP-epsinR constructs were prepared by amplifying either the full-length protein or the NH2-terminal and COOH-terminal domains by PCR and ligating the products into pEGFP-C1 (Clontech, Palo Alto, CA). The GFP-γ appendage construct has already been described (Lui et al., 2002). Constructs were transfected into COS cells using Fugene 6 (Roche, Nutley, NJ), and expression was monitored after 12–17 h. For immunofluorescence, both transfected and nontransfected COS cells were fixed either with 3% paraformaldehyde, followed by permeabilization with 0.1% saponin, or with methanol/acetone (Seaman et al., 1993). For some experiments, the cells were treated with 100 μg/ml BFA for 2 min or with 20 μg/ml nocodazole for 2 h before fixation. Recruitment experiments were carried out on permeabilized NRK cells using pig brain cytosol (Seaman et al., 1993; West et al., 1997), supplemented with 250 μg/ml recombinant His/Xpress-tagged epsinR ENTH domain prepared as described above, either with or without 100 μM GTPγS and/or 1 mM neomycin. Primary antibodies are described above; secondary antibodies were purchased from Molecular Probes (Eugene, OR). Cells were viewed using a Zeiss Axiophot fluorescence microscope (Thornwood, NY) equipped with a CCD camera (Princeton Instruments, Monmouth, NJ) and photographs were recorded using IP Labs software (Fairfax, VA). Live cell imaging of cells expressing full-length GFP-epsinR was carried out using a Zeiss Axiovert 135T4 microscope with a Perkin-Elmer Cetus (Norwalk, CT) spinning disk confocal attachment. Frames were acquired every 3.25 s for a total of 139 s.
For immunogold localization of GFP, COS cells transiently transfected with either full-length GFP-epsinR or GFP-epsinR ENTH domain were fixed for 1 h with 8% paraformaldehyde in 0.1 M sodium cacodylate, pH 7.2, at room temperature, pelleted, and embedded in gelatin. The cells were then prepared for ultrastructural immunocytochemistry as previously described (Hirst et al., 2000), using rabbit anti-GFP followed by protein A coupled to 15-nm gold. Sections were observed in a Philips CM100 transmission electron microscope (Mahwah, NJ).
RNA Interference
siRNA duplexes designed against conserved sequences of the target cDNAs were purchased from Dharmacon (Lafayette, CO). A μ2 sequence, which had previously been found to be ineffective for knockdown experiments, was used as a control. The sequences were as follows: AACACAGCAACCUCUACUUGG (control); AAGUGCCAGAGAACACAUUUA (epsinR); and AAGGCAUCAAGUAUCGGAAGA (μ1A). HeLa and COS cells were transfected using Oligofectamine (Invitrogen) as specified by the manufacturer. The transfection mixture was left on the cells for 4 h, after which 1 ml DMEM/20%FCS was added to each well. For μlA, two transfections 3 d apart were performed for efficient knockdown, and experiments were carried out 3 d after the second knockdown. For epsinR, experiments were carried out 3 d after a single transfection. Cathepsin D sorting was assayed essentially as described by Davidson (1995), using 5 μl 35S Promix (Amersham, Piscataway, NJ) per ml medium. The cells were pulse labeled for 15 min and chased for 2.5 h in the presence of 5 mM mannose 6-phosphate before immunoprecipitation using rabbit anti-human cathepsin D (DAKO, Carpinteria, CA). Quantifications were carried out using a Packard Cyclone phosphorimager (Downers Grove, IL).
RESULTS
Identification of New Binding Partners for the γ Appendage
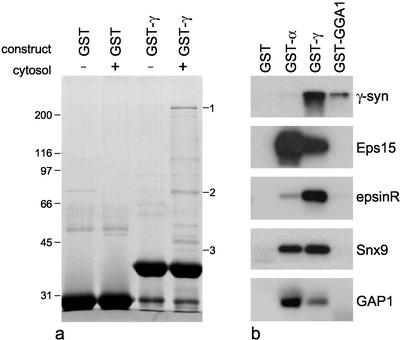
Figure 1a shows a Coomassie blue–stained gel of a GST pulldown from A431 cell cytosol using a γ appendage domain construct and GST alone as a control. Several bands come down with the fusion protein but not with the control, including some that have different mobilities from those that are pulled down from pig brain cytosol (see Hirst et al., 2000; Lui et al., 2002). By MALDI-TOF mass spectrometry, we found matches for bands 1, 2, and 3. Band 1 matched KIAA1414 or p200, a novel protein of unknown function that we have also found in pulldowns from pig brain cytosol (Lui et al., 2003). Band 2 was found to match two proteins: KIAA0171, a novel ENTH domain-containing protein; and Snx9 (or SH3PX1). Band 3 was matched with FLJ10767, the human ortholog of rat ARF GAP1.
Figure 1.
Identification of new binding partners for the γ appendage domain. (a) GST alone or a GST-γ appendage construct was incubated either with or without cytosol from A431 cells. Samples were subjected to SDS PAGE, the gel was stained with Coomassie blue, and the indicated bands were analyzed by MALDI-TOF mass spectometry. The matches were as follows: band 1, KIAA1414 or p200; band 2, two proteins, KIAA0171 or epsinR and Snx9; and band 3, FLJ10767 or human ARF GAP1. (b) Pulldowns were performed on A431 cell cytosol using either GST alone, GST fused to the α appendage, GST fused to the γ appendage, or GST fused to the GGA1 appendage. Blots were probed with the indicated antibodies.
Although both Snx9 and GAP1 have been implicated in membrane traffic, they have not previously been shown to bind to the γ appendage. However, because the physiological relevance of these interactions is not yet clear (see below), in the present study we focus mainly on the ENTH domain-containing protein, KIAA0171. This protein has also been entered into the database as epsin 4, although at the time this manuscript was submitted no papers had been published on it (but see DISCUSSION). Unlike epsins 1–3, however, KIAA0171 contains no NPF sequences, indicating that it does not interact with Eps15, and no UIMs. It also differs from epsins 1–3 in that it contains only a single DPW motif, which is within the ENTH domain and therefore unlikely to bind to the α appendage. Indeed, KIAA0171 is related to epsins 1–3 only in that it has an NH2-terminal ENTH domain and a COOH-terminal domain with low secondary structure predictions. Therefore, we feel that the name epsin 4 is misleading and propose that the protein should instead be called epsinR, for epsin-related protein. EpsinR is a highly conserved protein, with probable orthologues in Drosophila (AAF43421), Caenorhabditis elegans (C34E11) and Arabidopsis (AAL67001) as well as a possible ortholog in Saccharomyces cerevisiae, Ent3p.
To confirm the identities of the proteins identified in the pulldown and to determine whether they are also able to bind to the α or GGA appendage domains, we used GST fusions of all three appendages, together with a GST control, to pull down proteins from A431 cell cytosol, and then probed Western blots of the pulldowns with antibodies against Snx9 and GAP1 and with a new antibody that we have raised against recombinant epsinR. Eps15 and γ-synergin antibodies were included as positive controls. Figure 1b shows that all five of these proteins come down with the GST-γ appendage domain construct. γ-Synergin is also pulled down by the GGA1 appendage but not by the α appendage, as previously reported (Hirst et al., 2000). In contrast, Eps 15 interacts most strongly with the α appendage and not at all with the GGA1 appendage. The other three proteins identified in this study all interact to varying degrees with both the γ and the α appendage. EpsinR shows a strong preference for the γ appendage; similar amounts of Snx9 are pulled down with both the γ and α appendages, and GAP1 shows a preference for the α appendage (surprisingly, because AP-2 is thought to function independently of ARF).
