Abstract
Mammalian cDNA expression cloning was used to identify novel regulators of integrin-mediated cell-substratum adhesions. Using a focal adhesion morphology screen, we identified a cDNA with homology to a receptor for activated protein kinase C (RACK1) that induced a loss of central focal adhesions and stress fibers in CHO-K1 cells. The identified cDNA was a C-terminal truncated form of RACK1 that had one of the putative protein kinase C binding sites but lacked the region proposed to bind the β integrin cytoplasmic domain and the tyrosine kinase Src. To investigate the role of RACK1 during cell spreading and migration, we tagged RACK1, a C-terminal truncated RACK1 and a point mutant that does not bind Src (RACK Y246F) with green fluorescent protein and expressed them in CHO-K1 cells. We found that RACK1 regulates the organization of focal adhesions and that it localizes to a subset of nascent focal complexes in areas of protrusion that contain paxillin but not vinculin. We also found that RACK1 regulates cell protrusion and chemotactic migration through its Src binding site. Together, these findings suggest that RACK1 regulates adhesion, protrusion, and chemotactic migration through its interaction with Src.
INTRODUCTION
Cell migration involves dynamic regulation of cell-substratum adhesions. During migration, cells form adhesions to the extracellular matrix largely through the integrin family of adhesion receptors (Hynes, 1992). The mechanisms that govern adhesion formation, release, and their coordination during cell migration remain largely unknown (Huttenlocher et al., 1995; Lauffenburger and Horwitz, 1996; Cox and Huttenlocher, 1998). Focal adhesions are large protein complexes that form upon integrin ligation and adhesion to the extracellular matrix. Direct correlations between focal adhesion organization and the ability of cells to migrate have been made (Huttenlocher et al., 1996). Several studies have demonstrated that an absence of central adhesions is associated with inhibited cell migration. Inhibition of focal adhesion kinase (Sieg et al., 1999), Src kinases (Klinghoffer et al., 1999), or the calcium-dependent protease calpain (Huttenlocher et al., 1997) induces the loss of central focal adhesions and reduces cell migration rates. Accordingly, we used a focal adhesion morphology screen and cDNA expression cloning to identify novel proteins that regulate integrin-mediated adhesions and cell migration. Using this approach, we identified a cDNA with homology to the protein kinase C binding protein RACK1. Subsequent characterization of RACK1 has demonstrated that it is an important regulator of focal adhesion organization and cell migration. In this article, we examine how RACK1 and its interaction with Src modulate focal adhesions and cell migration.
RACK1 is a cytosolic, WD-40 repeat protein that belongs to a superfamily of proteins that includes the β subunit of G proteins (Ron et al., 1994). RACK1 was originally identified based on its ability to bind to the activated form of protein kinase C (PKC) (Ron et al., 1994). RACK1 stabilizes the active form of PKC and permits its translocation to different sites within the cell (Mochly-Rosen and Gordon, 1998). Interestingly, RACK1 can also bind and inhibit Src family kinases (Chang et al., 1998). Direct binding of Src to RACK1 has previously been demonstrated to require tyrosine 246 of RACK1 in CHO-K1 cells (Chang et al., 2001). Src family kinases have been shown to play a key role in regulating adhesion formation, adhesion release, and cell migration. RACK1, through its interaction with PKC and Src kinases, may function as a critical adaptor protein mediating cross talk between serine/threonine and tyrosine kinase signaling pathways. RACK1 has many other binding partners, including β1, β2, β3, and β5 integrins (Liliental and Chang, 1998; Buensuceso et al., 2001); the common β chain of the IL-5/IL-3/GM-CSF receptor (Geijsen et al., 1999); phospholipase C-γ and RasGAP (Chang et al., 2001); β-spectrin and dynamin (Rodriguez et al., 1999); the protein tyrosine phosphatase PTPμ (Mourton et al., 2001); and a phosphodiesterase isoform (Yarwood et al., 1999). This suggests that RACK1 may act as a scaffolding protein, recruiting proteins to various transmembrane receptors and providing a platform for protein–protein interactions. Much remains unknown concerning the functional role of RACK1; however, in vivo studies have indicated that RACK1 is up-regulated in human carcinomas and during tissue regeneration after ischemic renal injury (Padanilam and Hammerman, 1997; Berns et al., 2000). Furthermore, recent studies have demonstrated that overexpression of RACK1 in CHO-K1 cells inhibits haptotactic cell migration and increases focal adhesion number (Buensuceso et al., 2001), although the mechanism(s) involved have not been defined.
The purpose of this study was to characterize how RACK1 regulates integrin-mediated adhesion and cell migration and to determine whether the Src-binding site is involved. Our findings suggest that RACK1 regulates the organization of focal adhesions through its interaction with Src and that it localizes to a subset of nascent focal complexes in areas of protrusion that contain paxillin and α-actinin but not vinculin. Furthermore, we found that RACK1, via its Src binding site, regulates cell protrusion and chemotactic migration, but not random or haptotactic migration. These findings suggest that RACK1, through its interaction with Src, regulates focal adhesion organization, protrusion, and chemotactic cell migration.
MATERIALS AND METHODS
Vectors
pcDNA3.1 and pcDNA3.1hygro-lacZ were purchased from Invitrogen (Carlsbad, CA). pEGFP-N1 and pDsRed1-N1 were purchased from BD Biosciences Clontech (Palo Alto, CA). pPCR-Script AMP SK (+) was purchased from Stratagene (La Jolla, CA). pcDNA3.1 RACK 1-203 was created by digesting pcDNA3.1 RACK1 (Buensuceso et al., 2001) with NotI and BamHI and subcloning the resulting truncated RACK1 cDNA into pcDNA3.1 myc his A (Invitrogen). pEGFP-N1-zyxin was obtained from Irina Kaverina (Austrian Academy of Sciences, Austria) and was cloned into pDsRed1-N1 (Bhatt et al., 2002); pEGFP-N1-α-actinin was obtained from Carol Otey (University of North Carolina, Chapel Hill, NC). RACK1 and RACK 1-203 GFP-fusion constructs were constructed in the pEGFP-N1 and pDsRed1-N1 vectors as follows: polymerase chain reaction (PCR) was used to amplify RACK1 and RACK 1-203 from the pcDNA3.1 vectors. The primers were designed to introduce a HindIII site at the 5′ end and a KpnI site at the 3′ end of the PCR products, and two extra nucleotides were added to permit the insert to be cloned in frame with green fluorescent protein (GFP) or red fluorescent protein (RFP). The PCR-amplified inserts were blunt-end ligated into pPCR-Script AMP vector, removed by digestion with HindIII and KpnI, and ligated into pEGFP-N1 or pRsRed1. The pEGFP-N1-RACK Y246F vector was constructed by introducing a point mutation into the pEGFP-N1-RACK1 vector by using the Transformer site-directed mutagenesis kit according to the manufacturer's instructions (BD Biosciences Clontech).
Antibodies and Reagents
Fibronectin was purified from human plasma by affinity chromatography as described previously (Ruoslahti et al., 1982). Monoclonal anti-vinculin and monoclonal anti-Src antibodies were purchased from Sigma-Aldrich (St. Louis, MO). Rhodamine-labeled phalloidin and rabbit anti GFP antibody were obtained from Molecular Probes (Eugene, OR). Monoclonal anti-RACK1 antibody and monoclonal anti-paxillin antibody were purchased from Transduction Laboratories (San Diego, CA). Rabbit whole molecule IgG was purchased from Zymed Laboratories (South San Francisco, CA). Mouse whole molecule IgG, sheep anti-mouse antibody conjugated to fluorescein, and goat anti-mouse antibody conjugated to rhodamine red were obtained from Jackson Immunoresearch Laboratories (West Grove, PA).
