Abstract
The study of the nuclear pore in vertebrates would benefit from a strategy to directly identify new nucleoporins and interactions between those nucleoporins. We have developed a novel two-step “organelle trap” assay involving affinity selection and in vitro pore assembly. In the first step, soluble proteins derived from Xenopus egg extracts are applied to a column containing a ligand of interest. The bound proteins are then tagged by biotinylation and eluted. In the second step, potential nucleoporins are selected for by virtue of their ability to assemble into annulate lamellae, a cytoplasmic mimic of nuclear pores. The incorporated proteins are then recognized by their biotin tag. Here we use the lectin wheat germ agglutinin (WGA) as ligand; WGA inhibits nuclear transport and has been shown to directly bind three known nucleoporins from Xenopus extract, Nup62, Nup98, and Nup214, all of which contain N-acetylglucosamine residues. Under reduced-stringency conditions, three additional proteins bind to WGA–Sepharose and are revealed by the organelle trap assay. We identified all three as partner nucleoporins. Two were discovered to be Xenopus Nup93 and Nup205. The third is a novel vertebrate nucleoporin, Nup188. This new vertebrate protein, Xenopus Nup188, exists in a complex with xNup93 and xNup205. The Nup93-Nup188-Nup205 complex does not bind directly to WGA but binds indirectly via the N-acetylglucosamine–modified nucleoporins. A gene encoding human Nup188 was also identified. The discovery of vertebrate Nup188, related to a yeast nucleoporin, and its novel protein–protein interactions illustrates the power of the two-step organelle trap assay and identifies new building blocks for constructing the nuclear pore.
INTRODUCTION
The nuclear pore is essential for establishing the unique identity of the nuclear compartment of eukaryotic cells (Elliott et al., 1994; Izaurralde and Mattaj, 1995; Corbett and Silver, 1997; Ohno et al., 1998; Adam, 1999; Gorlich and Kutay, 1999; Stoffler et al., 1999). The pore acts as a highly specific gatekeeper regulating the traffic of macromolecules into and out of the nucleus. Although ions and small metabolites diffuse freely through the nuclear pore, larger molecules require specific signals and transport receptors for import. The vertebrate pore complex is a massive structure of ∼125 million Da composed of ∼50 distinct polypeptides present in multiple copies. Its structure consists of a set of three octagonally symmetric rings stacked one atop another, with the central ring being composed of eight spokes that hold in place the central transporter of the pore (Stewart et al., 1990; Hinshaw et al., 1992; Akey and Radermacher, 1993). Attached to the cytoplasmic ring are eight short filaments that project into the cytoplasm. Extending from the nuclear ring is a set of eight longer filaments connected to a ring at their termini to form a “nuclear basket.” This latter structure extends into the nucleoplasm. At present, ∼16 vertebrate pore proteins or nucleoporins have been identified (Adam, 1999; Stoffler et al., 1999). These include components of the nuclear basket (Nup153, Nup98), proteins of the central region of the pore (the Nup62-Nup58-Nup54-Nup45 complex), components of the cytoplasmic filaments (Nup214/CAN, Nup358), and integral membrane pore proteins (gp210, POM121), among others (Wozniak and Blobel, 1992; Pante et al., 1994; Söderqvist and Hallberg, 1994; Bastos et al., 1997; Grandi et al., 1997). A subset of the nucleoporins in metazoans contain N-acetylglucosamine (GlcNAc) residues and thus bind the lectin wheat germ agglutinin (WGA) (Davis and Blobel, 1987; Holt et al., 1987; Snow et al., 1987; Finlay and Forbes, 1990; Finlay et al., 1991; Guan et al., 1995). An overlapping set of pore proteins, both in metazoans and yeast, contain regions dominated by repeated phenylalanine–glycine or FG repeats (Davis and Blobel, 1986, 1987; Featherstone et al., 1988; Gorsch et al., 1995; Iovine et al., 1995; Powers et al., 1995; Neville et al., 1997; Bayliss et al., 1999). Even though the entire yeast genome has been sequenced, to date only a fraction of the known yeast pore proteins share extensive homology to vertebrate nucleoporins, so one cannot draw easy comparisons. Therefore, characterization of the pore complexes from higher eukaryotes has most often entailed taking a direct biochemical approach to elucidate the vertebrate nucleoporins.
One approach to the study of the molecular composition of the nuclear pore complex has involved the use of in vitro assembly assays. Several nuclear reconstitution systems have been used to examine the biology of the nuclear envelope and nuclear pores. A Chinese hamster ovary cell extract has been used to study both lamin and nuclear envelope assembly (Burke and Gerace, 1986; Suprynowicz and Gerace, 1986). Perhaps the most widely used assembly system is derived from extracts prepared from Xenopus eggs (Newmeyer and Wilson, 1991; Powers et al., 1995; Saitoh and Dasso, 1995). This system takes advantage of the large stockpiles of disassembled nuclear components present in the egg and has proven of great utility for examining the roles of a subset of nucleoporins in pore assembly and function. For example, removal of all of the WGA-binding proteins from the extract results in the assembly of nuclei with pores incompetent for import (Finlay and Forbes, 1990). Depletion of another nucleoporin, Nup93, causes a pronounced decrease in the number of pores formed per nucleus (Grandi et al., 1997). In theory, one could look for novel pore proteins by virtue of their incorporation into in vitro assembled nuclei. However, because there are other parallel biochemical processes, such as DNA replication and lamin assembly, occurring simultaneously, there is no simple method for detecting novel nucleoporins merely on the basis of their incorporation into assembled nuclei.
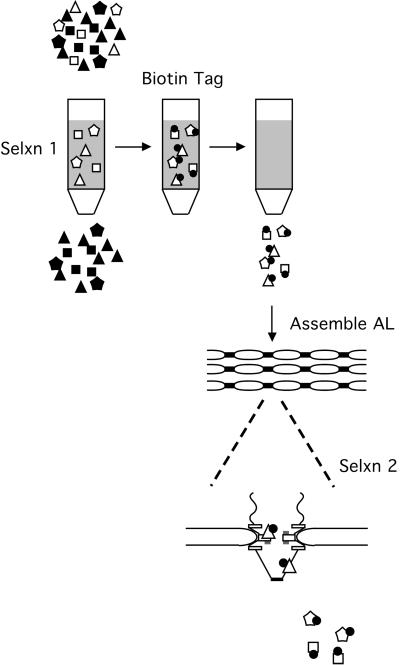
An alternative, simpler system for studying pore formation is that of in vitro annulate lamellae assembly. Annulate lamellae (AL) are cytoplasmic stacks of double membranes containing large numbers of tightly packed pore complexes (Dabauvalle et al., 1991; Kessel, 1992; Meier et al., 1995). In each double membrane, the pores are hexagonally packed and individual lamella are aligned in register via en face contacts between pores (see Figure 1). AL are commonly observed in rapidly growing or differentiating cells, such as tumor cells, male and female gametes, and virally infected cells (Kessel, 1992). Although the functional role of AL is not defined, it is surmised that they represent a storage form of excess pores for the rapid reassembly of daughter nuclei during cell division. It is thought that AL dissemble each cell cycle and that their components are used to assemble nuclear pores in daughter nuclei.
Figure 1.
The organelle trap assay for novel nucleoporins. In the ligand-binding/organelle trap assay, Xenopus egg cytosolic proteins (polygons) are applied to a ligand affinity matrix in the first selection step. In this study, immobilized WGA was used as the matrix. The column is washed, and bound polypeptides (open polygons) are then labeled with biotin (closed circles). The biotin-tagged proteins are eluted from the column with competing ligand, e.g., GlcNAc/TCT buffer in the case of WGA. In the second selection step, the mixture is added to an AL assembly reaction. Only nucleoporins and pore-associated polypeptides (tagged open triangles) will assemble into AL and are identified by Western blotting with SA–HRP. Nonspecific biotin-tagged proteins (tagged open hexagons and squares) are not incorporated and are removed from the AL during centrifugation.
The pores of AL are morphologically indistinguishable from nuclear pores at the electron microscopic level, and they contain the normal complement of the known nucleoporins. In fact, these pores have been shown to bind both nuclear transport substrates and their cognate receptors (Cordes et al., 1997; B.R. Miller and D.J. Forbes, unpublished data). The Xenopus nuclear reconstitution system mentioned above readily forms large numbers of AL when DNA or chromatin is omitted from a reconstitution assay (Dabauvalle et al., 1991; Meier et al., 1995). In addition to being a much simpler system for the study of pore formation, we have shown that assembled AL can be readily separated from the bulk of cellular membranes on the basis of their buoyant density (Meier et al., 1995). The greater density of AL compared to other membranous organelles derives from the high ratio of protein to lipid in AL that results from the numerous, almost hexagonally packed pores. Using AL reconstitution, we were able to show that removal of the WGA-binding nucleoporins from an assembly reaction results in a detectable reduction in the density of in vitro assembled AL (Meier et al., 1995).
Because incorporation of nucleoporins into AL can be monitored simply by Western blotting, in this study we have used AL reconstitution as part of a “organelle trap” assay for novel pore proteins. This approach acts as a double sieve to identify novel nucleoporins, with the first round of selection being based on the affinity of the protein(s) for a transport-related ligand and the second round of selection, or “trap,” based on incorporation into AL. In this way, one can select for interaction between nucleoporins and ligands of low affinity or specificity, because the second pore assembly reaction provides quite a stringent selection. The high background possible when reducing the stringency of the first step is eliminated in the second step. Therefore, we can identify nucleoporins that interact less strongly or perhaps only transiently with a given ligand.