Enrichment in Clathrin-coated Vesicles
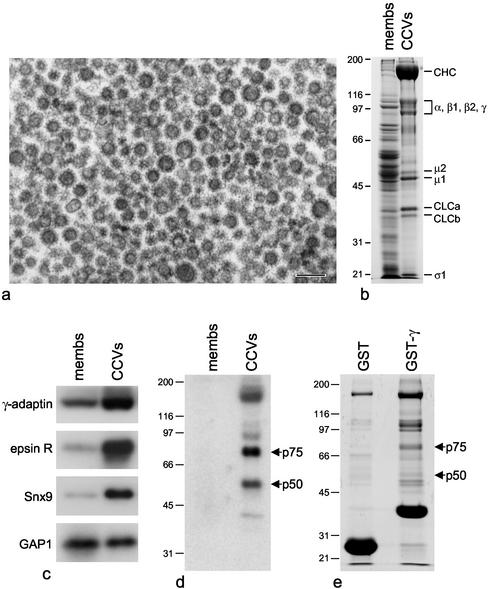
We next investigated whether epsinR, Snx9, and GAP1 are enriched in purified clathrin-coated vesicles and whether we could use coated vesicle preparations to find additional γ appendage binding partners. Figure 2a shows an electron micrograph of clathrin-coated vesicles purified from rat liver, and Figure 2b shows a gel of the purified coated vesicles next to an equal protein loading of crude membranes from an earlier stage in the preparation. Figure 2c shows blots of crude membranes and purified clathrin-coated vesicles probed with antibodies against epsinR, Snx9, and GAP1. Anti–γ-adaptin was included as a positive control. It is clear that both epsinR and Snx9 are strongly enriched in the purified coated vesicles, whereas GAP1 is neither enriched nor depleted.
Figure 2.
Binding partners for the γ appendage domain in rat liver clathrin-coated vesicles. (a) Electron micrograph of a conventional thin section of clathrin-coated vesicles purified from rat liver. Scale bar, 200 nm. (b) SDS PAGE showing the purified clathrin-coated vesicles next to an equal protein loading of crude membranes from an earlier stage in the preparation. Clathrin heavy and light chains (CHC and CLC) and subunits of the AP-1 and AP-2 adaptor complexes are indicated. (c) Crude membranes and purified clathrin-coated vesicles were analyzed by Western blotting, using antibodies against γ-adaptin, epsinR, Snx9, and ARF GAP1. γ-Adaptin, epsinR, and Snx9 are all enriched in the purified coated vesicles. (d) Blots of crude membranes and purified clathrin-coated vesicles were probed with the GST-γ appendage domain construct followed by anti-GST. Two labeled bands, p75 and p50, are strongly enriched in the purified coated vesicles. (e) GST pulldowns of coated vesicle extracts. By MALDI-TOF mass spectrometry, both p75 and p50 were found to match epsinR.
We also probed Western blots of coated vesicles and crude membranes with the γ appendage domain itself. Figure 2d shows that appendage-binding proteins of ∼75 and ∼50 kDa (p75 and p50) are highly enriched in clathrin-coated vesicles. To concentrate these proteins and to determine their identities, we carried out GST pulldowns on Tris-HCl extracts of the coated vesicles. Figure 2e shows that in addition to clathrin and the adaptor complexes, which also come down (although to a lesser extent) with GST alone, the γ appendage construct brings down bands of ∼75 and ∼50 kDa. Overlays confirmed that we had brought down most of the p75 and p50 from the starting material (unpublished data). When the two bands were subjected to MALDI-TOF mass spectrometry, the closest match for both of them was found to be epsinR. The 50-kDa band is presumably a breakdown product of the 75-kDa band. Thus, epsinR appears to be the strongest and/or most abundant γ appendage binding partner in clathrin-coated vesicles.
Demonstration of Direct Binding
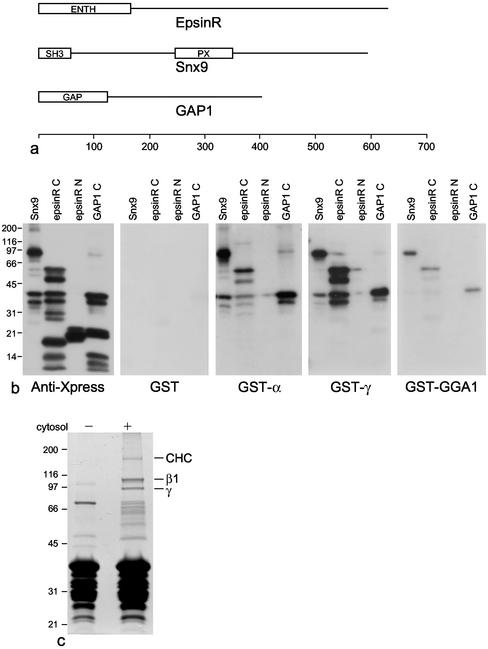
The overlay experiment shown in Figure 2d suggests that epsinR binds directly to the γ appendage; however, we cannot completely rule out the possibility that the labeling might be due to minor components of the 75- and 50-kDa bands. To show formally that epsinR binds directly to the γ appendage and to determine whether Snx9 and GAP1 bind directly as well, we expressed all three proteins in Escherichia coli with NH2-terminal His/Xpress tags and purified them by Ni-NTA chromatography for overlay studies. The full-length epsinR construct was found to be extremely protease sensitive, so the NH2-terminal ENTH domain (amino acids 1–165) and the COOH-terminal domain (amino acids 165–625) were expressed separately (Figure 3a). GAP1 was also made as a smaller protein, omitting the NH2-terminal GAP domain. The four constructs were subjected to SDS PAGE followed by Western blotting, and the blots were probed either with anti-Xpress or with various GST constructs followed by anti-GST (Figure 3b).
Figure 3.
Binding to appendage domains analyzed by ligand blotting and pulldown with an epsinR domain. (a) Schematic diagrams of epsinR, Snx9, and GAP1, showing the positions of the various domains. (b) Snx9, the epsinR COOH-terminal domain (epsinR C), the epsinR NH2-terminal ENTH domain (epsinR N), and the GAP1 COOH-terminal domain (GAP1 C) were expressed as NH2-terminally His/Xpress-tagged constructs and analyzed by Western blotting. Blots were probed either with anti-Xpress or with the indicated GST constructs. The ability of the appendage domains to bind on blots indicates that they all recognize short linear motifs. (c) A construct consisting of GST fused to amino acids 324–428 of epsinR was used to pull down proteins from A431 cell cytosol, the gel was stained with Coomassie blue, and selected bands were analyzed by MALDI-TOF mass spectrometry. The indicated bands were found to contain clathrin heavy chain (CHC) and the AP-1 β1 and γ subunits.
The anti-Xpress labeling shows that all of the constructs except for the NH2-terminal ENTH domain of epsinR are protease sensitive and appear as multiple bands. These same three constructs, but not the ENTH domain construct, all bind on overlays to the three fusion proteins. However, there are differences in the relative amounts of labeling. As predicted from the pulldowns, GST-γ labels the epsinR construct especially strongly, whereas GST-α labels this construct more weakly than the others. GST-GGA1 gives weak labeling of all three constructs. The proteolysis of the constructs, although unintentional, provides a rough map of where the appendage binding site(s) are located. It is interesting to note that the α and γ appendage domains label the same degradation products with similar (although not identical) relative intensities, suggesting that they may bind to either similar or adjacent motifs on the constructs.
Because of the strong interaction between the epsinR COOH-terminal domain and the γ appendage, we also constructed various GST-epsinR fusion proteins and used them to try to pull down γ-adaptin from A431 cell cytosol. The construct that produced the strongest signal by Western blotting consisted of amino acids 324–428. Clathrin heavy chain, β1, and μ1 could also be detected in the pulldowns by Western blotting but not GGA1 or GGA2 (unpublished data). When the pulldowns were stained with Coomassie blue, three prominent high-molecular-weight bands could be seen: a band of ∼170 kDa and two very strong bands of ∼110 and ∼95 kDa. These bands were subjected to analysis by MALDI-TOF mass spectometry and found to contain clathrin heavy chain, β1, and γ, respectively. Thus, AP-1 appears to be the major binding partner for this portion of epsinR.