Cell Culture and Transfection
Chinese hamster ovary fibroblast-like cells (CHO-K1 cells) were obtained from the American Type Culture Collection (Manassas, VA). CHO αiibβ3 and CHO β3 724 cells were obtained from Mark Ginsberg (The Scripps Research Institute, La Jolla, CA). Cells were cultured in complete growth media as described previously (Cox et al., 2001).
CHO-K1 cells were transiently transfected using LipofectAMINE (Invitrogen) as described previously (Cox et al., 2001). Cells were used in experiments 24–60 h posttransfection. To plate CHO-K1 cells for experiments, cells were removed with 0.02% EDTA in phosphate-buffered saline (PBS) and then washed twice with serum-free media (CCM1; Hyclone Laboratories, Logan, UT). The cells were plated in CCM1 onto nontissue culture plastic plates or glass coverslips that had been coated with fibronectin (1 h, 37°C) and blocked with 2% bovine serum albumin (BSA) (30 min, 37°C). Cells were seeded onto 35-mm plates at 1.5 × 105/plate for time-lapse videomicroscopy experiments. Cells were seeded onto 12-mm glass coverslips at 2 × 104 for immunofluorescence cell staining experiments. Cells were seeded onto 10-cm plates at 2 × 106 for coimmunoprecipitations and kinase assays.
Expression Cloning
CHO-K1 cells were plated in tissue culture-treated 12-well plates at 4 × 104 cells/well. Twenty-four hours after plating, cells were transfected either with a pcDNA3.1hygro-lacZ control plasmid (Invitrogen) or with plasmids purified from a HeLa cDNA library (constructed using pcDNA3.1; Invitrogen). Plasmids were purified from the cDNA library using a 96-well plasmid mini-preparation. CHO cells were transiently transfected using 1 μg of DNA (estimated) and 4 μl of LipofectAMINE according to the manufacturer's instructions with the following exception: after adding the DNA–lipid complexes to the cells, .75 ml of complete growth media was added and cells were incubated for 12 h before splitting 1:3 into a new 12-well plate. Twenty-four hours later, cells were split 1:10 and seeded in complete growth media onto silanated coverslips coated with 10 μg/ml fibronectin. Glass coverslips were acid washed, silanated, conjugated to fibronectin, and blocked with 2% BSA using previously described methods (Crowley and Horwitz, 1995). One hour after plating, cells were washed with PBS and 400 μl of CCM1 was added to each well. Twelve hours after plating, cells were fixed, permeabilized, and stained for vinculin and actin as described previously (Huttenlocher et al., 1996).
Ninety-Six–Well DNA Mini-Preparation
Individual plasmids were prepared from a HeLa cDNA library (Invitrogen) through 96-well plasmid mini-preparation using MultiScreen-FB plates according to the manufacturer's instructions (technical note TN004; Millipore, Bedford, MA). The DNA preparations were performed according to the protocol with the following exceptions: 1) 10 μl of chloroform was added after DNA precipitation with solution III; 2) the FB plate was washed with 80% ethanol supplemented with 20 mM Tris-HCl pH 7.0, 50 mM NaCl, and 1 mM EDTA pH 8.0; and 3) an extra wash step was included for the FB plate. The DNA concentration was approximated through averaging the value obtained for seven randomly selected samples from the preparation.
Immunofluorescence
Cells were prepared for use in immunofluorescence cell staining experiments as described above. Glass coverslips were acid washed, silanated, conjugated to fibronectin, and blocked with 2% BSA using previously described methods (Crowley and Horwitz, 1995). Coverslips were coated with poly-l-lysine as follows: coverslips were etched for 2 h with 2 N NaOH, washed three times with PBS, dipped into a poly-l-lysine solution (0.001% poly-l-lysine [Sigma-Aldrich], 0.01 mg/ml Collagen [Sigma] in 27% ethanol), UV irradiated for 45 min, and washed one time with PBS. Cells were plated as described above. After a 4-h incubation at 37°C, 10% CO2, cells were fixed and stained for vinculin or paxillin as described previously (Huttenlocher et al., 1996). Coverslips were observed using an inverted fluorescent microscope (Nikon, Kanayawa, Japan). Images for colocalization studies were obtained using a cooled charge-coupled device camera (Hammamatsu, Bridgewater, NJ) and ISee analytical imaging software (Inovision, Raleigh, NC). For quantification of focal adhesion localization, 50–100 GFP-positive cells in three separate experiments were scored by blinded observers. The percentage of cells exhibiting central focal adhesions (as demonstrated by the vinculin-stained cells in Figure 4A, a and c) was determined.
Figure 4.
RACK1 regulates focal adhesion organization. Transiently transfected cells were plated onto coverslips coated with 20 μg/ml fibronectin. (A) Representative pictures that show the distribution of focal adhesions in cells that expressed pEGFP-N1 (a and b), pEGFP-N1-RACK1 (c and d), pEGFP-N1-RACK 1-203 (e and f), or pEGFP-N1-RACK Y246F (g and h). Vinculin staining is shown in the left column (a, c, e, and g) and GFP fluorescence is shown in the right column (b, d, f, and h). Bar, 20 μm. (B) Quantification of focal adhesion distribution. Blinded observers scored 50–100 GFP-positive cells from three separate experiments. Error bars represent the SD. *p < 0.01 in a Student's t test (in comparison with control).
Cell Spreading Assay
CHO-K1 cells were transiently transfected and prepared for cell spreading experiments as described above. Twenty-four to 48 h after transfection 1 × 104 cells were plated on coverslips coated with 10 μg/ml fibronectin. Cells were fixed at 10-min intervals and prepared as described above. Cell areas were determined of GFP-positive cells stained with rhodamine-labeled phalloidin using MetaMorph software (Universal Imaging, Downingtown, PA). Cell areas were determined for a total of 20–30 GFP-positive cells from two separate experiments.
Transwell Assays
Transwell assays (modified Boyden chamber assays) were performed using 6.5-mm polycarbonate Transwell filter inserts with 8-μm pores (CoStar, Cambridge, MA). For random and chemotactic migration assays, both sides of the filters were coated with 10 μg/ml fibronectin. For haptotactic migration assays, only the bottom side of the filter was coated with 10 μg/ml fibronectin. Transwell filters were incubated with fibronectin for 1 h at 37°C, washed with PBS, and blocked with 2% BSA in PBS for 30 min at 37°C. Filters used for chemotactic assays were dried completely before use. Forty-eight hours before the assays, cell were cotransfected with a 1:4 ratio of pcDNA3.1-hygro-lacZ to pEGFP-N1, pcDNA3.1 cDNA#25, pEGFP-N1-RACK 1-203, pEGFP-N1-RACK1, or pEGFP-RACK Y246F. Two plates of each were made: one for use in the Transwell assay and one to score transfection efficiency. Cells were prepared for random and haptotactic Transwell assays as described above and 1 × 105 cells in 100 μl of CCM1 were seeded onto the top of the Transwell filters (with CCM1 in the bottom chamber). For chemotactic assays, CCM1 was added to the bottom chamber of the Transwell and serum-starved CHO-K1 cells were added to the top in DMEM supplemented with 2 mM glutamine, 1 mM sodium pyruvate, 1% nonessential amino acids, 100 U/ml penicillin, 100 μg/ml streptomycin, and .2% BSA (Sigma-Aldrich). Cells were serum starved as described previously (Cox et al., 2001). For all Transwell assays, cells were incubated for 3 h at 37°C, 10% CO2 and then fixed for 5 min with a solution of 2% formaldehyde, 0.05% gluteraldehyde in PBS. The Transwells were washed once with PBS and then incubated in β-galactosidase staining solution (5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 2 mM MgCl2, 1 mg/ml X-gal in PBS) overnight. The next day, the number of blue cells per 10× field was scored for six fields per Transwell filter. The average number of cells that migrated per field was normalized to the transfection efficiency (as determined by β-galactosidase staining of the second plate). At least three experiments were performed for each condition.