In the experiments described here, we were able to use the two-step organelle trap assay to identify a novel vertebrate nucleoporin, Xenopus Nup188, and further found that this new nucleoporin exists in a complex with two other pore proteins, Nup93 and Nup205. The Nup93-Nup188-Nup205 complex binds indirectly to WGA via known WGA-binding nucleoporins. Because this interaction is normally quite sensitive to both salt and detergent, the organelle trap assay was instrumental both in revealing Nup188 as a vertebrate nucleoporin and also in revealing the interactions that occur between separate subcomplexes of the pore.
MATERIALS AND METHODS
Reagents
The mouse monoclonal mAb414 was purchased from BAbCo (Berkeley, CA). Affinity-purified rabbit polyclonal antibodies to Xenopus Nup62, Nup98, and Nup214 have been described (Finlay and Forbes, 1990; Macaulay et al., 1995; Powers et al., 1995). Antibodies to recombinant human Nup93 were prepared as described by Grandi et al. (1997) and affinity purified against Xenopus Nup93 (purified as below) bound to polyvinylidene difluoride (PVDF) strips. Affinity-purified antibodies to human Nup205 were generated as follows. A PstI–HindIII fragment of the human cDNA (KIAA0225) encoding amino acids 673-1232 was cloned into pET28 A (Invitrogen, Carlsbad, CA). This construct was transformed into BL21, and the cells were grown to early log phase. Production of the protein was induced for 3 h by the addition of isopropylthio-β-galactoside to 1 mM. The insoluble fusion protein was extracted in urea and purified under denaturing conditions on Ni2+-NTA (nickel-nitrilotriacetic acid) agarose essentially as described by the manufacturer (Qiagen Inc., Valencia, CA). Antibodies to this fusion protein were generated in female New Zealand White rabbits. A Nup205 affinity column was prepared by dialyzing the fusion protein overnight against a large volume of 0.5 M NaCl, 0.1 M NaHCO3, pH 8.5. The dialyzed protein was coupled to cyanogen bromide–activated Sepharose in this buffer overnight at 4°C, and unreacted sites on the matrix were blocked by a 1-h incubation in 0.1 M ethanolamine, pH 8.0. The column was washed with three cycles of alternating low pH/high pH (0.5 M NaOAc, pH 4.0; 0.5 M NaBO3, pH 9.0), washed extensively with PBS, and stored in PBS containing 1 mM NaN3. Anti-Nup205 antibodies were prepared on this column essentially as described by Harlow and Lane (1988), dialyzed against PBS, and stored in small aliquots at −80°C. ELB buffer consisted of 20 mM HEPES, 50 mM KCl, 2.5 mM MgCl2, 250 mM sucrose, pH 7.4; ELBLS, i.e., ELB low-salt buffer, lacked the 50 mM KCl.
Preparation of Egg Extracts
High-speed Xenopus egg cytosolic and membrane fractions were prepared as described previously (Finlay and Forbes, 1990; Meier et al., 1995). For AL formation assays, extracts were prepared from high-quality eggs and stored in small aliquots at −80°C. Xenopus WGA-binding proteins (low-salt XE) were prepared essentially as described by Finlay and Forbes (1990), except that the salt concentration of the ELB buffer used was reduced from 50 mM KCl to 0 mM KCl to make ELBLS. In addition, the washing of the WGA-bound proteins was done with ELBLS rather than ELB plus 300 mM KCl, as used in previous studies of the Xenopus WGA-binding nucleoporins. These low-salt conditions were used to maintain the weaker protein–protein interactions present in some multiprotein complexes. Briefly, freshly prepared egg cytosol was clarified by centrifugation at 200,000 × g for 30 min and applied to one-tenth volume of WGA–Sepharose (EY Laboratories, San Mateo, CA) that had been equilibrated previously with ELBLS. After incubation at 4°C with gentle rotation for 2 h, the matrix was washed extensively with ELBLS. Bound proteins were eluted with two successive 45-min incubations with one bed volume of a high-sugar buffer with two times the concentration of triacetyltrichitobiose (TCT): 0.5 M GlcNAc, 16 mM TCT in ELB normal salt). The eluates were pooled and stored in small aliquots at −80°C. Xenopus egg glycogen was prepared as described by Hartl et al. (1994) and stored at 200 mg/ml in ELB at −20°C.
AL Formation Assay
In general, AL were formed from a mixture of Xenopus egg cytosol, membranes, and glycogen in an 8:1:1 ratio. In experiments in which the AL formation assays were supplemented with other reagents, the amount of input cytosol was reduced accordingly. A standard 25-μl reaction would thus contain 17.5 μl of cytosol, 2.5 μl of packed membranes, 2.5 μl of glycogen, and 2.5 μl of biotinylated low-salt Xenopus WGA eluate (bioXE), low-salt XE, or buffer. After a 3-h incubation at room temperature, the reaction was diluted with 75 μl of ELB and placed on ice. After a 10-min incubation, the AL were isolated by centrifugation through a 50-μl sucrose cushion (ELB containing 500 mM sucrose) at 30,000 × g for 25 min. The supernatant was carefully removed, and the AL pellet was processed as described below.
Immunoblot Analysis
Samples were resuspended in 2× sample buffer (Meier et al., 1995), heated to 95°C for 3 min, and electrophoresed on 7.5% SDS-PAGE gels. The separated proteins were transferred to PVDF, and the filters were stained with India ink in PBST (Harlow and Lane, 1988). After staining, filters were blocked with either 2% BSA in PBST (for antibodies or streptavidin [SA]–HRP) or 2% polyvinylpyrrolidone-40 in PBST (for WGA-HRP or concanavalin A–HRP; EY Laboratories). Blots were probed with primary antibodies for 1 h, and the filters were washed extensively in PBST and then probed with the appropriate secondary antibody–HRP conjugate (Jackson Laboratories, Bar Harbor, ME). In the case of SA–HRP- or concanavalin A–HRP-probed filters, no secondary reagent was needed for detection. Blots were developed with the use of ECL (Dupont, Wilmington, DE).
Biotinylation
Freshly prepared Xenopus egg extract was prepared and bound to WGA–Sepharose in low salt as described above. The beads were washed with 20 mM HEPES, 2 mM MgCl2, pH 8.0, and resuspended to the original volume of extract in this same buffer. Biotin–N-hydroxysuccinimide (NHS) (NHS-LC-Biotin II, Pierce, Rockford, IL) was added to a final concentration of 400 μg/ml (prepared as a 40 mg/ml stock in dry DMSO within 5 min of addition), and the mixture was tumbled at 4°C for 45 min. The reaction was terminated by washing sequentially with reaction buffer three times and with ELBLS three times. The bioXE were eluted with high-sugar buffer, as described above.
Sucrose Gradient Analysis of AL
A 250-μl AL assembly assay was performed in the presence of 25 μl of bioXE. After 3 h at room temperature, the reaction was diluted twofold with cold ELB and held on ice for 20 min to depolymerize microtubules, and the membrane fraction was recovered by centrifugation through a 0.5-ml sucrose cushion as described above. The AL were resuspended in 125 μl of ELB and homogenized in a ground-glass tissue grinder as described by Meier et al. (1995). The sample was adjusted to 65% sucrose and overlaid with 60–32.5% sucrose (in ELBS) in SW55 tubes. The sample was then centrifuged at 300,000 × g for 20 h, and 0.5-ml fractions were collected from the bottom of the tube. Each fraction was precipitated and run on four sets of SDS-PAGE gels and blotted with concanavalin A–HRP to detect gp210, a mAb against ribophorin I to detect endoplasmic reticulum (ER), mAb414 to detect soluble phenylalanine–glycine repeat nucleoporins, and SA-HRP to detect the biotinylated proteins. The resulting fluorographs were scanned and quantitated as described by Meier et al. (1995).
Purification of p170
One milliliter of egg cytosol was diluted with 2 volumes of ELBLS and clarified by centrifugation at 265,000 × g for 20 min. The supernatant was mixed with 100 μl of packed WGA–Sepharose (previously equilibrated with ELBLS) and incubated with tumbling at 4°C for 1.5 h. The beads were washed five times with 1 ml of ELBLS to remove unbound proteins. Ten percent of the beads were removed for analysis, and the remaining beads were incubated with 250 μl of ELB plus 50 mM KCl (100 mM KCl final concentration) for 15 min. The supernatant was removed to a fresh tube, and the beads were eluted sequentially with ELB plus 200 mM KCl and ELB plus 450 mM KCl. Each supernatant was supplemented with 5 μg of glycogen as a carrier, and the samples were precipitated by the addition of 500 μl of cold methanol. The precipitates were held at −20°C overnight and recovered by centrifugation at 16,000 × g for 30 min. The precipitates were washed with 80% methanol, air-dried, and resuspended in 200 μl of sample buffer. Ten percent of the remaining salt-washed WGA–Sepharose was washed with ELB, then ELBLS, and then resuspended in 100 μl of sample buffer. Aliquots of each fraction were run on a 7.5% SDS-PAGE gel and silver stained according to the manufacturer's directions (Bio-Rad, Richmond, CA).