Localization of the Proteins
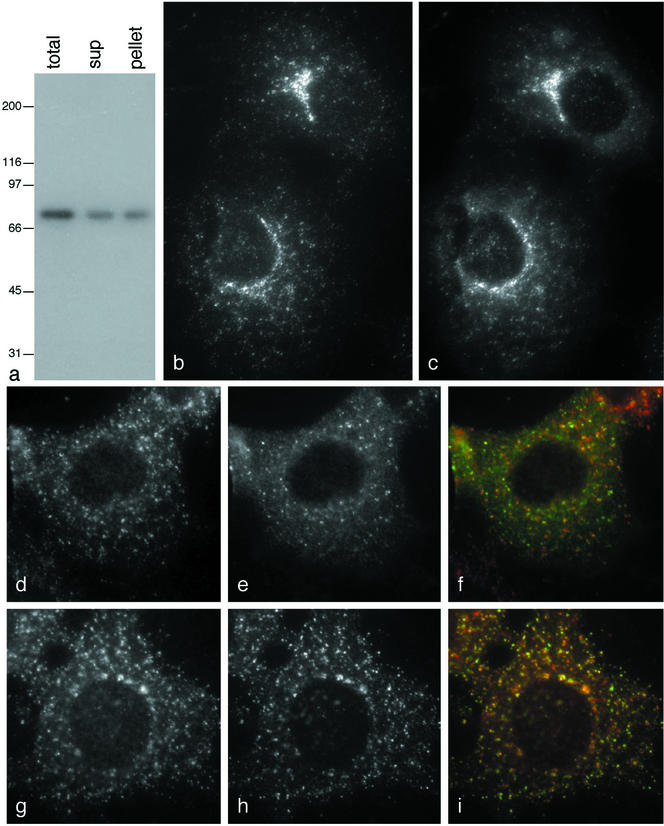
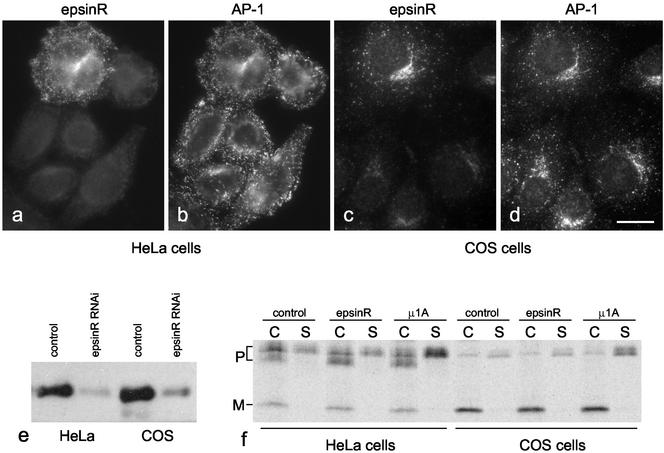
We have previously shown that binding to an appendage domain in vitro does not necessarily mean that a protein will also bind in vivo, so it was important to confirm the biochemical data by immunofluorescence, especially because epsinR, Snx9, and GAP1 all bind not only to the γ appendage, but also to the α appendage and weakly to the GGA1 appendage. We were unable to detect any convincing localization of Snx9 to membranes using either antibodies against the endogenous protein or tagged constructs (unpublished data), whereas GAP1 localized to the entire Golgi region, as previously reported (Cukierman et al., 1995, and unpublished observations). EpsinR showed the most convincing colocalization with AP-1. Figure 4a shows that our epsinR antibody specifically labels a band of the appropriate size on whole cell Western blots, which partitions ∼50:50 between cytosol and membranes. By immunofluorescence, epsinR (Figure 4b) and AP-1 (Figure 4c) were found to have nearly identical distributions, both in the perinuclear region and toward the cell periphery.
Figure 4.
Localization of epsinR. (a) Western blot of homogenized A431 cells, either untreated (total) or centrifuged at high speed (sup and pellet), probed with affinity-purified antiepsinR. (b and c) COS cells were double-labeled for epsinR (b) and the AP-1 γ subunit (c). The two patterns show very good colocalization. (d and e) COS cells transiently transfected with myc-tagged p56 were treated with nocodazole for 2 h and then double-labeled either for epsinR (d, red in f) and γ-adaptin (e, green in f) or for epsinR (g, red in i) and tagged p56 (h, green in i). To quantify the extent of overlap, six cells double-labeled for epsinR and γ-adaptin and six cells double-labeled for epsinR and tagged p56 were analyzed, and individual spots were scored. Out of 1607 spots analyzed from the cells double-labeled for epsinR and γ-adaptin, 9% were positive for epsinR only, 7% were positive for γ-adaptin only, and 84% were positive for both proteins. Out of 1192 spots analyzed from the cells double-labeled for epsinR and p56, 48% were positive for epsinR only, 16% were positive for p56 only, and 36% were positive for both proteins. Scale bar, 20 μm.
EpsinR clearly has a distinct distribution from AP-2 (unpublished data), but it was more difficult to compare its localization with that of the GGAs because both are associated with perinuclear membranes. To disperse the membranes and improve resolution, we treated cells with the microtubule-disrupting drug nocodazole. For these experiments we used myc-tagged p56 as a marker for the GGA compartment, because we have shown that it completely colocalizes with the GGAs (Lui et al., 2003), and it produces less background than tagged GGA constructs. Figure 4, d–f, shows that there is very good colocalization between epsinR (d) and AP-1 (e) in nocodazole-treated cells. However, the relative intensity of some of the spots labeled with the two antibodies varies, producing some spots that are more red and some that are more green. This is in contrast to nocodazole-treated cells double-labeled for AP-1 and γ-synergin, where the spots are all the same shade of yellow (Lui et al., 2003). Double-labeling for epsinR (g) and p56 (h) shows less colocalization, although again many of the spots coincide, suggesting that frequently the two proteins are associated with the same membranes.
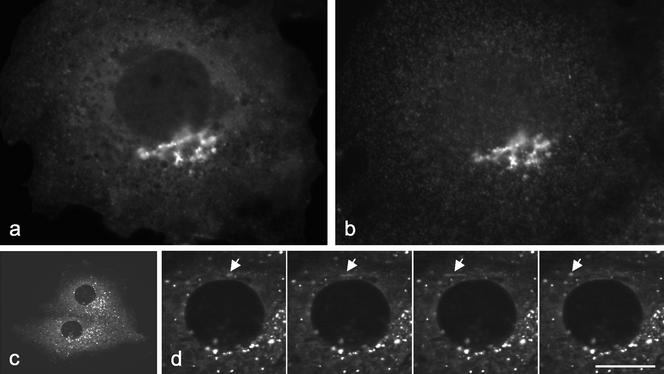
We also expressed NH2-terminally GFP-tagged epsinR (Figure 5a) and again saw very similar patterns when we double-labeled for AP-1 (Figure 5b), although the patterns were not quite so coincident as when we localized the endogenous protein. The GFP tag meant that we could observe the dynamics of the labeled membranes in living cells. Figure 5c links to a video (Figure 5c), from which enlarged still images, taken at 3.25-s intervals, are shown in Figure 5d. The labeled structures are highly motile, moving with speeds of up to ∼1 μm/s, indicating that they are being pulled along by microtubule motors. Similar types of movements have been seen in cells expressing the AP-1 μ1 subunit coupled to YFP (Huang et al., 2001). Interestingly, the labeled membranes often appear to be moving away from the Golgi region, toward the cell periphery. Although at first this might seem inconsistent with a role for AP-1 and epsinR in retrieval from a post-TGN compartment, the brightness of these membranes suggests that they are not individual vesicles but larger structures with coated buds associated with them. These structures may be analogous to the vesicular tubular transport complex (VTC), which carries secretory proteins from the ER to the Golgi apparatus and from which COPI-coated vesicles bud to bring escaped proteins back to the ER (Stephens and Pepperkok, 2001). Similarly, AP-1 and epsinR on the moving membranes may be helping to form clathrin-coated vesicles to bring proteins back to the TGN. The coated vesicles themselves are probably too small and/or short-lived to be easily resolved by live cell imaging.