Time-Lapse Videomicroscopy and Transient Protrusion analysis
CHO-K1 cells were plated as described above onto nontissue culture-treated 35-mm dishes coated with 3 μg/ml fibronectin and incubated for 1 h at 37°C in 10% CO2. The cells were placed onto a heated microscope stage with 10% CO2. A constant pH of 7.0–7.4 and temperature of 37°C was maintained during taping. Cells were allowed to equilibrate on the microscope stage for 1 h before images were captured for transient protrusion analysis. Positively transfected cells were identified through fluorescence, and transient protrusion analysis was performed using NIH Image software as described previously (Cox et al., 2001).
Immunoprecipitations and Src Kinase Assays
For immunoprecipitations, CHO-K1 cells were plated at 24 h posttransfection (as described above) onto nontissue culture-treated 10-cm dishes coated with 5 μg/ml fibronectin. Cells were incubated for 5 h at 37°C in 10% CO2, washed one time with Hanks' balanced saline solution, and lysed with ice cold radioimmunoprecipitation assay buffer (10% glycerol, 1% Triton X-100, 0.5% NP-40, 1% deoxycholate, 20 mM Tris-Cl pH 7.4, 150 mM NaCl, 2 mM EGTA, 2 mM EDTA, 40 mM NaF, 30 mM NaPPi) supplemented with 1 μg/ml pepstatin A, 10 μg/ml leupeptin, 20 μg/ml aprotinin, and 200 nM phenylmethanesulfonyl fluoride. Lysates were cleared of insoluble material by centrifugation at 180 kG for 40 min at 4°C. For the GFP coimmunoprecipitations, 400 μg of lysate was used; for kinase assays 600 μg of lysate was used. Next, 0.05% SDS, 10 μl of protein G-Sepharose (Amersham Biosciences, Piscataway, NJ), and either 1 μl of rabbit anti-GFP antisera, 2 μg of mouse antisera, 10 μg of rabbit IgG, or 10 μg of mouse IgG were added to the cleared lysates. After a 12-h incubation at 4°C with tilting, samples were washed three times with radioimmunoprecipitation assay buffer supplemented with 0.05% SDS and then extracted with Laemmli SDS sample buffer. Extracts were either used in a kinase assay or resolved in 5–18% gradient SDS-PAGE gels, transferred to 0.2-μm nitrocellulose, and blotted for GFP, vinculin, paxillin, or Src. Staining was detected using enhanced chemiluminescence. Kinase assays were performed as described previously (Chang et al., 1998).
RESULTS
Identification of cDNA#25, a C-terminal Truncated RACK1, by Using Expression Cloning
To identify novel proteins that regulate focal adhesions and cell migration, a mammalian cDNA expression cloning scheme was used. Putative proteins involved in regulating integrin-mediated cell-substratum adhesions were identified using a single cDNA screening method and an adhesion morphology screen as described under MATERIALS AND METHODS. A HeLa cell line cDNA library was used as a source of cDNAs and individual plasmids from the cDNA library (each encoding a single cDNA) were prepared and transiently transfected into CHO-K1 cells. Thirty-six hours posttransfection, cells were plated onto fibronectin-coated coverslips and were fixed and stained for vinculin and actin. In the initial feasibility screen 125 cDNAs were analyzed. We were interested in identifying cDNAs that induced a loss of central focal adhesions because this phenotype is induced by the inhibition or depletion of several proteins that regulate cell migration, including Src kinases (Klinghoffer et al., 1999), focal adhesion kinase (Sieg et al., 1999), and the calcium-dependent protease calpain (Huttenlocher et al., 1997).
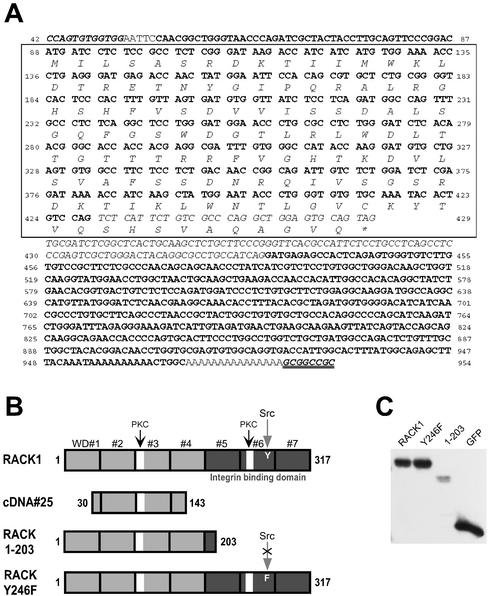
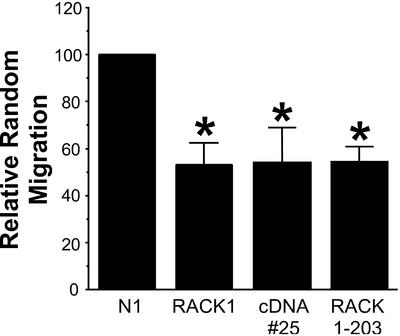
Two of the 125 cDNAs screened reproducibly induced the loss of central adhesive complexes and stress fibers. One of the cDNAs encoded a transcript with 99% sequence homology to the human lysosomal hyaluronidase and will be the focus of future investigation. The other cDNA, cDNA#25 (Figure 1), had 100% homology to the coding sequence for amino acids 30–143 of RACK1 (Figure 2A). The predicted protein product of cDNA#25 is a truncated form of RACK1 that contains one of the putative PKC binding sites but lacks the C-terminal sequences reported to bind integrin β cytoplasmic tails and Src (Liliental and Chang, 1998; Chang et al., 2001). The homology of the open reading frame of cDNA#25 with RACK1 is interrupted by an intervening sequence that has homology to repetitive human Alu sequences and introduces a stop codon into the reading frame (Figure 2A). We hypothesize that this truncated form of RACK1 may act as a dominant negative through binding to active PKC but failing to interact with other binding partners, including the integrin cytoplasmic domain and Src. A previous study reported a role for RACK1 during cell migration (Buensuceso et al., 2001). To determine whether cDNA#25 also modulates cell motility random Transwell migration assays were performed. We found that transient expression of cDNA#25, a C-terminal truncated RACK1 (RACK 1-203), or full-length RACK1 (Figure 2B) inhibited random cell migration, supporting an important role for RACK1 during cell migration (Figure 3). RACK 1-203 has a similar C-terminal deletion as cDNA#25, but lacks the N-terminal deletion (Figure 2B). Because cDNA#25 and RACK 1-203 induced similar effects on cell migration and on the organization of focal adhesions (Figure 4A, e), the C-terminal truncation of RACK1 (RACK 1-203) was used in subsequent experiments to characterize how RACK1 modulates adhesion and cell migration.