Large-scale purification of p170 was performed as follows. Egg lysate (50 ml) was diluted to 100 ml with ELBLS and centrifuged at 100,000 × g for 30 min. The cleared cytosol was mixed with 5 ml of WGA–Sepharose beads and incubated for 3 h. The beads were washed 10 times with 50 ml of ELBLS and twice with 10 ml of ELB plus 50 mM KCl. The Nup188 complex was then eluted with 10 ml of ELB plus 200 mM KCl and 10 ml of ELB plus 450 mM KCl. The eluates were concentrated 20-fold by centrifugal ultrafiltration (Biomax 50k, Millipore, Bedford, MA) and precipitated overnight at −20°C by the addition of methanol (1 ml). The samples were washed with 80% methanol and air-dried before being resuspended in 0.5 ml of sample buffer. One-third of the sample was run on an SDS-PAGE gel and stained with Coomassie blue. Comparison with molecular weight standards indicated that the total yield was ∼10 μg of p170. The remainder of the sample was run on a separate SDS-PAGE gel and used for sequencing.
Peptide Sequencing
Approximately 150–200 μl of a large-scale preparation of p170 was separated on preparative SDS-PAGE gels. The samples were transferred to PVDF, and the band corresponding to p170 was identified by staining with amido black, as described by Harlow and Lane (1988). The band corresponding to p170 was excised and digested with endoproteinase Asp-N, and the peptides obtained were fractionated by reverse phase HPLC, as described (Fischer et al., 1991). Individual peaks were subjected to automated Edman degradation on a Procise 494 sequenator (Perkin Elmer-Cetus–Applied Biosystems, Foster City, CA).
Two of the peptides, DHKVIST and EVKANK, obtained from the protein sequences were exact matches to sequences found within a human expressed sequence tag (EST), KIAA0169 (GenBank accession number 1136398). This EST encodes a single ORF of 1745 amino acids. The KIAA0169 sequence was then compared with the nr database at the National Center for Biotechnology Information with the use of PSI-BLAST. In addition to a number of hits to short mouse ESTs, a 443-amino acid sequence was found from amino acids 552 to 1011 of Saccharomyces cerevisiae Nup188 (GenBank accession number AAA88904/P52593) and from the Schizosaccharomyces pombe Nup188 homologue (GenBank accession number AAD43830). This region displays 21% identity and 40% homology with the corresponding sequence of human KIAA0169.
Immunoprecipitations
Immunoprecipitations were performed in either RIPA buffer (50 mM Tris, 150 mM NaCl, 1% NP40, pH 8.0; see Figure 6A) or in buffer A (20 mM HEPES, 100 mM NaCl, 2.5 mM MgCl2, pH 7.4). In general, samples to be immunoprecipitated were diluted into buffer supplemented with 5 mg/ml BSA at 4°C and precleared over protein A–Sepharose for 30–60 min at 4°C with gentle tumbling. Where indicated, 1-μl aliquots of bioXE were first diluted to 50 μl with buffer containing 0.5% SDS at room temperature to denature the proteins. Samples were then diluted 10-fold with either RIPA buffer or buffer A plus 0.1% 3-([3-chloramidopropyl]dimethylammonio)-2-hydroxy-1-propanesulfonate (CHAPS) before preclearing to reduce the SDS concentration. Control reactions were diluted on ice in buffer minus SDS. Antibodies (1 μg) were incubated with protein A–Sepharose (10 μl; Pharmacia) in 500 μl of PBS containing 5 mg/ml BSA and 1% Triton X-100 for 2.5 h to overnight. These antibody beads were washed extensively with either RIPA buffer or buffer A before incubation with the precleared samples for 2 h. Immune complexes were centrifuged, then washed extensively with the appropriate buffer, and eluted with 25 μl of sample buffer. One-fifth of the samples were run on SDS-PAGE gels and processed for blotting with SA–HRP. Control blots were probed with a mixture of anti-Nup98, mAb414, and anti-Nup93 to verify the specificity of the immunoprecipitations to compare the location of the biotin-labeled bands.
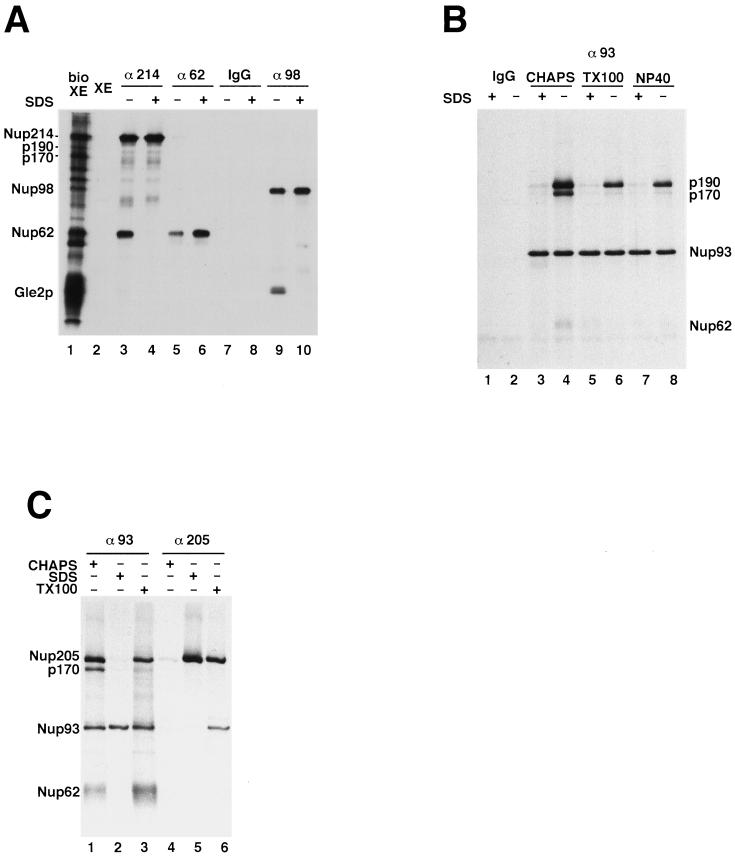
Figure 6.
Identification of p90 and p190 as nucleoporins. (A) Neither p170 nor p190 is derived from any of the previously identified WGA-binding nucleoporins. Affinity-purified antibodies to Nup62, Nup98, and Nup214 were incubated with bioXE under native (−SDS) conditions or with denatured bioXE (+SDS), as described in MATERIALS AND METHODS. Immunoprecipitated proteins were washed extensively, and the bound biotinylated proteins were detected by Western blotting with the use of SA–HRP. Rabbit immunoglobulin G (IgG) was used as a negative control. From the data in A and from companion immunoblots probed with antibodies to Nup62, Nup98, Nup214, and Gle2, the bands in lanes 3–10 could be identified. p90, p170, and p190 did not immunoprecipitate with antibodies to Nup214, Nup62, or Nup98. (B) Both p170 and p190 coimmunoprecipitate with Nup93 under native conditions. BioXE was diluted with buffer A or buffer A plus 0.5% SDS for 10 min at room temperature. The samples were then diluted 10-fold with buffer A containing 0.1% CHAPS, 1% Triton X-100, or 1% NP40, as described in MATERIALS AND METHODS. The diluted samples were incubated with either rabbit IgG–protein A beads (IgG) or with affinity-purified anti-Nup93–protein A beads. Bound proteins were detected by SA–HRP. A small amount of Nup62 also coimmunoprecipitates with the complex (lane 4). (C) p190 is the Xenopus homologue of human Nup205. BioXE was incubated in buffer A containing 0.1% CHAPS, 0.5% SDS, or 1% Triton X-100, as described above, and then diluted 10-fold with buffer A plus 0.1% CHAPS. The samples were incubated with anti-Nup93 antibody–protein A beads or with anti-Nup205 antibody–protein A beads and washed. The bound proteins were detected as described above. Anti-Nup205 antibodies immunoprecipitated p190 under denaturing conditions (lane 5), indicating that p190 is the Xenopus homologue of human Nup205.
Assay for Indirect Binding of p90, p170, and p190 to WGA
AL (100-μl assembly reaction volume) were formed in the presence of bioXE as described above. After centrifugation through the sucrose cushion, the biotinylated AL–containing membrane pellet was treated with 100 μl of buffer A containing 0.5 M NaCl to disassemble the biotinylated AL pores that had formed. The sample was incubated on ice for 20 min and centrifuged at 30,000 × g for 25 min. The supernatant, containing disassembled nuclear pore proteins, was applied to WGA–Sepharose (25 μl; previously equilibrated in buffer A plus 0.5 M NaCl) and tumbled for 2 h at 4°C. Nucleoporins Nup62, Nup98, and Nup214 should bind quantitatively under these high-salt conditions, but p90, p170, and p190 should not. The WGA beads were discarded, and the unbound protein supernatant containing p90, p170, and p190 was diluted fourfold in buffer A and applied to either WGA–Sepharose or WGA–Sepharose to which Xenopus WGA-binding proteins (i.e., Nup62, Nup98, Nup214) had been prebound. The two sets of reactions were incubated at 4°C for 1.5 h, washed four times with buffer A plus 0.1 M NaCl, eluted with gel sample buffer, electrophoresed, transferred to PVDF, and probed with SA–HRP to reveal whether the biotinylated nucleoporins bound directly or indirectly to WGA. To prepare these two sets of beads, a 25-μl aliquot of WGA–Sepharose was incubated with 500 μl of egg cytosol diluted 1:1 in buffer A plus 0.5 M NaCl, whereas a second aliquot of WGA—Sepharose was incubated with 5 mg/ml BSA in buffer A plus 0.5 M NaCl. After 1.5 h, the WGA–Sepharose beads were washed extensively with buffer A plus 0.5 M NaCl, followed by three washes with buffer A plus 0.1 M NaCl, and then used in the experiment described above.