Figure 5.
(a and b) COS cells were transfected with GFP-tagged full-length epsinR (a) and then double-labeled for the AP-1 γ subunit (b). (c) Link to a video (Figure 5c.mov) showing two cells expressing GFP-tagged full-length epsinR. Frames were acquired every 3.25 s, and the total acquisition time for the entire sequence was 139 s. (d) A series of frames from the video showing an enlarged region of one of the cells. The arrow indicates a structure that is moving away from the Golgi region. Although the structure appears to tubulate, this is probably because it was moving while the pictures were taken. Scale bar: a and b, 12 μm; c, 40 μm; d, 10 μm.
Expression of the Two epsinR Domains
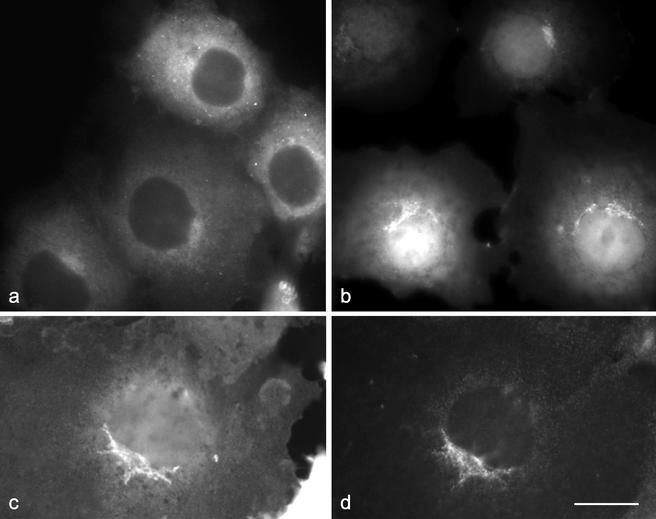
How is epsinR targeted to AP-1–positive membranes? One possibility is that it is recruited via its COOH-terminal domain, by binding to the γ appendage. Alternatively, because there is evidence that ENTH domains can bind to lipids (Itoh et al., 2001), it may be recruited independently via its NH2-terminal ENTH domain. To investigate the relative importance of the two domains in epsinR localization, each was expressed separately as a GFP-tagged construct. Figure 6a shows that the COOH-terminal domain of epsinR, when expressed on its own, is cytosolic and essentially featureless. In contrast, the GFP-ENTH domain construct (Figure 6, b and c) produces flat-looking labeling out to the cell periphery, indicating that at least some of the protein is associated with the plasma membrane as well as a strong signal in the perinuclear region. Double-labeling for AP-1 (Figure 6d) shows that the two perinuclear patterns are similar but not identical.
Figure 6.
Localization of individual epsinR domains. The COOH-terminal γ appendage binding domain (a) and the NH2-terminal ENTH domain (b and c) were each expressed as GFP constructs and transfected into COS cells. The COOH-terminal domain is cytosolic; however, the ENTH domain is localized to the plasma membrane and to a perinuclear compartment that shows partial overlap with the AP-1 γ subunit (d). Scale bar, 20 μm.
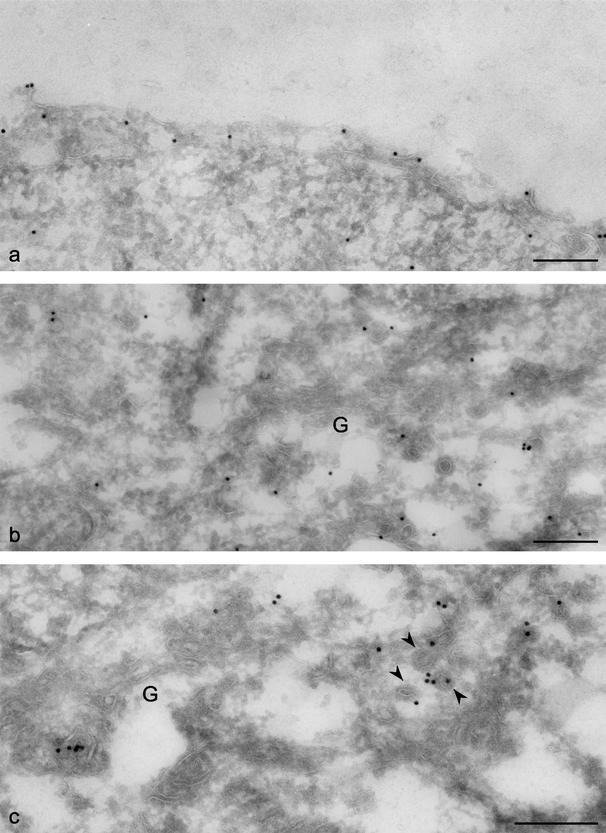
To confirm the plasma membrane association of the ENTH domain construct and to investigate the perinuclear pattern at higher resolution, immunogold EM was carried out. Figure 7a shows that although there is some labeling of the cytosol, consistent with what we see by light microscopy, a significant proportion of the label is at the cell surface. When we looked at the Golgi region (Figure 7b), we saw labeling on tubulovesicular membranes in the vicinity of the Golgi stack but not on the stack itself (labeled G). We also investigated the distribution of the full-length GFP-tagged epsinR construct by immunogold EM. Figure 7c shows that the full-length protein is also found on tubulovesicular membranes in the Golgi region and furthermore that it is frequently associated with coated budding profiles (arrowheads).
Figure 7.
EM localization of the epsinR constructs. Cells expressing either GFP-tagged ENTH domain (a and b) or GFP-tagged full-length epsinR (c) were processed for immunogold EM. (a) Much of the ENTH domain construct is associated with the plasma membrane. (b) The ENTH domain construct is also associated with tubulovesicular membranes near the Golgi stack (labeled G). (c) Full-length epsinR has a similar distribution; in addition, it is often found on membranes covered with a thick coat that is probably clathrin (arrowheads). Scale bars, 500 nm.
Does epsinR Require AP-1 for Its Localization?
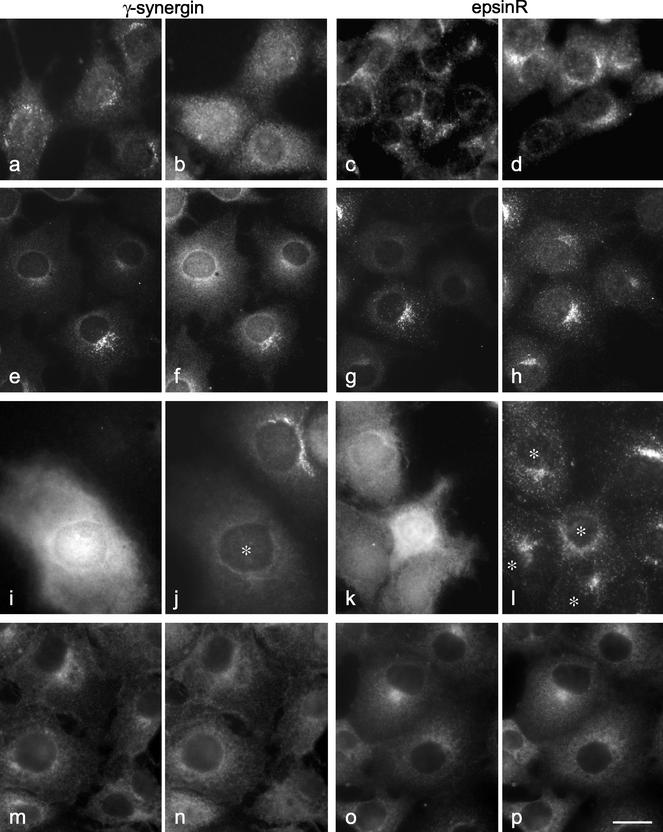
Although the epsinR COOH-terminal domain is unable to be recruited onto membranes on its own, its interaction with AP-1 may still be essential for the correct localization of the full-length protein. Thus, we investigated the distribution of epsinR in a cell line made from a μ1A knockout mouse. These cells have been shown to assemble partial AP-1 complexes, but the complexes are cytosolic rather than membrane-associated. Stably transfecting the cells with wild-type μ1A enables AP-1 complexes to assemble and localize correctly (Meyer et al., 2000). Figure 8a shows the distribution of γ-synergin in μ1A-transfected cells, and Figure 8b shows the distribution of γ-synergin in the mutant cells. It is clear that γ-synergin is membrane-associated in the transfected cells but cytosolic in the mutant cells (see also Lui et al., 2003). However, epsinR is membrane-associated not only in the transfected cells (Figure 8c) but also in the mutant cells (Figure 8d). Similarly, we can mimic the μ1A knockout phenotype in COS cells using μ1A siRNA. Immunoprecipitation with anti–γ-adaptin reveals that the cells form partial complexes which lack the β1 and μ1 subunits (unpublished data), and by immunofluorescence we find that most of the cells have diffuse rather than membrane-associated γ-adaptin (Figure 8, e and g). The γ-synergin in these cells also becomes diffuse (Figure 8f), whereas the epsinR stays membrane-associated (Figure 8h). Another way of shifting the distribution of γ-synergin into the cytosol is by overexpressing the γ appendage as a GFP construct (Figure 8, i and j; Lui et al., 2003). However, overexpressing the γ appendage has little or no effect on the distribution of epsinR (Figure 8, k and l).