Figure 1.
Expression of cDNA#25 induces the loss of central focal adhesions and stress fibers. Vinculin staining is shown in green and actin staining is shown in red. (A) Representative cell from a population transfected with pcDNA3.1 lacZ (control vector). (B) Representative cells from a population transfected with pcDNA3.1 cDNA#25.
Figure 2.
Sequence of cDNA#25, schematic of constructs and transient expression levels. (A) Sequence of cDNA#25 is shown. cDNA#25 was cloned into pcDNA3.1 between a BstXI site (bold italics) and a NotI site (bold italics with double underline). Plain bold lettering indicates nucleotides that have homology to human RACK1. Homology starts 42 nucleotides downstream of the RACK1 start codon and spans the entire remaining sequence of RACK1. Numbers indicate the corresponding nucleotide of the full-length RACK1 sequence (starting with 42 and ending with 954 at the stop codon). Italicized lettering indicates an intervening sequence that introduces a stop codon into the open reading frame of cDNA#25. The predicted open reading frame is boxed and the amino acid translation is given. The open reading frame corresponds to amino acids 30–143 of RACK1. (B) Constructs used in experiments. Full-length RACK1 consists of seven tandemly repeated WD-40 motifs. Features of RACK1 include two putative PKC binding sites (white boxes), a Src binding site (tyrosine 246), and an integrin-binding domain (dark shading). The predicted open reading frame of cDNA#25 has homology to amino acids 30–143 of RACK1. RACK 1-203 is a truncated RACK1 construct and RACK Y246F is a construct with a point mutation at the Src binding site. (C) Representative blot showing transient expression levels for constructs used in experiments. Cells were transfected with pEGFP-N1-RACK1 (RACK1), pEGFP-N1-RACK Y246F (Y246F), pEGFP-N1-RACK 1-203 (1-203), or pEGFP-N1 (GFP). Lysates were collected at 24 h posttransfection, run on an SDS-PAGE gel, and blotted for GFP.
Figure 3.
Cell migration is inhibited by expression of full-length RACK1, cDNA#25, or C-terminal–truncated RACK1. Data from Transwell random migration experiments. To determine the relative random migration, the number of positively transfected cells that migrated for each condition was normalized to the transfection efficiency and then determined as a percentage of the control cells that migrated (set to a default of 100). Data represents the average from at least three separate experiments, with error bars representing the SEM. *p < 0.05 in a Student's t test (in comparison with control).
RACK1 Regulates Focal Adhesion Organization through Its Src Binding Site
To characterize the role of RACK1 and the RACK1–Src interaction on the regulation of focal adhesion organization, we generated GFP fusion proteins to full-length RACK1, truncated RACK1 (RACK 1-203), and a point mutant that cannot bind to Src (RACK Y246F) (Chang et al., 2001) (Figure 2B). The GFP-fusion constructs were transiently transfected into cells that were subsequently plated onto coverslips coated with 20 μg/ml fibronectin, a concentration that promotes the formation of both central and peripheral adhesion complexes in wild-type CHO-K1 cells. Cells were fixed and stained for vinculin to visualize adhesion complexes. Positively transfected cells were identified through GFP fluorescence and blinded observers assessed the localization of focal adhesions. We found equivalent expression (80–90% positive) of the constructs after transient expression by flow cytometry (our unpublished data), but interestingly we found less RACK 1-203 in the soluble fraction by Western immunoblotting (Figure 2C). In agreement with previously published results (Buensuceso et al., 2001), cells that expressed full-length RACK1 had strong peripheral and central adhesions and stress fibers (Figure 4A, c). We found that, similar to cDNA#25, RACK 1-203 reproducibly induced a loss of central focal adhesions (Figure 4A, e). These findings suggest that the C-terminal region of RACK1 plays a central role in regulating the organization and distribution of focal adhesions and stress fibers.
To determine whether the Src-binding site of RACK1 was critical for the effects of RACK1 on focal adhesion organization, we examined the effects of expressing a point mutation (Y246F) that lacks Src binding on adhesion complex organization. Previous studies have demonstrated in CHO-K1 cells that wild-type RACK1, but not the point mutant Y246F, binds to Src (Chang et al., 2001). We therefore used the Y246F mutation to characterize how RACK1–Src interactions modulate focal adhesion organization. Strikingly and in contrast to wild-type RACK1, we found that the point mutant, RACK Y246F induced the loss of central adhesive complexes (Figure 4A, g), similar to the effects observed with RACK 1-203 (Figure 4A, e). Only 19.3 ± 4.5% of RACK Y246F-transfected cells exhibited central focal adhesions, in comparison with 74.0 ± 3.6% of control transfected cells (Figure 4B). The effects of RACK Y246F on focal adhesion organization were independent of expression level because both high and low expressers displayed a similar phenotype (our unpublished data). Together, the data suggest that RACK1 modulates the organization of focal adhesions through its interaction with Src.
RACK1 Regulates Cell Spreading
The effects of RACK1 on focal adhesions suggest that RACK1 may play an important role during cell spreading. To determine whether cell spreading is RACK1 dependent, the effects of transient expression of wild-type RACK1, RACK 1-203, and RACK Y246F on cell spreading were examined. Cells were fixed at different time points after adhesion to a fibronectin substratum and the cell area of GFP-positive cells was determined. Control cells displayed an initial delay in cell spreading with a rapid increase in cell area after 20 min (Figure 5A). By 50 min, control cells were well spread with visible central and cortical actin filaments. In contrast, transient expression of any of the RACK1 constructs inhibited cell spreading. Cells that expressed the RACK1 constructs had an average cell area of ∼60% of control cells at 1 h, but by 4 h all cell types exhibited similar cell spreading (our unpublished data). Furthermore, with expression of the RACK1 constructs the actin cytoskeleton remained in punctate complexes without the visible organization observed in control cells at 50 min (Figure 5B). Interestingly, the kinetics of cell spreading varied in cells expressing the different RACK1 constructs. Initial cell spreading was rapid in cells that expressed RACK 1-203 or RACK Y246F, whereas cells that expressed wild-type RACK1 had a delay in initial spreading. However, by 50 min a similar inhibition in cell spreading was observed with expression of any of the RACK1 constructs. Together, these findings suggest that RACK1 plays a central role in regulating both the organization of adhesion complexes and cell spreading.
Figure 5.
Cell spreading is inhibited by expression of RACK1, RACK 1-203, and RACK Y246F. (A) Graph showing cell area plotted as a function of time after plating onto fibronectin-coated coverslips. Data are shown for cells transiently transfected with pEGFP-N1 (closed triangles), pEGFP-N1-RACK1 (open circles), pEGFP-N1-RACK 1-203 (open squares), and pEGFP-N1-RACK Y246F (open triangles). (B) Cells stained for actin 50 min after plating onto fibronectin. Representative GFP-positive cells transfected with pEGFP-N1 (a), pEGFP-N1-RACK1 (b), pEGFP-N1-RACK 1-203 (c), and pEGFP-N1-RACK Y246F (d) are shown. Bar, 20 μm.