RESULTS
An Organelle Trap Assay for Novel Nucleoporins
To gain a better understanding of the mechanism of transport through the nuclear pore, identification of the molecular constituents of the massive pore is required. In the yeast S. cerevisiae, this work has been facilitated by the genetic tractability of the organism. Starting from known nucleoporin genes, it is relatively straightforward to use molecular genetic approaches (synthetic lethality, high-copy suppression, etc.) to identify novel pore proteins (see Damelin and Silver, 2000). Moreover, the entire genome of this yeast has been sequenced, making it possible to identify potential nucleoporins based on their homology to other nucleoporins. Finally, biochemical purification of the pore in yeast has also been achieved, allowing for direct identification of potential nucleoporins (Rout and Blobel, 1993; Rout et al., 2000).
The situation in higher eukaryotes is more complicated. To date, known metazoan nucleoporins have been identified biochemically, e.g., via binding to the transport inhibitor WGA, via mAbs raised to the nuclear pore–lamina complex, or based on the abundance of such proteins in specific nuclear envelope fractions (Davis and Blobel, 1987; Finlay et al., 1987; Radu et al., 1993). Because genetic analysis is difficult or impossible in the most widely used higher eukaryotic systems, new biochemical assays to identify novel nuclear pore proteins would be of great utility. Having shown that we could follow the incorporation of known nucleoporins into AL (Meier et al., 1995), we designed an assay that we predicted could be used to identify new nucleoporins. In this two-step organelle trap assay, a ligand predicted to bind nucleoporins is first chosen for an affinity column. Xenopus egg cytosol is passed over the ligand matrix, and the bulk of the cytosolic constituents is washed away (Figure 1). In theory, affinity chromatography can be used as a one-step purification of interacting proteins. However, unless the interaction between a ligand and target are extremely specific and strong enough that highly stringent conditions can be used, both specific and nonspecific proteins will be retained on the affinity matrix. Not all of the retained proteins (Figure 1, open polygons) would be expected to be nucleoporins (open triangles). A large percentage of the bound polypeptides might be nonspecifically interacting species. In our strategy, the mixture of ligand-bound proteins, once bound, is biotin-labeled on the column with the use of an activated biotin derivative. This allows for later detection of the labeled protein by SA conjugates. For the second step of the assay, the biotin-labeled proteins are eluted and added to an in vitro AL formation assay. Only nucleoporins and potential transport factors are expected to be specifically incorporated into AL. The pore formation step acts to discriminate between nonspecific proteins that had spurious interactions with the ligand and specific nucleoporins “trapped” by assembly into the newly formed AL pores.
Detection of Three Novel Potential Nucleoporins
Previous work in the laboratory had identified the lectin WGA as an inhibitor of nuclear import (Finlay et al., 1987). Further analysis revealed that Xenopus egg cytosol contains three major proteins capable of binding WGA under relatively stringent conditions. All three of these proteins—Nup62, Nup98, and Nup214—proved to be nucleoporins and to bind to WGA because of covalently added GlcNAc modification (Davis and Blobel, 1987; Finlay et al., 1987; Holt et al., 1987; Snow et al., 1987; Finlay and Forbes, 1990). We reasoned that there could be additional novel nucleoporins present in the WGA-binding fraction of cytosol. Because WGA binds strongly to proteins labeled with large amounts of clustered GlcNAc moieties, weakly glycosylated polypeptides might not be detectable if their GlcNAc residues were far apart. Alternatively, nonglycosylated nucleoporins could be retained on the WGA–Sepharose by virtue of their interaction with a GlcNAc-modified nucleoporin. Therefore, we followed the labeling approach outlined in Figure 1 with the use of WGA–Sepharose as the ligand.
When egg cytosol was applied to WGA–Sepharose in a low-salt buffer chosen to stabilize protein–protein complexes, a silver stain of the proteins retained by WGA–Sepharose revealed a complex mixture of proteins retained in low salt (Figure 2, lane 6). To test the organelle trap strategy, egg cytosol proteins were bound to WGA–Sepharose under conditions of relatively low stringency, washed, labeled with NHS-biotin, and eluted with competing sugar. As shown in Figure 2, WGA–HRP staining of retained proteins detects primarily the three previously identified nucleoporins Nup62, Nup98, and Nup214 (lane 1). However, when the composition of bioXE was probed with SA–HRP, a much more complex pattern of proteins was observed (lane 7). We asked which of these biotinylated proteins became incorporated into AL pores.
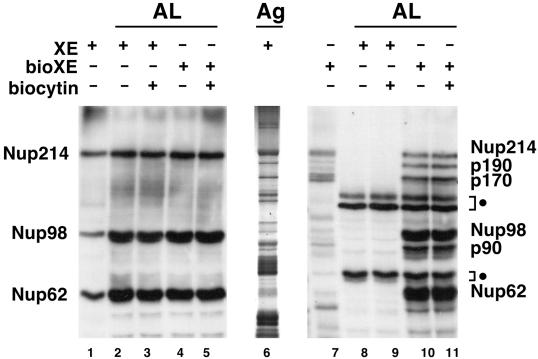
Figure 2.
Detection of three novel potential nucleoporins. AL assembly reactions, set up as described in MATERIALS AND METHODS, were supplemented either with untagged Xenopus proteins that bind to WGA in low salt (XE; lanes 2, 3, 8, and 9) or with biotinylated XE (bioXE; lanes 4, 5, 10, and 11). After a 3-h incubation, the assembled AL were pelleted, electrophoresed on SDS-PAGE gels, and blotted. Duplicate blots were probed either with WGA–HRP (lanes 1–5) to detect the GlcNAc-modified nucleoporins Nup214, Nup98, and Nup62 or with SA–HRP to detect biotinylated proteins (lanes 7–11). Samples of low-salt XE and bioXE were run in lanes 1 and 7, respectively. The total polypeptide composition of low-salt XE is shown via a silver-stained gel (lane 6). Positions of the known nucleoporins Nup214, Nup98, and Nup62 are indicated. AL reactions use Xenopus extract that contains endogenous Nup214, Nup98, and Nup62, as seen in lanes 2–5. In the AL assembly reactions performed in the presence of bioXE, three new major species are detected and are labeled on the right of the figure according to their apparent molecular weights: p90, p170, and p190. Also indicated are the endogenous biotinylated proteins (closed circles) that are present in the membrane fraction. Biocytin, a biotin analogue, was added with the bioXE to some AL assembly reactions in lanes 3, 5, 9, and 11.
For this, the labeled bioXE was added to an AL formation assay and allowed to assemble for 3 h. Incorporation of the biotin-labeled proteins into AL pores was assayed on Western blots with the use of SA-HRP to detect the biotin-tagged proteins. Parallel reactions were performed with the use of unlabeled eluate (XE). Addition of bioXE for Xenopus eluate did not inhibit AL pore assembly (compare lanes 4 and 5 with lanes 2 and 3), indicating that the biotinylated proteins did not have an inhibitory effect on the assembly of endogenous Nup62, Nup98, and Nup214 into AL pores. It should be noted that all AL reactions, even those to which no bioXE was added contained three endogenously biotinylated polypeptides (Figure 2, lanes 8 and 9), indicated by the closed circle to the right of the figure. These bands represent naturally biotin-containing proteins from the mitochondria that invariably are present in the Xenopus egg membrane fraction (B.R. Miller, unpublished observations). We believe that these are carboxylases from the mitochondrial matrix, which are known to contain covalently attached biotin groups.
Interestingly, when AL reactions to which bioXE had been added were probed with SA, six additional strongly labeled bands were detected (Figure 2, lanes 10 and 11). Three of these correspond to the WGA-binding nucleoporins Nup62, Nup98, and Nup214, as determined by mobility (Figure 2, lanes 10 and 11) and by immunoprecipitation with the use of specific antibodies (our unpublished observations). The other three bands, labeled by their apparent molecular weights, were designated p90, p170, and p190. It was possible that these polypeptides were spuriously associated with the AL membrane fraction via the introduced biotin tag. However, addition of the water-soluble biotin analogue biocytin, which should compete for any such nonspecific interactions between the biotin tag and the membranes, had no effect on the incorporation of the biotin-tagged proteins into AL (Figure 2, lanes 3, 5, 9, and 11), discounting this possibility. Thus, p90, p170, and p190 were potential novel candidate nucleoporins.