Figure 8.
The distribution of epsinR does not depend upon AP-1. (a–d) Cells from a μ1A knockout mouse, either transfected with wild-type μ1A (a and c) or nontransfected (b and d), were labeled with antibodies against either γ-synergin (a and b) or epsinR (c and d). (e–h) COS cells were treated with siRNA directed against μ1A and then double-labeled for either γ-adaptin (e) and γ-synergin (f) or for γ-adaptin (g) and epsinR (h). In both cases, γ-synergin becomes cytosolic in the affected cells, whereas epsinR stays membrane associated. (i–l) COS cells were transiently transfected with GFP coupled to the γ appendage domain (i and k) and double-labeled for either γ-synergin (j) or epsinR (l). Expression of this construct causes γ-synergin to become cytosolic; however, it has little effect on epsinR (see asterisks). (m–p) Cells were treated with 100 μg/ml BFA for 2 min and then double-labeled for either AP-1 (m) and γ-synergin (n) or for AP-1 (o) and epsinR (p). All three proteins redistribute in response to BFA. Scale bar: a–d, 14 μm; e–h, 25 μm; i–l, 18 μm; m–p, 20 μm.
We also looked at the effect of brefeldin A (BFA) on the localization of epsinR. BFA has been shown to cause ARF to dissociate rapidly from membranes, leading to the dissociation of other proteins whose localization is ARF dependent, such as AP-1. Because the experiments shown in Figure 8, a–l, indicate that epsinR does not require AP-1 for its localization, we expected it to be resistant to BFA. Surprisingly, we found that epsinR is BFA sensitive (Figure 8p; Figure 8o shows the same cells double-labeled for AP-1). γ-Synergin is also BFA sensitive (Figure 8n; m shows AP-1), as we have previously shown (Page et al., 1999). However, although the BFA sensitivity of γ-synergin can be explained by its association with AP-1, the effect of BFA on epsinR must be more direct. Together, these observations indicate that the association of epsinR with membranes is dependent on ARF but is independent of AP-1.
Recruitment of the epsinR ENTH Domain
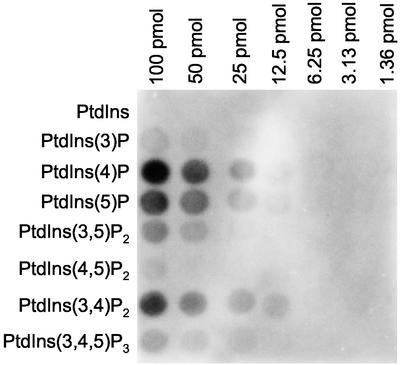
How is epsinR recruited onto membranes in an ARF-dependent manner, if not by binding to AP-1? One of ARF's many activities is to stimulate PIP2 production on Golgi-enriched membranes (Godi et al., 1999). Therefore, one possibility is that the ENTH domain of epsinR, like the ENTH domain of epsin 1, may be recruited onto membranes by binding to PIP2. However, PIP2 is not the only phosphoinositide to be generated on such membranes in an ARF-dependent manner: PtdIns(4)P production is also stimulated by ARF (Godi et al., 1999). To investigate the possible role of phosphoinositides in epsinR recruitment, we probed an array of phosphoinositides spotted onto nitrocellulose with a GST-ENTH domain construct. Figure 9 shows that this construct gives little or no labeling of PtdIns(4,5)P2. However, PtdIns(4)P was strongly labeled, and there was also significant labeling of PtdIns(5)P and of PtdIns(3,4)P2.
Figure 9.
Binding of the epsinR ENTH domain to phosphoinositides spotted onto a nitrocellulose filter. A phosphoinositide array was probed with a GST-ENTH domain construct. PtdIns(4)P is most strongly labeled, followed by PtdIns(5)P and PtdIns(3,4)P2. There is little or no significant labeling of PtdIns(4,5)P2.
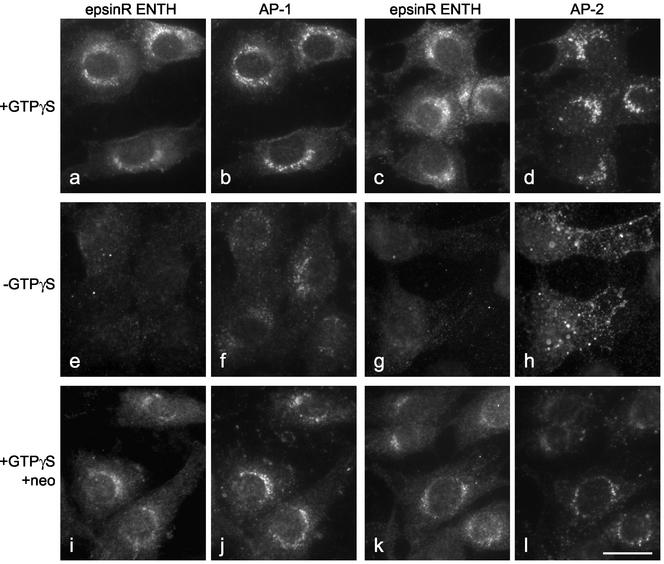
As a more physiological alternative to protein-lipid overlays, we also addressed the question of ENTH domain recruitment by making use of an in vitro system that we originally developed to study the recruitment of coat proteins. The system involves incubating permeabilized tissue culture cells with pig brain cytosol and then assaying for de novo recruitment using either species- or tissue-specific antibodies that recognize proteins in the cytosol but not endogenous proteins left behind on the membranes. By adding recombinant Xpress-tagged epsinR ENTH domain to the cytosol, we can monitor ENTH domain recruitment as well as adaptor recruitment. Figure 10, a and c, shows that in the presence of ATP, an ATP-regenerating system, and GTPγS, tagged ENTH domain is recruited onto a perinuclear membrane compartment in permeabilized NRK cells. Double-labeling with an antibody that recognizes both endogenous and newly recruited γ-adaptin shows that the two proteins have similar, although not identical, distributions (Figure 10b). We also double-labeled with an antibody that recognizes a brain-specific splice variant of the AP-2 α subunit. We have previously shown that in the presence of GTPγS, AP-2 is mistargeted to a perinuclear endosomal compartment (Seaman et al., 1993; West et al., 1997). Although the ENTH domain construct (Figure 10c) and mistargeted AP-2 (Figure 10d) both localize to the perinuclear region, the patterns are less similar than in cells double-labeled for the ENTH domain construct and AP-1.