RACK1 Localizes to Peripheral Adhesion Complexes That Contain Paxillin
To provide insight into RACK1 function, the subcellular localization of RACK1 was examined using immunofluorescence cell staining and GFP-fusion proteins. On concentrations of fibronectin that support the formation of both central and peripheral adhesive complexes (i.e., 20 μg/ml), we generally found that RACK1 displayed a diffuse cytoplasmic distribution with some localization to peripheral adhesive contact sites. In contrast, CHO-K1 cells plated on lower concentrations of fibronectin (5 μg/ml), which do not support the formation of central adhesive contact sites, showed more prominent localization of RACK1 in peripheral adhesion-like complexes (Figure 6). Staining was particularly evident in areas of membrane protrusion and at the periphery of spreading cells. We also examined the localization of the RACK1-GFP constructs in CHO-K1 cells adherent to fibronectin. The full-length RACK1-GFP colocalized with endogenous RACK1 in peripheral adhesion-like complexes (Figure 6B). To determine whether RACK1 localization was integrin dependent, RACK1-GFP localization was studied in cells plated on poly-l-lysine. In contrast to cells plated on fibronectin, RACK1-GFP was distributed diffusely and did not localize to peripheral complexes in cells plated on poly-l-lysine (Figure 6D), suggesting that RACK1 localization to adhesive complexes may be integrin dependent. Furthermore, RACK 1-203-GFP, which lacks the region proposed to bind to β integrin cytoplasmic domains, did not localize to adhesion sites (Figure 6E). In addition, we found that localization of RACK1 to adhesive complex sites was dependent on the β3 integrin cytoplasmic domain. We found that RACK1 localized normally to complexes at the cell periphery in cells that expressed wild-type αIIbβ3 integrin plated on fibrinogen, but not in cells that expressed a truncated β3 integrin (β3 724) that fails to localize to focal adhesions (our unpublished data) (Huttenlocher et al., 1996). Together, these findings suggest that RACK1 localization to adhesion-like structures is an integrin-dependent process. Interestingly, the RACK Y246F-GFP fusion protein exhibited a similar distribution as the wild-type RACK1-GFP (Figure 6F). These findings demonstrate that the C terminus of RACK1 (containing the integrin and Src binding domains) was necessary for localization to adhesion-like sites but the Src binding site was not required.
Figure 6.
RACK1 localization to peripheral adhesion complexes requires C-terminal sequences but not the Src-binding site. Cells were plated onto 5 μg/ml fibronectin except for D. (A) Cells stained for endogenous RACK1. The enlarged area is magnified two times from the original. (B) Cells that expressed RACK1-GFP (green) stained for endogenous RACK1 (red). Bar, 10 μm. (C) Cells that expressed RACK1-GFP plated on fibronectin. (D) Cells that expressed RACK1-GFP plated on poly-l-lysine. (E) Cell that expressed RACK 1-203-GFP. (F) Cell that expressed RACK Y246-GFP. Images show representative results observed in at least three separate experiments. Bar, 20 μm.
To characterize the peripheral complexes that contain RACK1, we determined whether RACK1 colocalized with specific focal adhesion components in these peripheral complexes. Recent studies have shown that adhesive complex sites can differ in composition, with some complexes containing only distinct components (Zamir et al., 1999, 2000). Interestingly, we found that RACK1 colocalized in the peripheral complex sites with only specific focal adhesion proteins. RACK1 localized to a subset of adhesions that contained paxillin or α-actinin, but did not colocalize with adhesions containing vinculin or zyxin (Figure 7A). Furthermore, full-length RACK1 was found to coimmunoprecipitate with paxillin but not with vinculin (Figure 7B). The colocalization of RACK1 with paxillin was not dependent on the Src binding site, because RACK Y246F was also found to colocalize with paxillin in a subset of nascent peripheral adhesive complex sites (our unpublished data). In addition, RACK 1-203 was found to coimmunoprecipitate with paxillin, suggesting that the C-terminal region of RACK1 was not required for paxillin binding (Figure 7B). Together, the data indicate that RACK1 localizes to a subset of peripheral adhesions that contain paxillin and α-actinin. These findings suggest that RACK1 may have a role in regulating nascent adhesions that form in areas of protrusion.
Figure 7.
RACK1 localizes to a subset of peripheral complexes that contain paxillin and α-actinin but not vinculin or zyxin. (A) Cells were plated onto coverslips coated with 5 μg/ml fibronectin. Images were taken with a 60× objective and enlarged areas were magnified two times in comparison with the original. a, Cell that expressed RACK1-GFP (green) stained for vinculin (red). b, Cell that expressed RACK1-GFP (green) stained for paxillin (red). c, Cell that expressed RACK1-GFP (green) and zyxin-RFP (red). d, Cell that expressed RACK-RFP (red) and α-actinin-GFP (green). Images show representative results observed in at least three separate experiments. Bar, 20 μm. (B) RACK1 and RACK 1-203 coimmunoprecipitate with paxillin, but not vinculin. Cells were transiently transfected with pEGFP-N1 (GFP), pEGFP-N1-RACK 1-203 (1-203), or pEGFP-N1-RACK1 (RACK1). Lysates were immunoprecipitated with either an anti-GFP antibody or a nonspecific rabbit IgG and blotted for vinculin, paxillin, or GFP. Total cell lysate was stained for vinculin and paxillin. Blots show a representative result from three separate experiments.
RACK1 Regulates Protrusive Activity via Its Src Binding Site
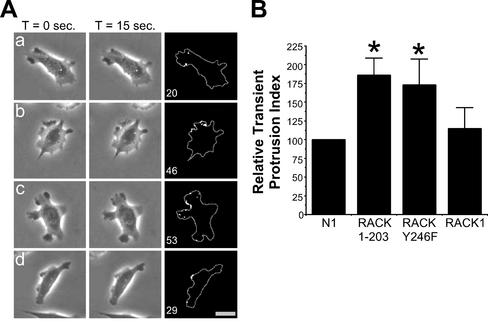
The localization of RACK1 to protrusion sites and its effects on cell spreading suggest that RACK1 may be involved in regulating protrusive activity. We therefore studied the effects of transient expression of control vector, RACK1, RACK 1-203, and RACK Y246F on transient protrusive activity. We analyzed protrusive activity using time-lapse videomicroscopy and transient protrusion analysis as described previously (Cox et al., 2001). This analysis quantifies changes in gray levels on the cell surface and can detect membrane movements that occur over short 15-s intervals. Cells were transiently transfected with either pEGFP-N1 (control), pEGFP-N1-RACK1, pEGFP-N1-RACK 1-203, or pEGFP-N1-RACK Y246F, and GFP-positive cells were analyzed. Examples demonstrating the transient protrusion analysis are shown in Figure 8A. Cells that expressed RACK 1-203 or RACK Y246F typically exhibited multiple protrusions around the cell periphery (Figure 8A, b and c), in contrast to control cells and RACK1-expressing cells that typically had a defined leading edge (Figure 8A, a and d). Transient protrusion analysis demonstrated that cells that expressed RACK 1-203 or RACK Y246F, but not wild-type RACK1, exhibited increased transient protrusive activity in comparison with control cells (Figure 8B). Together, the data suggest that RACK1, through its interaction with Src, may regulate protrusive activity.