Association of p90, p170, and p190 with AL Is Blocked by the Addition of BAPTA
The AL incorporation assay, in its simplest form, measures incorporation of nucleoporins from the soluble portion of the egg extract into the membrane fraction. Therefore, the association of the p90, p170, and p190 proteins could represent proteins irrelevant to nuclear pores that are instead involved in some other membrane-associated process. For example, addition of the membrane fraction to the soluble egg extract results in a large amount of membrane–membrane fusion. It remained a formal possibility that the three polypeptides were not pore proteins but instead were involved in vesicle targeting or fusion. Previous work in this and other laboratories has shown that the addition of the Ca2+ chelator bis-(o-aminophenoxy)-N,N,N′,N′-tetraacetic acid (BAPTA) allows nuclear membrane fusion but causes a profound defect in nuclear pore assembly in Xenopus nuclear reconstitution assays. Nuclei reconstituted in the presence of BAPTA are surrounded by a complete double nuclear envelope devoid of nuclear pores (Sullivan et al., 1993; Macaulay and Forbes, 1996; Goldberg et al., 1997; Shumaker et al., 1998). The addition of BAPTA to AL formation assays completely blocked the incorporation of the known nucleoporins Nup358, Nup214, Nup153, Nup98, and Nup62 (Figure 3A, lane 3) (Shah and Forbes, 1998). BAPTA similarly blocked the incorporation of the biotin-tagged nucleoporins and of p90, p170, and p190 (detected with SA–HRP; Figure 3B, lane 6). This indicates that incorporation of the p90, p170, and p190 proteins into the membrane fraction is similar to that of known nucleoporins with respect to BAPTA.
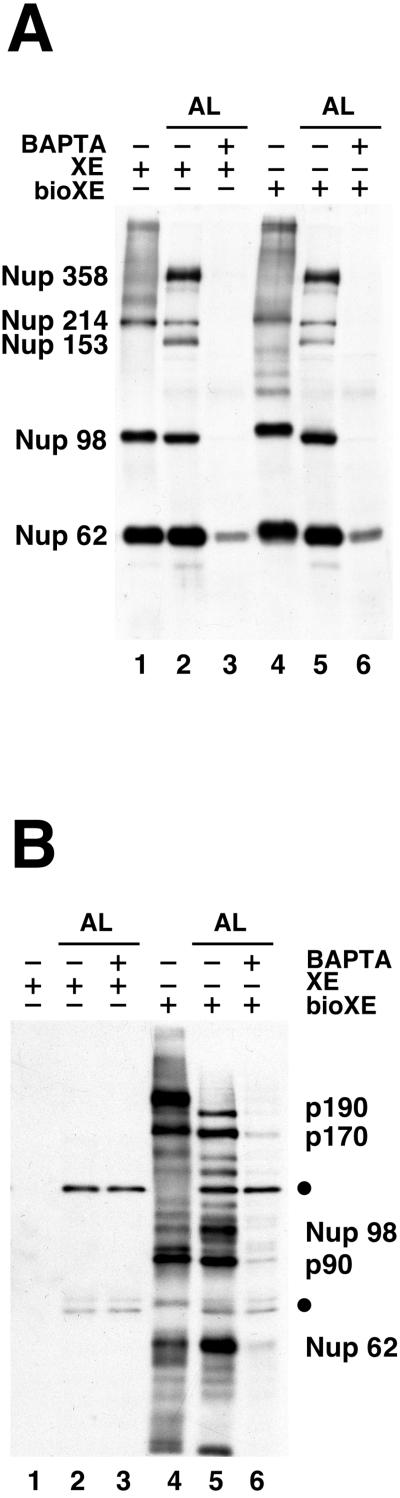
Figure 3.
Association of p90, p170, and p190 with AL is blocked by the addition of BAPTA. Addition of BAPTA to Xenopus nuclear assembly assays blocks the assembly of pore complexes into the nuclear membrane (Macaulay and Forbes, 1996). Parallel AL assembly reactions (50 μl) were set up containing either low-salt XE (lanes 2 and 3) or bioXE (lanes 5 and 6). Each reaction was then split in half. One half received buffer (1.25 μl of 100 mM HEPES, pH 7.4; lanes 2 and 5), and the other half was supplemented with BAPTA (5 mM final concentration; lanes 3 and 6). After a 3-h incubation, the AL were recovered by centrifugation and analyzed in duplicate by Western blotting with the use of either a mixture of mAb414 andanti-Nup98 (A) or SA–HRP (B). For reference, small amounts of low-salt XE and bioXE were also run on the gels (lanes 1 and 4). The blots are labeled as in Figure 2. BAPTA prevents the incorporation of the known nucleoporins Nup62, Nup98, Nup153, Nup214, and Nup358 into AL (A, lane 6) and also prevents the incorporation of p90, p170, and p190 (B, lanes 3 and 6). The presence of the endogenously biotinylated proteins (closed circles) are not altered by BAPTA (B, compare lanes 5 and 6).
The experiment described above rules out one possible candidate for p170 or p190, i.e., the nucleoporin Nup153. By SDS-PAGE, the apparent molecular mass of Nup153 is known to be 180 kDa (Sukegawa and Blobel, 1993; Shah et al., 1998; Nakielny et al., 1999) (Figure 3A, lanes 2 and 5), which is close to that of either p170 or p190. However, as shown in Figure 3A, no detectable Nup153 is present in low-salt XE (lane 1) or bioXE (lane 4) from WGA–Sepharose. Use of a specific polyclonal antibody to Xenopus Nup153 in Western analysis also did not detect any Nup153 protein in either low-salt XE or bioXE (our unpublished results). Thus, Nup153 can be ruled out as being either p170 and p190.
p90, p170, and p190 Cofractionate with AL
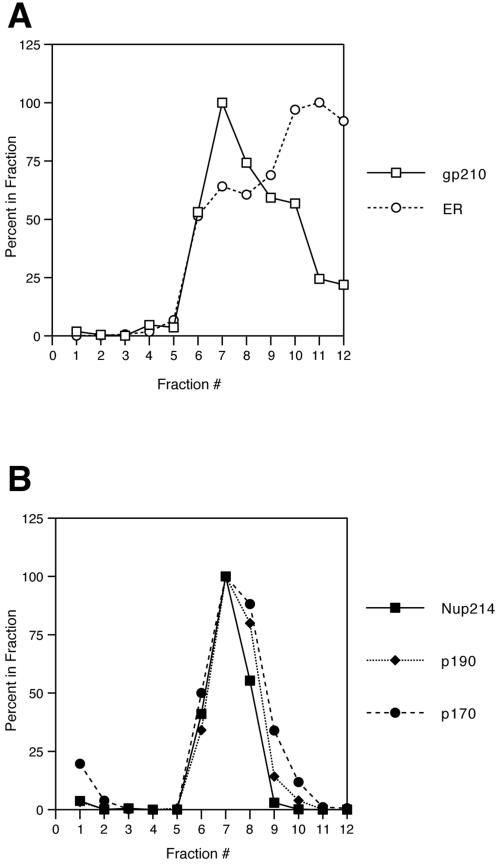
We further analyzed the behavior of the incorporated proteins, fractionating the AL-containing membrane pellet by floatation through a sucrose gradient. Previously, we showed that the AL appear to assemble into, and to be physically contiguous with, the ER (Meier et al., 1995). By gently shearing the membrane fraction and floatation through a sucrose gradient, we were able to separate the AL from the bulk of the ER. In this study, AL were assembled on a large scale in the presence of bioXE, pelleted, sheared, and floated through a 65–32.5% linear sucrose gradient. Fractions were collected, and the positions of individual marker proteins were quantitated, as described in MATERIALS AND METHODS. In Figure 4A, the position of the integral membrane pore protein gp210 is plotted in comparison with that of the ER integral membrane protein ribophorin. As in our previous study, the peak of gp210 reactivity was found in the bottom third of the gradient (for clarity, only the bottom 12 of 20 fractions are plotted here), and this peak is separated from the bulk of the ER. Figure 4B shows the positions of the proteins p170, p190, and the nucleoporin Nup214. As can be seen, the p170 and p190 polypeptides display an identical distribution to that of the nucleoporins Nup214 and gp210. The biotinylated p90 protein showed an identical pattern to that of these nucleoporins (our unpublished observations). Therefore, on the basis of biochemical criteria, we conclude that p90, p170, and p190 do in fact behave like nucleoporins.
Figure 4.
p90, p170, and p190 cofractionate with AL. A large-scale AL reaction (0.5 ml) was assembled in the presence of bioXE, and the membranous organelles were analyzed by flotation through a 65–32.5% sucrose gradient, as described by Meier et al. (1995). One-half-milliliter fractions were collected from the bottom of the gradient, and the presence of different organelle proteins was assayed by Western blotting; the amount of each polypeptide was quantified by densitometry. For clarity, only the bottom two-thirds of the gradient is shown. In A, the positions of the fractions containing the integral pore membrane protein, gp210, and the ER integral membrane protein, ribophorin, are plotted. The gp210-containing membranes peak in fraction 7 at a density of 1.2 g/cm3, in agreement with our previous report for AL. In contrast, the peak of ER reactivity is much higher in the gradient. B shows the positions of the p190-, p170-, and Nup214-containing fractions from the same flotation gradient. All three polypeptides cofractionate with gp210, peaking in fraction 7, indicative of nucleoporins or pore-associated proteins.