Figure 10.
In vitro recruitment of the epsinR ENTH domain. NRK cells were permeabilized by freezing and thawing and then incubated with pig brain cytosol containing recombinant Xpress-tagged epsinR ENTH domain, either with or without GTPγS and/or neomycin. Recruitment of the ENTH domain construct only occurs in the presence of GTPγS, indicating that it is ARF dependent. It is slightly blocked by neomycin, but the effect is less severe than on AP-2. Scale bar, 20 μm.
When we omitted the GTPγS, epsinR ENTH domain recruitment was effectively blocked (Figure 10, e and g), consistent with the BFA data indicating that epsinR localization is ARF dependent. The amount of membrane-associated AP-1 is also reduced (Figure 10f), although not quite so dramatically as the ENTH domain construct; however, because (for technical reasons) the antibody we were using cannot distinguish between endogenous and exogenous AP-1, it is possible that much of the labeling is due to preexisting AP-1 that was already on the membrane when the cells were permeabilized. In contrast, exogenous AP-2 is still efficiently recruited, but to the plasma membrane instead of to the endosomal compartment (Figure 10h).
We have previously shown that the drug neomycin, which sequesters PIP2, inhibits the recruitment of AP-2, both onto the plasma membrane in the absence of GTPγS and onto the endosomal compartment in the presence of GTPγS, without affecting the recruitment of AP-1 (West et al., 1997). Figure 10, i and k, shows that neomycin also causes a very slight inhibition of epsinR ENTH domain recruitment. We have so far been unable to quantify the extent of inhibition because the construct tends to precipitate when centrifuged at high speed, which is necessary for our biochemical assay. However, the effect of neomycin on AP-2 (Figure 10l) is clearly much more profound than its effect on the epsinR ENTH domain. This suggests that, although PIP2 may contribute to ENTH domain recruitment, it is not a major player, consistent with the protein-lipid overlay data shown in Figure 9.
Depletion of epsinR by RNAi
As a first step toward establishing the precise function of epsinR, we used RNA interference to knock down expression of the protein in both HeLa cells and COS cells. Figure 11, a–d, shows HeLa and COS cells treated with epsinR siRNA and then double-labeled with anti-epsinR and anti–AP-1. At least 90% of the cells were strongly affected, although AP-1 labeling looks essentially normal. Western blotting (Figure 11e) was used to quantify the efficiency of knockdown. EpsinR expression in the siRNA-treated HeLa cells was found to be 13.3% of control, and in the COS cells it was found to be 17.5% of control.
Figure 11.
EpsinR depletion by RNAi. (a–d) HeLa cells (a and b) or COS cells (c and d) were incubated with epsinR siRNA for 3 d and then double-labeled for immunofluorescence with anti-epsinR (a and c) and anti–γ-adaptin (b and d). In both cases >90% of the cells had greatly reduced epsinR labeling. Scale bar, 20 μm. e, Western blots of HeLa and COS cells treated with either a control siRNA or epsinR siRNA were probed with anti-epsinR. (f) HeLa and COS cells, treated with either control, epsinR, or μ1A siRNA, were pulse chased with 35S, and both cell-associated (C) and secreted (S) cathepsin D were immunoprecipitated. The positions of the precursor (P) and mature (M) forms of the enzyme are marked. In the HeLa cells the ratio of secreted cathepsin D precursor to total cathepsin D is 0.158 in the control, 0.161 in the epsinR knockdown, and 0.377 in the μ1A knockdown. In the COS cells the ratios are 0.191 in the control, 0.196 in the epsinR knockdown, and 0.504 in the μ1A knockdown.
The best characterized consequence of AP-1 deficiency is the missorting of lysosomal enzymes like cathepsin D, due to an inability of the mannose 6-phosphate receptors to cycle normally between TGN and endosomes (Meyer et al., 2000). To assay for cathepsin D sorting, cells were treated with either control siRNA, epsinR siRNA, or μ1A siRNA and then pulse labeled with 35S for 15 min and chased for 2.5 h. Cell-associated (C) and secreted (S) cathepsin D were immunoprecipitated and quantified. Figure 11f shows that in both HeLa and COS cells, knocking down μ1A causes ∼2.5-fold more cathepsin D precursor (P) to be secreted. Surprisingly, however, knocking down epsinR does not significantly affect cathepsin D sorting. Thus, although epsinR is a highly conserved component of the AP-1 network, it does not appear to be required for AP-1–mediated sorting of lysosomal enzymes.
DISCUSSION
Until recently, it was assumed that clathrin and adaptor complexes could facilitate coated vesicle formation and cargo recruitment essentially single-handedly. This idea was challenged by the discovery of a large number of accessory proteins that participate in clathrin-mediated endocytosis. Most of these proteins have been shown to bind either directly or indirectly to the appendage domain of the α subunit of the AP-2 complex, and many of them also make connections with other molecules including lipids, cargo proteins, and components of signaling pathways. For instance, epsin 1 interacts not only with the α appendage; it also binds to PIP2 via its ENTH domain (Itoh et al., 2001); it can interact with ubiquitinated cargo via its UIM (Riezman, 2002); and its Drosophila homolog, liquid facets, has been shown to participate in signaling events during eye development (Cadavid et al., 2000). The identification of binding partners for some of the other adaptor appendage domains indicates that the AP-2 complex is not unique and that protein–protein interaction networks are set up during vesicle formation from intracellular membranes as well as from the plasma membrane. We have previously shown that the AP-1 γ appendage interacts with γ-synergin and that the GGA appendage interacts with p56 (Page et al., 1999; Lui et al., 2003). Both of these proteins are likely to interact with other molecules as well. Other proteins that bind to the γ and GGA appendage domains in vitro and that may also bind in vivo include rabaptin-5 and p200 (Hirst et al., 2000; Lui et al., 2002). Here we identify three more proteins that can interact with the γ appendage in vitro: Snx9, GAP1, and a novel protein, epsinR.
Although there is still some question as to whether Snx9 and GAP1 interact with AP-1 under physiological conditions, epsinR is clearly an AP-1 binding partner in vivo as well as in vitro. It shows excellent colocalization with AP-1 by immunofluorescence, and it also localizes to coated budding profiles in the Golgi region by immunogold EM. It is highly enriched in purified clathrin-coated vesicle preparations from rat liver, where it seems to be the major γ appendage binding partner. While our manuscript was under review, McPherson and colleagues published an article about the same protein, which they found in a proteomics analysis of clathrin-coated vesicles and named “enthoprotin” (Wasiak et al., 2002). Like us, they found that epsinR/enthoprotin binds to the γ appendage in vitro, colocalizes with AP-1 by immunofluorescence, and is highly enriched in purified clathrin-coated vesicles. Thus, most of our data are consistent with theirs. However, they also detected a strong interaction in vitro between epsinR and GGA2, while we detected only a weak interaction between epsinR and GGA1. This probably reflects the different binding preferences of the various GGA appendage domains. In addition, Wasiak et al. reported strong colocalization between epsinR and GGA2, using a tagged epsinR construct. In contrast, we find that endogenous epsinR shows partial colocalization with the various GGAs and their binding partner p56 but much better colocalization with AP-1. Here the discrepancy may be due to differences between the tagged and endogenous proteins.