Figure 8.
RACK1 regulates transient protrusion via Src. (A) Examples of cells used in the transient protrusion analysis. Representative cells that expressed pEGFP-N1 (a), pEGFP-N1-RACK 1-203 (b), pEGFP-N1-RACK Y246F (c), or pEGFP-N1-RACK1 (d) are shown. Videos are available at www.molbiolcell.org (Figure 7Aa.mov–Figure 7Ad.mov). Analysis was performed by collecting images at 15-s intervals for 5 min as described under MATERIALS AND METHODS. Pixels undergoing more than a 5% change in gray level intensity were scored as positive. The number in the lower left corner of the right column indicates the number of positive pixels detected for the 15-s interval shown. Bar, 20 μm. (B) Data from the transient protrusion analysis. The transient protrusion index for cells transfected with RACK 1-203, RACK Y246F, or RACK1 was determined as a percentage of control (relative transient protrusion index). The transient protrusion index is the average number of pixels per cell undergoing a ≥5% change in gray level value per 15-s interval, over a 5-min time period. Data represent the average of at least three separate experiments (at least 10 cells were analyzed per experiment) with the error bars representing the SEM. *p < 0.05 in a Student's t test.
Because RACK1 has previously been shown to decrease Src kinase activity (Chang et al., 1998), these results suggest that RACK1 may suppress protrusion by inhibiting Src. Accordingly, expression of RACK Y246F may enhance protrusive activity by increasing Src activity. To determine whether expression of RACK Y246F affects Src activity, kinase activity assays were performed. We found that transient expression of RACK Y246F enhanced Src activity in comparison with cells that expressed control vector or wild-type RACK1 (Figure 9). We consistently observed a small decrease in Src activity in cells that expressed RACK1 compared with control vector; however, the baseline Src activity was low in CHO-K1 cells under the conditions of the assay. These results suggest that RACK1 may suppress protrusive activity through inhibiting Src kinase activity.
Figure 9.
Transient expression of RACK Y246F results in elevated Src activity. Cells were transiently transfected with either pEGFP-N1 (GFP), pEGFP-N1-RACK Y246F (Y246F), or pEGFP-N1-RACK1 (RACK1). Immunoprecipitations were performed with either mouse IgG or a Src-specific antibody. Kinase activity assays were performed as described under MATERIALS AND METHODS. Top, blot showing the relative amounts of Src that were pulled down in the immunoprecipitation step. Bottom, autoradiograph showing results from the kinase assay. Expression of RACK Y246F increased Src kinase activity in comparison with cells expressing the control vector or wild-type RACK1. Shown is a representative result from three separate experiments.
RACK1 Regulates Chemotactic Cell Migration via Its Src Binding Site
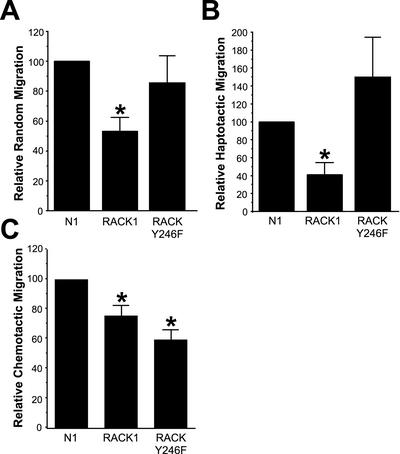
The localization of RACK1 to nascent adhesions in areas of protrusion and the effects of RACK Y246F expression on membrane protrusion suggest that RACK1 may modulate directional migration via its Src binding site. To test this possibility, the random, haptotactic, and chemotactic migration of CHO-K1 cells was examined using Transwell migration assays. We found that expression of RACK Y246F, unlike wild-type RACK1, did not affect random cell migration (Figure 10A). Transient expression of RACK Y246F also did not significantly inhibit haptotactic migration toward fibronectin; however, similar to previously published results (Buensuceso et al., 2001), expression of wild-type RACK1 did inhibit haptotactic migration (Figure 10B). Interestingly, transient expression of RACK Y246F significantly inhibited the directional migration of serum-starved CHO-K1 cells to growth factor-containing media (Figure 10C), supporting an important role for the Src-binding site of RACK1 in regulating chemotactic migration. Together, the data indicate that RACK1 regulates random, haptotactic, and chemotactic cell migration and that the Src binding site is necessary for optimal chemotactic cell migration.
Figure 10.
RACK1 regulates directional cell migration via Src. (A) Data from random Transwell migration experiments. (B) Data from haptotactic Transwell migration experiments. (C) Data from chemotactic Transwell migration experiments. To determine the relative migration, the number of positively transfected cells that migrated for each condition was normalized to the transfection efficiency and then determined as a percentage of the control cells that migrated (set to a default of 100). Data represent the average from at least three separate experiments, with error bars representing the SEM. *p < 0.05 in a Student's t test (in comparison with control).
DISCUSSION
This study demonstrates that RACK1 is an important regulator of focal adhesion organization, cell spreading, and migration. The effects of RACK1 on cell migration are at least in part mediated through the Src binding site of RACK1. Expression of RACK1 Y246F, a point mutant that does not bind to Src, induced a loss of central adhesions, increased protrusiveness, and inhibited chemotactic cell migration. The localization of RACK1 with the Src substrate paxillin in adhesion complexes suggests that RACK1 may play an important role in regulating paxillin-containing adhesions. Together, the results indicate that RACK1 regulates focal adhesion organization, protrusion, and chemotactic cell migration through its Src binding site.
Our findings demonstrate the utility of using mammalian cDNA expression cloning and a functional screen to identify novel proteins that regulate focal adhesion organization and cell migration. Despite recent progress, we still have a limited understanding of the key players that regulate the organization of adhesive complex sites and cell migration. Mammalian cDNA expression cloning has been used successfully by others to identify a novel regulator of adhesion to collagen (Pullman and Bodmer, 1992) and signaling molecules that regulate integrin affinity (Fenczik et al., 1997; Hughes et al., 1997; Ramos et al., 1998). We now provide evidence that a single cDNA expression cloning approach can be used with a functional screen to identify novel cDNAs that regulate the organization of focal adhesions and cell migration. In an initial feasibility screen of 125 cDNAs using a low-throughput approach, we identified two cDNAs that reproducibly induced a loss of central adhesion complexes, a phenotype that has been associated with inhibited cell migration (Horwitz and Parsons, 1999). This suggests that a large percentage of expressed genes may regulate focal adhesion composition and organization. One of the identified cDNAs (#25) was found to encode a C-terminal truncated form of the PKC and Src-binding protein RACK1 (Figures 1 and 2). Our characterization of RACK1 has demonstrated that it is an important regulator of integrin-mediated adhesion, protrusion, and cell migration. Together, the results demonstrate the feasibility of using mammalian cDNA expression cloning to identify novel regulators of focal adhesion organization and cell migration.