The Three Novel Pore Proteins Bind Indirectly to WGA
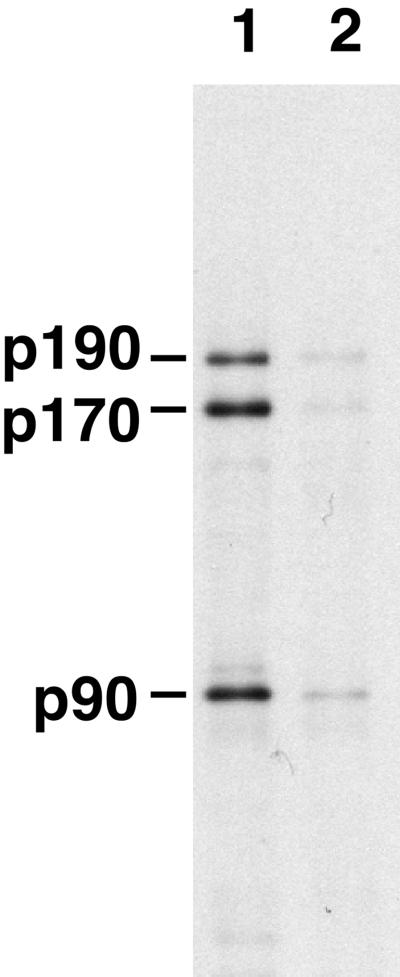
One explanation for the retention of p90, p170, and p190 on WGA–Sepharose was that these proteins might interact indirectly with the lectin through an interaction with one or more of the glycosylated nucleoporins. Our preliminary studies gave some indication that this was the case, because washing the WGA–Sepharose-containing bound proteins with 300 mM KCl before biotinylation greatly decreased the amount of biotinylated p90, p170, and p190 obtained. To test the hypothesis of indirect binding, AL were assembled in the presence of bioXE and the assembled AL were then recovered by centrifugation. These were then incubated in high-salt buffer to disassemble the AL pores, i.e., release the nucleoporins from the membranes. After centrifugation to remove the membranes, the supernatant containing the disassembled pore proteins, both biotin-tagged and untagged, was incubated with WGA–Sepharose in buffer containing 0.5 M NaCl. Under these conditions, binding of the glycosylated pore proteins Nup62, Nup98, and Nup214 to the WGA–Sepharose was quantitative. The unbound proteins, which contained the biotin-tagged p90, p170, and p190, were diluted with buffer to reduce the salt concentration and then incubated at 0.1 M NaCl with WGA–Sepharose that had been preloaded with either WGA-binding nucleoporins (Nup62, Nup98, and Nup214) or BSA alone. Binding of the biotin-tagged p90, p170, and p190 to these columns was assessed on Western blots by SA–HRP. We found that p90, p170, and p190 bound to the column preloaded with WGA-binding pore proteins (Figure 5, lane 1) but did not bind to WGA–Sepharose preincubated with BSA alone (lane 2). Thus, p90, p170, and p190 must bind to WGA indirectly and appear to do so through one or more of the glycosylated nucleoporins Nup62, Nup98, or Nup214.
Figure 5.
p90, p170, and p190 bind to WGA–Sepharose indirectly. AL was assembled in the presence of bioXE and isolated. The nucleoporins were extracted from the membranes with the use of high salt. This high-salt extract containing disassembled pores was passed over WGA–Sepharose to remove the direct WGA-binding proteins Nup62, Nup98, and Nup214 and then diluted with buffer A to 100 mM NaCl final concentration. The diluted mixture containing disassembled p90, p170, and p190 was then applied to either WGA–Sepharose preloaded with Nup62, Nup98, and Nup214 (lane 1) or to WGA–Sepharose prewashed with BSA (lane 2). After washing, the bound proteins were eluted with SDS sample buffer, electrophoresed, transferred to PVDF, and probed with SA–HRP. The p90, p170, and p190 proteins bind to WGA containing prebound GlcNAc-modified Nup62, Nup98, and Nup214 but not to WGA directly.
Coimmunoprecipitation of p170 and p190 with Nup93
One possibility for the apparent association of p90, p170, and p190 with O-glycosylated nucleoporins is that the former might be breakdown products of the canonical O-glycosylated nucleoporins and bind to them by homotypic interactions. For example, p170 and p190 could be proteolytic fragments of Nup214, and p90 could be a fragment of Nup98. Indeed, in yeast a potential homologue of Nup98 is proteolytically clipped into two fragments (Fabre et al., 1994; Dockendorff et al., 1997; Teixeira et al., 1997; Fontoura et al., 1999). To test the relationship of p90, p170, and p190 to Nup98 and Nup214, we performed immunoprecipitations from bioXE with the use of affinity-purified antibodies to Nup214 and Nup98 under both native (Figure 6A, −SDS) and denaturing (Figure 6A, +SDS) conditions. Native immunoprecipitations with anti-Nup214 immunoprecipitated both biotinylated Nup214 and biotinylated Nup62 (Figure 6A, lane 3); a complex between these nucleoporins was reported by us previously (Macaulay et al., 1995). After SDS denaturation, only biotin-tagged Nup214 was immunoprecipitated (lane 4). It should be noted that there are less abundant Nup214 breakdown products seen in these experiments; however, upon further examination, none of these comigrated exactly with either p170 or p190 (our unpublished observations). Antibodies to Nup98 immunoprecipitated Nup98 and its partner protein, Gle2p (lane 9). The Gle2p association with Nup98 disappeared after SDS denaturation (lane 10) (Matsuoka et al., 1999).
Importantly, neither the anti-Nup214 nor the anti-Nup98 antibody cross-reacted with or immunoprecipitated p90, p170, or p190 under any condition tested (Figure 6A, lanes 2–10, and our unpublished observations). However, overexposure of the blots revealed a small amount of p90 associating with the Nup62 immunoprecipitated under native conditions (our unpublished observations). Previous work had identified a novel nonglycosylated nucleoporin of approximately the size of p90 called Nup93, some of which is associated with Nup62 in HeLa nuclear extracts (Grandi et al., 1997). Based on molecular weight alone, we reasoned that Nup93 could be p90. To test this possibility, affinity-purified antibodies raised to human Nup93 were used for a series of immunoprecipitations from bioXE. Anti-Nup93 antibodies under denaturing conditions (+SDS) specifically immunoprecipitated the 90-kDa biotinylated protein (Figure 6B, lanes 3, 5, and 7). Therefore, p90 is indeed Nup93. Strikingly, in the presence of the nonionic detergent CHAPS, a more gentle detergent than SDS, biotinylated p170 and p190 both coimmunoprecipitated with this Nup93 (lane 4). A small amount of Nup62 was similarly associated with Nup93 under these conditions (lane 4). Increasing the stringency of the immunoprecipitations by the substitution of either 1% Triton X-100 (lane 6) or 2% NP40 (lane 8) removed the p170 band from the anti-Nup93 immune complexes but retained p190. These results indicate that p90 is the known nucleoporin Nup93 and that p170 and p190 are in complex with Nup93. These results also indicate that the association of p170 with Nup93 is weaker than the association between Nup93 and p190.
When analyzing Nup93 in HeLa extracts previously, we found it to be associated with a novel human nucleoporin, Nup205 (Grandi et al., 1997). Based on the predicted molecular mass, Nup205 could potentially account for either or both of the p190 and p170 bands observed in Xenopus. For example, p190 could represent intact Nup205, with p170 being a proteolytic breakdown product. To test this possibility, we raised a polyclonal antiserum to a 60-kDa recombinant fragment of human Nup205. The anti-Nup205 antibody recognizes a protein of ∼200 kDa in rat liver nuclei, HeLa cells, and Xenopus extracts (our unpublished results). Because the antibody was raised against the central third of full-length hNup205, it would be expected to react with any proteolytic fragment of the protein of >100 kDa and thus could be used to examine whether p190 and/or p170 are derived from Nup205. We performed immunoprecipitations from bioXE with the anti-Nup205 antibody (Figure 6C). Under native conditions, the anti-Nup205 antibodies immunoprecipitated only trace amounts of p190 (Figure 6C, lane 4). However, when the input bioXE was denatured with SDS before immunoprecipitation, the anti-Nup205 antibodies immunoprecipitated large amounts of p190 (lane 5). Thus, the anti-Nup205 antibody recognizes p190, but only in its denatured form. This cross-reactivity, together with the apparent molecular weight of p190 and its association with Nup93, indicate that p190 is in fact the Xenopus homologue of Nup205. However, no p170 was found in Nup205 immunoprecipitations under any conditions (lanes 4–6), which strongly suggests that p170 is not derived from Nup205 but in fact represents a distinct novel nucleoporin.
Identification of the Vertebrate Nucleoporin, Nup188
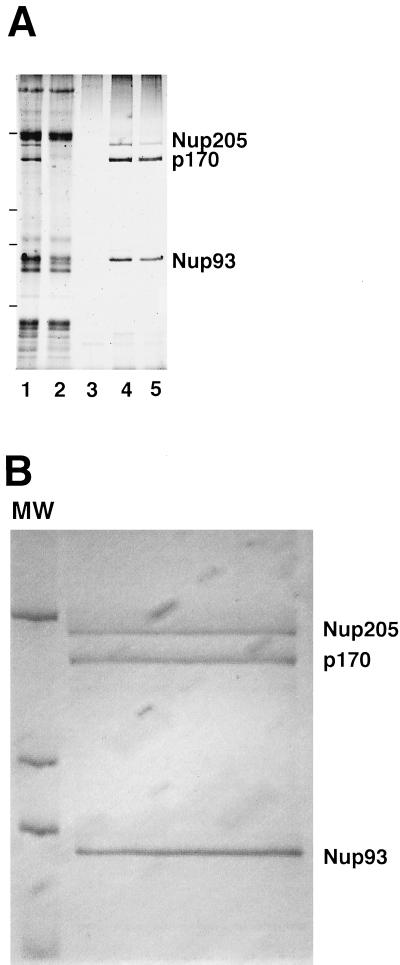
To identify p170, we purified p170 protein from large-scale preparations of low-salt XE. We found that Nup93 and its associated proteins, Nup205 and p170, once bound to a WGA–Sepharose column in low salt, could be purified rapidly and quantitatively simply by increasing the salt concentration. Specifically, when 1 ml of clarified Xenopus egg cytosol was applied to WGA–Sepharose in low-salt buffer containing 0.1% CHAPS, followed by extensive washing, Nup93, p170, and Nup205 could be specifically eluted with buffer containing 0.25 and 0.5 M NaCl, as shown by silver staining of a gel of the different eluates (Figure 7A, lanes 4 and 5). The identity of the three proteins was confirmed by repeating the experiment with the use of cytosol spiked with a small amount of bioXE and by probing Western blots with either specific antibodies (Nup93 and Nup205) or SA–HRP (our unpublished observations).