What is the motif on epsinR that is recognized by the γ appendage? One possible clue comes from the observation that like Snx9 and GAP1, epsinR interacts in vitro not only with the γ appendage but also with the α appendage. This suggests that, although structurally dissimilar, the γ and α appendage domains may interact with similar motifs. One possible candidate for such a motif is DLF, which fits the DΦF consensus sequence for binding to the α appendage domain and is present in three copies in epsinR. However, the DLF sequences in epsinR also fit the consensus for clathrin binding (Doray and Kornfeld, 2001), and both our study and that of McPherson and colleagues show that epsinR constructs containing these motifs can bring down clathrin as well as AP-1 (Wasiak et al., 2002; Figure 3c). Another candidate motif for γ appendage binding is DDFXDF, a sequence that we previously noted in γ-synergin where it is present in five copies (Page et al., 1999). EpsinR contains at least two sequences related to this motif (GGFADF and GDFGDW), which are also present in the epsinR constructs that pull down AP-1 from cytosol (see Figure 3c and Wasiak et al., 2002). Homologues of epsinR in flies, worms, and plants all contain both DLF and DDFXDF-type sequences, indicating that both are important for function. However, other proteins that can bind to the γ appendage in vitro, including Snx9 and GAP1, contain no obvious examples of either motif. Thus, it is clear that further studies will be needed to establish the precise requirements for γ appendage binding.
The nearly complete colocalization between epsinR and AP-1 suggested to us initially that one might recruit the other. We have previously shown that γ-synergin follows AP-1 onto membranes, so we investigated whether the same might be true for epsinR. We found that epsinR takes a much more active role in its own localization than γ-synergin. First, although γ-synergin becomes cytosolic in μ1A-deficient cells, epsinR looks nearly normal. Second, overexpressing the γ appendage domain pulls γ-synergin into the cytosol, but epsinR remains membrane associated. Third, the epsinR ENTH domain, which has no detectable AP-1 binding activity, is able to localize to membranes on its own. So if AP-1 is not recruiting epsinR, might epsinR be recruiting AP-1? Again, the answer seems to be no. In a previous study we showed that AP-1 complexes containing truncated γ-adaptin with no appendage domain, or chimeric γ-adaptin with the α appendage domain, have a nearly normal distribution (Robinson, 1993). In the present study, we show that knocking down epsinR to undetectable levels does not prevent AP-1 from associating with membranes. Together, these observations indicate that epsinR and AP-1 do not need each other to get onto membranes in the first place; however, once they are on the membrane they associate with each other. This would ensure that they get into the same coated vesicle, and it might also enable both proteins to recruit additional components into the same network.
What does the ENTH domain interact with, to get it onto the membrane? The epsin 1 ENTH domain has been shown to bind to PIP2; however, some of the amino acids in epsin 1 that contribute to PIP2 binding are not conserved in epsinR (Ford et al., 2002), and the two proteins localize to different compartments. More formal evidence against a role for PIP2 in epsinR recruitment is our observation that in protein-lipid overlay assays, the epsinR ENTH domain binds to a subset of phosphoinositides, which does not include PtdIns(4,5)P2. We also addressed the question of ENTH domain recruitment by making use of a permeabilized cell system. This system enables the ENTH domain to select its preferred target membrane in a more physiological context. The construct was faithfully recruited onto perinuclear membranes in a manner that was strongly dependent on GTPγS, consistent with a requirement for ARF. However, recruitment was only weakly affected by neomycin, which sequesters PIP2 and blocks AP-2 recruitment. So far the best candidate for a phosphoinositide that might recruit epsinR in vivo as well as in vitro is PtdIns(4)P. It produces the strongest signal in our overlay assay, and it has been localized to the TGN, where its synthesis is stimulated by ARF (Godi et al., 1999). Using our permeabilized cell system, we should now be able to test whether recruitment of the epsinR ENTH domain is blocked by different phosphoinositide-modifying enzymes or by PH domain constructs with different phosphoinositide preferences. This approach may also lead to a better understanding of how the various adaptors are recruited onto the appropriate membrane. AP-2 and epsin 1 have both been shown to bind to PIP2, so it is possible that AP-1 and epsinR might also have similar phosphoinositide-binding specificities.
What is the function of epsinR? Unlike γ-synergin and p56, which only appear to be expressed in mammals, epsinR is highly conserved, indicating that it plays a more fundamental role. Surprisingly, we can deplete epsinR in both HeLa and COS cells using RNAi, and under these conditions cathepsin D sorting still proceeds normally. This is in contrast to dominant negative studies carried out on several of the AP-2 binding partners, including epsin 1. Overexpressing mutagenized versions of such proteins has been shown to block clathrin-mediated endocytosis, as assayed by transferrin uptake (e.g., see Chen et al., 1998). However, overexpression may lead to indirect effects; for example, the proteins may saturate binding sites on the α appendage and prevent other proteins from interacting. RNAi is more specific because it removes just one protein from the cell, so it will be important to see what happens when this approach is used on some of the α appendage binding partners.
There are two possible explanations for the phenotype of cells treated with epsinR siRNA. One is that there may be other proteins in the cell that can substitute for epsinR. Although there are no obvious candidates in the database, there may be proteins with functions similar to epsinR that do not show any apparent sequence homology. Alternatively, epsinR may not be involved in lysosomal enzyme sorting, but in some other pathway. For instance, it may be a novel cargo adaptor for such a pathway. Epsins 1–3 are potential cargo adaptors, using their UIMs to bring ubiquitinated proteins into AP-2–coated vesicles, and although epsinR has no UIM, it may have as yet unidentified motifs to bring another type of cargo into AP-1–coated vesicles. AP-1 has been shown to be involved not only in the sorting of lysosomal enzymes, but also in a number of other events, including granule maturation in regulated secretory cells and targeting of proteins to the basolateral plasma membrane in polarized epithelial cells. We intend to investigate the consequences of epsinR depletion in these more specialized types of cells. The high degree of conservation of epsinR means that we can also perform targeted gene disruptions in whole organisms such as Drosophila. Another approach toward establishing the function of epsinR will be to search for additional binding partners for its COOH-terminal domain. A comparison of the mammalian epsinR sequence with that of other organisms, in particular Drosophila, reveals that although their COOH-terminal domains are divergent, there are several stretches of conserved amino acids, and these are good candidates for protein–protein interaction motifs. Our current working model is that epsinR, like epsin 1 (Kalthoff et al., 2002), uses its NH2-terminal ENTH domain as a membrane anchor and uses its extended, flexible COOH-terminal domain to “rope in” other proteins to the site of coated vesicle formation.
Supplementary Material
ACKNOWLEDGMENTS
We are particularly indebted to Peter Schu for the μ1A-deficient cell line. We are also grateful to Ian Mills and Harvey McMahon for sharing unpublished data. We thank Carl Blobel and Linda Howard for the anti-Snx9, Dan Cassel for the anti-GAP1, Folma Buss for help with the live cell imaging, Abi Stewart for performing the immunogold EM, and Nicola Berg for technical assistance. We also thank Paul Luzio, David Owen, Matthew Seaman, Rainer Duden, Howard Davidson, John Kilmartin, and members of the Robinson lab for helpful discussions. This work was supported by grants from the Wellcome Trust and the Medical Research Council
Note added in proof. While this article was in print, the identification of a protein identical to epsinR was reported and given the name Clint. (C. Kalthoff, S. Groos, R. Kohl, S. Mahrhold, and E.J. Ungewickell. [2002]. Clint: a novel clathrin-binding ENTH-domain protein at the Golgi. Mol. Biol. Cell 13, 4060–4073.)
Footnotes
Online version of this article contains video material. Online version is available at www.molbiolcell.org.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–09–0552. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–09–0552.