Recent evidence indicates that RACK1 functions as a scaffolding protein that binds to many intracellular proteins, including PKC (Ron et al., 1994), Src (Chang et al., 1998), β integrin cytoplasmic tails (Liliental and Chang, 1998; Buensuceso et al., 2001), RasGAP and phospholipase C-γ (Chang et al., 2001). RACK1 is therefore poised to serve as a critical intermediate during integrin-mediated signaling events. In support of this, our findings demonstrate that RACK1 localizes to peripheral adhesive complex sites that contain specific focal complex components, including paxillin and α-actinin (Figure 7A) and that this localization is integrin dependent (Figure 6). Furthermore, we find that RACK1 is an important regulator of focal adhesion organization (Figure 4). In contrast to the full-length RACK1, expression of a truncated form of RACK1 that lacks the integrin and Src binding sites reduced focal adhesion number and promoted the formation of only peripheral adhesive contacts (Figure 4A, e). These findings suggest that the truncated RACK1 may be acting as a dominant negative, by binding to protein kinase C but failing to bind to other key partners such as integrin and Src. Further studies demonstrated that perturbing the Src binding site of RACK1 was sufficient to induce a loss of central adhesions (Figure 4A, g). Together, the results suggest that the interaction of RACK1 with Src seems to be important for the regulation of focal adhesion organization.
The localization of RACK1 to a subset of nascent adhesions in areas of protrusion that contain paxillin and α-actinin suggested that RACK1 may modulate the stabilization or dynamics of these early adhesive contact sites. Interestingly, paxillin has recently been shown to be one of the first proteins enriched in nascent adhesions that form in areas of protrusion (Laukaitis et al., 2001). Recent evidence suggests that factors that affect the disassembly or dynamics of these early peripheral complexes may affect the formation and distribution of central adhesive complex sites (Horwitz and Parsons, 1999; Laukaitis et al., 2001; Bhatt et al., 2002). Our findings suggest that RACK1 localizes to these nascent peripheral complexes and may be a critical regulator of the subsequent formation and organization of central adhesive complex sites.
In addition to having a role in focal adhesion organization, we found that the Src binding site was critical for RACK1-mediated effects on protrusion and chemotactic cell migration, but not for random or haptotactic motility (Figures 8 and 10). This indicates that the Src binding site of RACK1 is not necessary for basal cell motility but is required specifically for directional motility during chemotaxis. This points to the interesting possibility that RACK1 may mediate cross talk between growth factor receptors and integrins to promote directional migration toward chemotactic gradients. Additionally, RACK1 also seems to be an important regulator of basal cell motility. We found that expression of full-length RACK1, cDNA#25, or RACK 1-203 reduced random cell migration by ∼50% in comparison with control cells (Figure 3). Together, the data indicate that sequences in the C terminus of RACK1 are important for basal cell motility, but the Src-binding site is not required. Because the C terminus contains the integrin-binding domain, it is possible that RACK1 localization to adhesion sites is necessary for optimal cell migration. In addition, the interaction of RACK1 with protein kinase C is also likely to be important for the regulation of cell motility (Buensuceso et al., 2001).
We believe that the RACK Y246F mutation is likely to exert its effects through perturbing the interaction of RACK1 with Src family kinases. Although it is possible that Y246 also mediates interaction of RACK1 with other Src-family members such as Fyn, the direct effect of the Y246F mutation on Src activity (Figure 9) suggests that Src is likely to be an important mediator of the observed effects. Src phosphorylates many adhesion-associated proteins, including paxillin and focal adhesion kinase, and modulates the incorporation of signaling molecules into adhesion complex sites (reviewed in Richardson and Parsons, 1995; Yamada and Miyamoto, 1995). Expression of activated v-Src causes cell rounding and loss of focal adhesions (Burridge et al., 1988; Chambers and Tuck, 1988; Fincham et al., 1995; Fincham and Frame, 1998). Therefore, it is possible that RACK1 regulates adhesive complexes through affecting the activity and/or localization of Src. Interestingly, expression of v-Src, like RACK Y246F, has been shown to induce increased membrane protrusiveness and inhibited directional cell migration (Chambers and Tuck, 1988; Kundra et al., 1994). Therefore, it is possible that the Y246F mutation prevents RACK1 binding to Src, causing elevated Src activity, enhanced membrane ruffling, and decreased chemotactic migration. In summary, our results demonstrate that RACK1 regulates cell-substratum adhesion organization, protrusion, and directional cell migration through its interaction with Src. Future investigations will focus on determining the downstream mechanisms by which RACK1 and Src family kinases regulate adhesive structures, protrusion, and directional cell migration.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alison Holub and Stacy Schommer for excellent technical assistance. We also thank Rick Horwitz for useful discussions and Amit Bhatt for critical reading of the manuscript. This research was support by National Institutes of Health grant R01-CA85862-01.
Footnotes
Online version of this article contains video material for some figures. Online version available at www.molbiolcell.org.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–03–0142. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–03–0142.
REFERENCES
- Berns H, Humar R, Hengerer B, Kiefer FN, Battegay EJ. RACK1 is up regulated in angiogenesis and human carcinomas. FASEB J. 2000;14:2549–2558. doi: 10.1096/fj.99-1038com. [DOI] [PubMed] [Google Scholar]
- Bhatt A, Kaverina I, Otey C, Huttenlocher A. Regulation of focal complex composition and disassembly by the calcium-dependent protease calpain. J Cell Sci. 2002;115:3415–3425. doi: 10.1242/jcs.115.17.3415. [DOI] [PubMed] [Google Scholar]
- Buensuceso CS, Woodside D, Huff JL, Plopper GE, O'Toole TE. The WD protein RACK1 mediates protein kinase C and integrin-dependent cell migration. J Cell Sci. 2001;114:1691–1698. doi: 10.1242/jcs.114.9.1691. [DOI] [PubMed] [Google Scholar]
- Burridge K, Fath K, Kelly T, Nuckolls G, Turner C. Focal adhesions: transmembrane junctions between the extracellular matrix and the cytoskeleton. Annu Rev Cell Biol. 1988;4:487–525. doi: 10.1146/annurev.cb.04.110188.002415. [DOI] [PubMed] [Google Scholar]
- Chambers AF, Tuck AB. Oncogene transformation and the metastatic phenotype. Anticancer Res. 1988;8:861–871. [PubMed] [Google Scholar]
- Chang BY, Chiang M, Cartwright CA. The interaction of Src and RACK1 is enhanced by activation of protein kinase C and tyrosine phosphorylation of RACK1. J Biol Chem. 2001;276:20346–20356. doi: 10.1074/jbc.M101375200. [DOI] [PubMed] [Google Scholar]