Figure 7.
Purification of p170. (A) Purification protocol for p170. One milliliter of Xenopus egg cytosol was diluted with ELB, clarified by centrifugation, and incubated with WGA–Sepharose, as described in MATERIALS AND METHODS. Unbound polypeptides were removed by extensive washing. Ten-microliter of beads were removed and eluted in sample buffer to probe by silver staining for the total proteins bound (lane 1; 0.01 volume of the original beads). The p170 protein was tested for elution from WGA–Sepharose by successive elutions with ELB plus 50 mM KCl (lane 3; 1/50th of the elution), ELB plus 200 mM KCl (lane 4; 1/50th of the elution), and ELB plus 450 mM KCl (lane 5; 1/50th of the elution). The remainingWGA-binding proteins were eluted with SDS sample buffer (lane 2; 1/100th of the remainder). The proteins present in each lane were detected by silver staining. The positions of molecular mass markers (205, 116, 98, and 66 kDa) are indicated by bars at the left of the figure. p170 is quantitatively removed from the column by the higher-salt washes. (B) Large-scale purification of p170. Xenopus egg cytosol (50 ml, ∼2 g of total protein) was diluted with ELB, clarified by centrifugation, and incubated with WGA–Sepharose, as described in MATERIALS AND METHODS. After extensive washing with ELB, the WGA beads were eluted with ELB containing 500 mM KCl. The eluates were pooled and precipitated. One-third was run on an SDS-PAGE gel and stained with Coomassie blue; a scan of the wet gel is shown in B. Comparison with molecular weight standards indicated that the total yield was ∼10 μg of p170. The remainder of the sample was run on a separate SDS-PAGE gel and transferred to PVDF. The p170 band was excised for sequencing.
To obtain large amounts of p170 for peptide sequence analysis, the preparation was scaled up 50-fold and the high-salt eluates were pooled and precipitated by ethanol. The pelleted proteins were resuspended in SDS sample buffer and separated by SDS-PAGE. Coomassie staining of the purified proteins from 10 ml of cytosol consisted almost exclusively of Nup93, Nup205, and p170 (Figure 7B). After electrophoretic transfer of the proteins to PVDF membranes, the p170 band was excised and subjected to peptide sequence analysis. The longest Xenopus p170 peptide sequence obtained, DLVHKVIST, was searched against the nr GenBank database and was an exact match of a sequence within a human EST cDNA clone. This sequence, KIAA0169, also contained an exact match to another peptide derived from the sequencing of Xenopus p170 (EKVKANK), strongly suggesting that KIAA0169 represents the human homologue of p170.
Conceptual translation of the KIAA0169 clone revealed a 1885-amino acid (without a starting methionine) ORF capable of giving rise to a protein of 196 kDa. A BLAST search of nr GenBank revealed strong homology of human KIAA0169 to the S. cerevisiae gene that encodes the yeast nucleoporin Nup188p as well as to the S. pombe Nup188 homologue. BLAST searches with either S. cerevisiae Nup188 or S. pombe Nup184 reciprocally identified KIAA0169 as the most closely related protein, after one another. A 443-amino acid central segment of the KIAA0169 ORF displayed the highest degree of homology, showing 40% homology to the equivalent region of the S. cerevisiae Nup188 coding sequence (Figure 8). Further analysis with the use of PSI-BLAST also revealed several mouse ESTs that encode protein fragments almost identical to portions encoded by KIAA0169. We conclude that human KIAA0169 encodes human Nup188 and that Xenopus p170 is the Xenopus Nup188 homologue. This conclusion was further strengthened by the fact that the yeast homologue of Nup93 is a member of a multiprotein complex that contains yeast Nup188p (Zabel et al., 1996; Grandi et al., 1997). Based on these results, we believe that Xenopus p170 and KIAA0169 represent the Xenopus and human homologues of yeast Nup188p, respectively. We have thus renamed p170 Xenopus Nup188.
Figure 8.
Comparison of human Nup188 with yeast Nup188. Conceptual translation of clone KIAA0169 revealed a 1745-amino acid ORF encoding a 196-kDa polypeptide, which is now renamed hNup188. An alignment of the most homologous portions of S. cerevisiae Nup188 and human Nup188, as described in the text, is shown.
DISCUSSION
In this paper, we present a two-step organelle trap assay for the identification of novel nucleoporins via ligand binding and AL incorporation. Because incorporation into AL pores represents a rather stringent selection step, one can use low-stringency binding conditions for the ligand affinity step. For the studies presented here, the ligand chosen was the lectin WGA, which inhibits nuclear import both in vivo and in vitro. In Xenopus extracts, the three major WGA-reactive species are all nucleoporins, Nup62, Nup98, and Nup214 (Finlay et al., 1989; Powers et al., 1997). In contrast to the simple pattern of three O-glycosylated proteins bound to WGA in high salt, the pattern of proteins observed on WGA–Sepharose at the low-salt concentration used here is quite complex. Because the pore complex consists of a large number of interacting proteins, it seemed likely that this complexity could be explained in part by other nucleoporins being retained on the WGA–Sepharose matrix as a result of interaction with one or more of the glycosylated pore proteins.
When biotin-labeled WGA-binding proteins were assembled into AL, ∼8–10 SA-reactive bands were observed. Three of these corresponded to endogenous biotinylated proteins present in the membrane fraction of egg extracts. Another three bands represented biotinylated forms of the known WGA-binding nucleoporins Nup62, Nup98, and Nup214. The major remaining bands, p90, p170, and p190, were potentially novel nucleoporins.
We found that incorporation of all of the known and candidate pore nucleoporins into AL was completely blocked by BAPTA, a compound that inhibits the assembly of nuclear pores in Xenopus egg extracts without blocking membrane fusion (Macaulay and Forbes, 1996; Goldberg et al., 1997). In another test, the assembled AL were homogenized and fractionated by flotation through a 65–32.5% sucrose gradient. Under the conditions used here, AL have a characteristic buoyant density, more dense than that exhibited by mitochondria and ER (Meier et al., 1995). All of the candidate nucleoporins displayed the density profile of AL, consistent with their being either nuclear pore or pore-associated proteins.
Further characterization of the novel biotin-labeled bands revealed that they were distinct from the N-acetylglucosaminylated pore proteins. The protein p90 proved to be Nup93. This polypeptide interacts weakly with Nup62 and strongly with a nucleoporin, Nup205, in both HeLa and Xenopus extracts (Grandi et al., 1997). The p190 band was identified as Nup205, as revealed by immunoprecipitation with anti-human Nup205 antiserum that we raised in this study for the purpose of this identification. Antibodies directed against Nup93 precipitated Nup205 as well as p170 and a small amount of Nup62, suggesting that these proteins are in a large complex. Gel filtration indicates that Nup205 and Nup93 fractionate in a complex of ∼500 kDa (S. Shah and D.J. Forbes, unpublished results), which would be consistent with a complex of all three proteins (with or without Nup62).
To identify the p170 protein, we purified sufficient quantities of the protein from egg cytosol and performed peptide sequencing. Two of the peptides obtained proved to be exact matches to sequences contained within a human cDNA clone in GenBank, KIAA0169, encoding a protein of hitherto unknown function. BLAST searches revealed homology of KIAA0169 with S. cerevisiae Nup188 and its S. pombe homologue, Nup184. A 450-amino acid stretch in the center of the KIAA0169 ORF shows 40% similarity to the equivalent sequence found in S. cerevisiae Nup188. Together, these data were compelling evidence that KIAA0169 encodes the human homologue of Xenopus p170 and S. cerevisiae Nup188. This is especially interesting in that S. cerevisiae Nup188 has been shown to interact directly with the yeast homologue of Nup93 (ScNic96) and the yeast homologue of hNup205 (ScNup192). Here we find that Xenopus p170 interacts with two known nucleoporins, xNup93 and xNup205, copurifies with AL, and shares homology with yeast and human Nup188, both in sequence and in protein–protein interaction. We conclude that p170 is indeed a nucleoporin and have named this protein Xenopus Nup188. Similarly, we have named the KIAA0169 gene as the gene encoding human Nup188. Because the homology between human Nup188 and yeast Nup188 is strongest in the central 450 amino acids of each protein, one possibility is that this domain is responsible for the interaction with either the cognate Nup93 or Nup205 partner.