REFERENCES
- Cadavid AL, Ginzel A, Fischer JA. The function of the Drosophila fat facets deubiquitinating enzyme in limiting photoreceptor cell number is intimately associated with endocytosis. Development. 2000;127:1727–1736. doi: 10.1242/dev.127.8.1727. [DOI] [PubMed] [Google Scholar]
- Chen H, Fre S, Slepnev VI, Capua MR, Takei K, Butler MH, Di Fiore PP, De Camilli P. Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature. 1998;394:793–797. doi: 10.1038/29555. [DOI] [PubMed] [Google Scholar]
- Collins, B.M., McCoy, A.J., Kent, H.M., Evans, P.R., and Owen, D.J. Molecular architecture and functional model of the endocytic APZ complex. Cell 109, 523–535. [DOI] [PubMed]
- Cukierman E, Huber I, Rotman M, Cassel D. The ARF1 GTPase-activating protein: zinc finger motif and Golgi complex localization. Science. 1995;270:1999–2002. doi: 10.1126/science.270.5244.1999. [DOI] [PubMed] [Google Scholar]
- Davidson HW. Wortmannin causes mistargeting of procathepsin D. Evidence for the involvement of a phosphatidylinositol 3-kinase in vesicular transport to lysosomes. J Cell Biol. 1995;130:797–805. doi: 10.1083/jcb.130.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doray B, Kornfeld S. γ subunit of the AP-1 adaptor complex binds clathrin: implications for cooperative binding in coated vesicle assembly. Mol Biol Cell. 2001;12:1925–1935. doi: 10.1091/mbc.12.7.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidarov I, Keen JH. Phosphoinositide-AP-2 interactions required for targeting to plasma membrane clathrin-coated pits. J Cell Biol. 1999;146:755–764. doi: 10.1083/jcb.146.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford MGJ, Mills IG, Peter BJ, Vallis Y, Praefcke GJK, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- Godi A, Pertile P, Meyers R, Marra P, Di Tullio G, Iurisci C, Luini A, Corda D, De Matteis MA. ARF mediates recruitment of PtdIns-4-OH kinaseβ and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat Cell Biol. 1999;1:280–287. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- Hirst J, Lui WWY, Bright NA, Totty N, Seaman MNJ, Robinson MS. A family of proteins with γ-adaptin and VHS domains that facilitate trafficking between the TGN and the vacuole/lysosome. J Cell Biol. 2000;149:67–79. doi: 10.1083/jcb.149.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard L, Nelson KK, Maciewicz RA, Blobel CP. Interaction of the metalloprotease disintegrins MDC9 and MDC15 with two SH3 domain-containing proteins, endophilin I and SH3PX1. J Biol Chem. 1999;274:31693–31699. doi: 10.1074/jbc.274.44.31693. [DOI] [PubMed] [Google Scholar]
- Huang F, Nesterov A, Carter RE, Sorkin A. Trafficking of yellow-fluorescent-protein-tagged μ1 subunit of clathrin adaptor AP-1 complex in living cells. Traffic. 2001;2:345–357. doi: 10.1034/j.1600-0854.2001.25020506.x. [DOI] [PubMed] [Google Scholar]
- Itoh T, Koshiba S, Kikuchi A, Yokoyama S, Takenawa T. Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science. 2001;291:1047–1051. doi: 10.1126/science.291.5506.1047. [DOI] [PubMed] [Google Scholar]
- Jensen ON, Podtelejnikov AV, Mann M. Identification of the components of simple protein mixtures by high-accuracy peptide mass mapping and database searching. Anal Chem. 1997;69:4741–4750. doi: 10.1021/ac970896z. [DOI] [PubMed] [Google Scholar]
- Kalthoff C, Alves J, Urbanke C, Knorr R, Ungewickell EJ. Unusual structural organization of the endocytic proteins AP180 and epsin 1. J Biol Chem. 2002;277:8209–8216. doi: 10.1074/jbc.M111587200. [DOI] [PubMed] [Google Scholar]
- Kent HM, McMahon HT, Evans PR, Benmerah A, Owen DJ. γ-Adaptin appendage domain: structure and binding site for Eps15 and γ-synergin. Structure. 2002;10:1139–1148. doi: 10.1016/s0969-2126(02)00801-8. [DOI] [PubMed] [Google Scholar]
- Lui, W.W.Y., Collins, B.M., Hirst, J., Motley, A., Millar, C., Schu, P., Owen, D.M., and Robinson, M.S. (2002). Binding partners for the COOH-terminal appendage domains of the GGAs and γ-adaptin. J. Cell Biol. (in press). [DOI] [PMC free article] [PubMed]
- Meyer C, Eskelinen EL, Guruprasad MR, von Figura K, Schu P. μ1A deficiency induces a profound increase in MPR300/IGF-II receptor internalization rate. J Cell Sci. 2001;114:4469–4476. doi: 10.1242/jcs.114.24.4469. [DOI] [PubMed] [Google Scholar]
- Meyer C, Zizioli D, Lausmann S, Eskelinen EL, Hamann J, Saftig P, von Figura K, Schu P. μ1A-adaptin-deficient mice: lethality, loss of AP-1 binding and rerouting of mannose 6-phsophate receptors. EMBO J. 2000;19:2193–2203. doi: 10.1093/emboj/19.10.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noji T, et al. Structural basis for the accessory protein recruitment by the gamma-adaptin ear domain. Nat Struct Biol. 2002;9:527–531. doi: 10.1038/nsb808. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Vallis Y, Pearse BM, McMahon HT, Evans PR. The structure and function of the β2-adaptin appendage domain. EMBO J. 2000;19:4216–4227. doi: 10.1093/emboj/19.16.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DJ, Vallis Y, Noble ME, Hunter JB, Daftora TR, Evans PR, McMahon HT. A structural explanation for the binding of multiple ligands by the alpha-adaptin appendage domain. Cell. 1999;97:805–815. doi: 10.1016/s0092-8674(00)80791-6. [DOI] [PubMed] [Google Scholar]
- Page LJ, Sowerby PJ, Lui WWY, Robinson MS. γ-Synergin: an EH domain-containing protein that interacts with γ-adaptin. J Cell Biol. 1999;146:993–1004. doi: 10.1083/jcb.146.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riezman H. Cell biology: the ubiquitin connection. Nature. 2002;416:451–455. doi: 10.1038/416381a. [DOI] [PubMed] [Google Scholar]
- Robinson MS. Assembly and targeting of adaptin chimeras in transfected cells. J Cell Biol. 1993;123:67–77. doi: 10.1083/jcb.123.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS, Bonifacino JS. Adaptor-related proteins. Curr Opin Cell Biol. 2001;13:444–453. doi: 10.1016/s0955-0674(00)00235-0. [DOI] [PubMed] [Google Scholar]
- Rosenthal JA, Chen H, Slepnev VI, Pellegrini L, Salcini AE, Di Fiore PP, De Camilli P. The epsins define a family of proteins that interact with components of the clathrin coat and contain a new protein module. J Biol Chem. 1999;274:33959–33965. doi: 10.1074/jbc.274.48.33959. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning, a Laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Seaman MNJ, Ball CL, Robinson MS. Targeting and mistargeting of plasma membrane adaptors in vitro. J Cell Biol. 1993;123:1093–1105. doi: 10.1083/jcb.123.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepnev VI, De Camilli P. Accessory factors in clathrin-dependent synaptic vesicle endocytosis. Nat Rev Neurosci. 2000;1:161–172. doi: 10.1038/35044540. [DOI] [PubMed] [Google Scholar]
- Spradling KD, McDaniel AE, Lohi J, Pilcher BK. Epsin 3 is a novel extracellular matrix-induced transcript specific to wounded epithelia. J Biol Chem. 2001;276:29257–29267. doi: 10.1074/jbc.M101663200. [DOI] [PubMed] [Google Scholar]
- Stephens DJ, Pepperkok R. Illuminating the secretory pathway: when do we need vesicles? J Cell Sci. 2001;114:1053–1059. doi: 10.1242/jcs.114.6.1053. [DOI] [PubMed] [Google Scholar]
- Valdivia RH, Baggott D, Chuang JS, Schekman RW. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev Cell. 2002;2:283–294. doi: 10.1016/s1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- Wasiak S, et al. Enthoprotin: a novel clathrin-associated protein identified through subcellular proteomics. J Cell Biol. 2002;158:855–862. doi: 10.1083/jcb.200205078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B, Steece KE, Emr SD. Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin, and are required for endocytosis. EMBO J. 1999;18:4383–4393. doi: 10.1093/emboj/18.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MA, Bright NA, Robinson MS. The role of ADP-ribosylation factor and phospholipase D in adaptor recruitment. J Cell Biol. 1997;138:1239–1254. doi: 10.1083/jcb.138.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.