- Chang BY, Conroy KB, Machleder EM, Cartwright CA. RACK1, a receptor for activated C kinase and a homolog of the beta subunit of G proteins, inhibits activity of Src tyrosine kinases and growth of NIH 3T3 cells. Mol Cell Biol. 1998;18:3245–3256. doi: 10.1128/mcb.18.6.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox EA, Huttenlocher A. Regulation of integrin-mediated adhesion during cell migration. Microsc Res Tech. 1998;43:412–419. doi: 10.1002/(SICI)1097-0029(19981201)43:5<412::AID-JEMT7>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Cox EA, Sastry SK, Huttenlocher A. Integrin-mediated adhesion regulates cell polarity and membrane protrusion through the Rho family of GTPases. Mol Biol Cell. 2001;12:265–277. doi: 10.1091/mbc.12.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley E, Horwitz AF. Tyrosine phosphorylation and cytoskeletal tension regulate the release of fibroblast adhesions. J Cell Biol. 1995;131:525–537. doi: 10.1083/jcb.131.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenczik CA, Sethi T, Ramos JW, Hughes PE, Ginsberg MH. Complementation of dominant suppression implicates CD98 in integrin activation. Nature. 1997;390:81–85. doi: 10.1038/36349. [DOI] [PubMed] [Google Scholar]
- Fincham VJ, Wyke JA, Frame MC. V-src-induced degradation of focal adhesion kinase during morphological transformation of chicken embryo fibroblasts. Oncogene. 1995;10:2247–2252. [PubMed] [Google Scholar]
- Fincham VJ, Frame MC. The catalytic activity of Src is dispensable for translocation to focal adhesions but controls the turnover of these structures during cell motility. EMBO J. 1998;17:81–92. doi: 10.1093/emboj/17.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geijsen N, Spaargaren M, Raaijmakers JA, Lammers JW, Koenderman L, Coffer PJ. Association of RACK1 and PKC beta with the common beta-chain of the IL-5/IL-3/GM-CSF receptor. Oncogene. 1999;18:5126–5130. doi: 10.1038/sj.onc.1202896. [DOI] [PubMed] [Google Scholar]
- Horwitz AR, Parsons JT. Cell migration–movin'on. [letter; comment] Science. 1999;286:1102–1103. doi: 10.1126/science.286.5442.1102. [DOI] [PubMed] [Google Scholar]
- Hughes PE, Renshaw MW, Pfaff M, Forsyth J, Keivens VM, Schwartz MA, Ginsberg MH. Suppression of integrin activation: a novel function of a ras/raf-initiated MAP kinase pathway. Cell. 1997;88:521–530. doi: 10.1016/s0092-8674(00)81892-9. [DOI] [PubMed] [Google Scholar]
- Huttenlocher A, Ginsberg MH, Horwitz AF. Modulation of cell migration by integrin-mediated cytoskeletal linkages and ligand-binding affinity. J Cell Biol. 1996;134:1551–1562. doi: 10.1083/jcb.134.6.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher A, Palecek SP, Lu Q, Zhang W, Mellgren RL, Lauffenburger DA, Ginsberg MH, Horwitz AF. Regulation of cell migration by the calcium-dependent protease calpain. J Biol Chem. 1997;272:32719–32722. doi: 10.1074/jbc.272.52.32719. [DOI] [PubMed] [Google Scholar]
- Huttenlocher A, Sandborg RR, Horwitz AF. Adhesion in cell migration. Curr Opin Cell Biol. 1995;7:697–706. doi: 10.1016/0955-0674(95)80112-x. [DOI] [PubMed] [Google Scholar]
- Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- Klinghoffer RA, Sachsenmaier C, Cooper JA, Soriano P. Src family kinases are required for integrin but not PDGFR signal transduction. EMBO J. 1999;18:2459–2471. doi: 10.1093/emboj/18.9.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundra V, Soker S, Zetter BR. Excess early signaling activity inhibits cellular chemotaxis toward PDGF-bb. Oncogene. 1994;9:1429–1435. [PubMed] [Google Scholar]
- Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- Laukaitis CM, Webb DJ, Donais K, Horwitz AF. Differential dynamics of α5 integrin, paxillin, and alpha-actinin during formation and disassembly of adhesions in migrating cells. J Cell Biol. 2001;153:1427–1440. doi: 10.1083/jcb.153.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liliental J, Chang DD. RACK1, a receptor for activated protein kinase C, interacts with integrin β subunit. J Biol Chem. 1998;273:2379–2383. doi: 10.1074/jbc.273.4.2379. [DOI] [PubMed] [Google Scholar]
- Mochly-Rosen D, Gordon AS. Anchoring proteins for protein kinase C: a means for isozyme selectivity. FASEB J. 1998;12:35–42. [PubMed] [Google Scholar]
- Mourton T, Hellberg CB, Burden-Gulley SM, Hinman J, Rhee A, Brady-Kalnay SM. The PTPμ protein-tyrosine phosphatase binds and recruits the scaffolding protein RACK1 to cell-cell contacts. J Biol Chem. 2001;276:14896–14901. doi: 10.1074/jbc.M010823200. [DOI] [PubMed] [Google Scholar]
- Padanilam BJ, Hammerman MR. Ischemia-induced receptor for activated C kinase (RACK1) expression in rat kidneys. Am J Physiol. 1997;272:F160–F166. doi: 10.1152/ajprenal.1997.272.2.F160. [DOI] [PubMed] [Google Scholar]
- Pullman WE, Bodmer WF. Cloning and characterization of a gene that regulates cell adhesion. Nature. 1992;356:529–532. doi: 10.1038/356529a0. [DOI] [PubMed] [Google Scholar]
- Ramos JW, Kojima TK, Hughes PE, Fenczik CA, Ginsberg MH. The death effector domain of PEA-15 is involved in its regulation of integrin activation. J Biol Chem. 1998;273:33897–33900. doi: 10.1074/jbc.273.51.33897. [DOI] [PubMed] [Google Scholar]
- Richardson A, Parsons JT. Signal transduction through integrins: a central role for focal adhesion kinase? Bioessays. 1995;17:229–36. doi: 10.1002/bies.950170309. [DOI] [PubMed] [Google Scholar]
- Rodriguez MM, Ron D, Touhara K, Chen CH, Mochly-Rosen D. RACK1, a protein kinase C anchoring protein, coordinates the binding of activated protein kinase C and select pleckstrin homology domains in vitro. Biochemistry. 1999;38:13787–13794. doi: 10.1021/bi991055k. [DOI] [PubMed] [Google Scholar]
- Ron D, Chen CH, Caldwell J, Jamieson L, Orr E, Mochly-Rosen D. Cloning of an intracellular receptor for protein kinase C: a homolog of the β subunit of G proteins. Proc Natl Acad Sci USA. 1994;91:839–843. doi: 10.1073/pnas.91.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoslahti E, Hayman EG, Pierschbacher M, Engvall E. Fibronectin: purification, immunochemical properties, and biological activities. Methods Enzymol. 1982;82:803–831. doi: 10.1016/0076-6879(82)82103-4. [DOI] [PubMed] [Google Scholar]
- Sieg DJ, Hauck CR, Schlaepfer DD. Required role of focal adhesion kinase (FAK) for integrin-stimulated cell migration. J Cell Sci. 1999;112:2677–2691. doi: 10.1242/jcs.112.16.2677. [DOI] [PubMed] [Google Scholar]
- Yamada KM, Miyamoto S. Integrin transmembrane signaling and cytoskeletal control. Curr Opin Cell Biol. 1995;7:681–689. doi: 10.1016/0955-0674(95)80110-3. [DOI] [PubMed] [Google Scholar]
- Yarwood SJ, Steele MR, Scotland G, Houslay MD, Bolger GB. The RACK1 signaling scaffold protein selectively interacts with the cAMP-specific phosphodiesterase PDE4D5 isoform. J Biol Chem. 1999;274:14909–14917. doi: 10.1074/jbc.274.21.14909. [DOI] [PubMed] [Google Scholar]
- Zamir E, Katz BZ, Aota S, Yamada KM, Geiger B, Kam Z. Molecular diversity of cell-matrix adhesions. J Cell Sci. 1999;112:1655–1669. doi: 10.1242/jcs.112.11.1655. [DOI] [PubMed] [Google Scholar]
- Zamir E, et al. Dynamics and segregation of cell-matrix adhesions in cultured fibroblasts. Nat Cell Biol. 2000;2:191–196. doi: 10.1038/35008607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.