Given that the vertebrate Nup93-Nup188-Nup205 and the yeast Nic96-Nup188-Nup192 proteins can be isolated in very similar multiprotein complexes, we predict that they may play similar roles in either the structure or the function of the nuclear pore complex. In yeast, five nucleoporins, Nic96, Nup157, Nup170, Nup188, and POM152, account for >20% of the mass of the pore (Rout and Wente, 1994; Stoffler et al., 1999; Rout et al., 2000). This suggests that the Nic96-Nup188-Nup192 complex may be an important structural building block of the pore complex. Indeed, certain mutations in yeast Nup188 cause a profound perturbation in pore structure at increased temperatures (Nehrbass et al., 1996; Zabel et al., 1996). Under these conditions, the outer nuclear envelope appears to have fused over the pore, forming membranous herniations in the nuclear envelope. Nup188p may be involved in the mechanical stability of the nuclear pore complex, although other nucleoporin mutations also give this herniated phenotype (Wente and Blobel, 1993, 1994; Heath et al., 1995; Siniossoglou et al., 1996). In addition, yeast Nup188 deletion mutants are synthetically lethal with deletion of the gene for the major yeast pore membrane protein, POM152 (Nehrbass et al., 1996). Although such synthetic lethal interactions between deletion mutants do not necessarily indicate a direct physical interaction between two proteins, these data do suggest that Nup188 may be involved in somehow linking the pore to the nuclear envelope. Yeast Nup188p and its partners, Nic96 and Nup192, have been shown to be located on both the nucleoplasmic and cytoplasmic faces of the pore (Grandi et al., 1995; Fahrenkrog et al., 1998). S. cerevisiae pores lack the nuclear and cytoplasmic ring structures found on vertebrate pores, but they do have the large central spoke ring. Thus, the octagonally symmetric spoke ring region of the pore, a structure that is common to both S. cerevisiae and vertebrate pores and that makes up the essential scaffold of the nuclear pore, likely contains the Nup93-Nup188-Nup205 complex (Akey, 1989; Hinshaw et al., 1992; Akey and Radermacher, 1993; Rout and Blobel, 1993; Fahrenkrog et al., 1998; Yang et al., 1998) Although speculative, this presumed localization is consistent with both the abundance of the nucleoporins and the phenotypes observed in the yeast mutants. Moreover, we previously found that when Xenopus nuclear reconstitution extracts are quite depleted of Nup93 and its associated proteins before assembly of nuclei, the nuclei formed are much reduced in pore number (Grandi et al., 1997). This finding reinforces the arguments that the vertebrate Nup93-Nup188-Nup192 complex isolated here is a structural building block of the pore.
The proteins of the Nup93-Nup188-Nup205 complex(es) in Xenopus apparently do not bind to WGA by virtue of being modified by O-glycosylation. Even when microgram quantities of the purified proteins are electrophoresed on SDS-PAGE gels and blotted, no significant reactivity with WGA–HRP is observed (B.R. Miller, unpublished observations). Instead, the Nup93 complex must bind to one or more glycosylated nucleoporins. One obvious candidate is Nup62, because ∼5% of the Nup62 present in HeLa extracts coimmunoprecipitates with Nup93 (Grandi et al., 1997). In our hands, the amount of Nup62 associated with Nup93 in egg extracts is much lower. The small quantity of Nup62 recovered in Nup93 immunoprecipitations may be due to a low affinity between the two proteins in extracts, a low affinity overcome on the column by the high density of Nup62 bound to the WGA–Sepharose. This hypothesis is supported by the observation that the Nup93-Nup188-Nup205 complex can be eluted from the WGA-binding proteins at relatively low-salt concentrations (≥250 mM). It may be the case that Nup93 interacts in a more stable manner with Nup62 only when they are assembled into intact pore complexes.
Upon repeated searches of the database with the most conserved region of hNup188 (Figure 8), we found that there was also a limited degree of homology between hNup188 and hNup205 in this region (our unpublished results). Although the homology is weak, it suggests that amino acids 500-1000 of the two proteins may play a similar role in the function of Nup188 and Nup205. One possibility is that these sequences are required for interaction with one another or with Nup93. Alternatively, this region of Nup188 and Nup205 may be involved in interactions with another nucleoporin. Finally, it is also possible that Nup188 and Nup205 represent extremely divergent homologues of one another and that the two proteins serve redundant purposes within the pore.
In summary, we have developed a novel method for the identification of nucleoporins based on affinity selection, chemical labeling, and subsequent organelle assembly or “trap.” In theory, any ligand can be chosen for the affinity selection. The organelle trap approach, if known nucleoporins are used for the affinity step rather than WGA, could potentially identify the large number of protein–protein interactions that occur within the vertebrate pore. In addition, it should be possible to use recombinant transport factors as the ligand in the first affinity selection step to identify their targets within the pore complex. The assay thus holds great promise for the identification of the proteins of the nuclear pore, interactions between individual nucleoporins, and interactions between nucleoporins and transport factors. Indeed, the organelle trap assay has already revealed a novel nucleoporin, i.e., vertebrate Nup188, and its presence in a vertebrate pore subcomplex, Nup93-Nup188-Nup205.
Finally, although the organelle trap approach described in this report was designed specifically to detect new pore proteins, it is generally applicable. Its advantage is that relatively nonstringent conditions for the affinity binding and labeling step can be used to select weakly interacting species. In this report, we used organelle reconstitution as the trap. However, it could be used with any in vitro assembly assay for the trap selection, including microtubule or microfilament assembly or assembly of clathrin-coated vesicles. Thus, the trap assay should be of great value in revealing the molecular components of a wide variety of subcellular pathways.
ACKNOWLEDGMENTS
The authors thank the members of the Forbes laboratory for many helpful discussions regarding this work. In particular, we thank Sundeep Shah for critical reading of the manuscript. This work was supported by National Institutes of Health grant RO1 GM33279 awarded to D.J.F., an American Cancer Society postdoctoral fellowship (PF-74507) to B.R.M., and an American Cancer Society–California senior postdoctoral fellowship (PF-4129) to B.R.M. W.F. was supported by a National Institutes of Health shared equipment grant (S10 RR 11404-01A1) and a grant from the Foundation for Medical Research.
Abbreviations used:
- AL
annulate lamellae
- BAPTA
bis-(o-aminophenoxy)-N,N,N′,N′-tetraacetic acid
- bioXE
biotinylated low-salt Xenopus wheat germ agglutinin eluate
- ELB
egg lysis buffer
- ELBS
egg lysis buffer salts
- EST
expressed sequence tag
- GlcNAc
N-acetylglucosamine
- NHS
N-hydroxysuccinimide
- PVDF
polyvinylidene difluoride
- SA
streptavidin
- TCT
triacetyltrichitobiose
- WGA
wheat germ agglutinin
REFERENCES
- Adam SA. Transport pathways of macromolecules between the nucleus and the cytoplasm. Curr Opin Cell Biol. 1999;11:402–406. doi: 10.1016/S0955-0674(99)80056-8. [DOI] [PubMed] [Google Scholar]
- Akey CW. Interactions and structure of the nuclear pore complex revealed by cryo-electron microscopy. J Cell Biol. 1989;109:955–970. doi: 10.1083/jcb.109.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey CW, Radermacher M. Architecture of the Xenopusnuclear pore complex revealed by three-dimensional cryo-electron microscopy. J Cell Biol. 1993;122:1–19. doi: 10.1083/jcb.122.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R, Ribas de Pouplana L, Enarson M, Bodoor K, Burke B. Nup84, a novel nucleoporin that is associated with CAN/Nup214 on the cytoplasmic face of the nuclear pore complex. J Cell Biol. 1997;137:989–1000. doi: 10.1083/jcb.137.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss R, Ribbeck K, Akin D, Kent HM, Feldherr CM, Gorlich D, Stewart M. Interaction between NTF2 and xFxFG-containing nucleoporins is required to mediate nuclear import of RanGDP. J Mol Biol. 1999;293:579–593. doi: 10.1006/jmbi.1999.3166. [DOI] [PubMed] [Google Scholar]
- Burke B, Gerace L. A cell free system to study reassembly of the nuclear envelope at the end of mitosis. Cell. 1986;44:639–652. doi: 10.1016/0092-8674(86)90273-4. [DOI] [PubMed] [Google Scholar]
- Corbett AH, Silver PA. Nucleocytoplasmic transport of macromolecules. Microbiol Mol Biol Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes VC, Rackwitz HR, Reidenbach S. Mediators of nuclear protein import target karyophilic proteins to pore complexes of cytoplasmic annulate lamellae. Exp Cell Res. 1997a;237:419–433. doi: 10.1006/excr.1997.3806. [DOI] [PubMed] [Google Scholar]
- Dabauvalle MC, Loos K, Merkert H, Scheer U. Spontaneous assembly of pore complex-containing membranes (“annulate lamellae”) in Xenopusegg extract in the absence of chromatin. J Cell Biol. 1991;112:1073–1082. doi: 10.1083/jcb.112.6.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damelin M, Silver PA. Mapping interactions between nuclear transport factors in living cells reveals pathways through the nuclear pore complex. Mol Cell. 2000;5:133–140. doi: 10.1016/s1097-2765(00)80409-8. [DOI] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 1986;45:699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc Natl Acad Sci USA. 1987;84:7552–7556. doi: 10.1073/pnas.84.21.7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockendorff TC, Heath CV, Goldstein AL, Snay CA, Cole CN. C-terminal truncations of the yeast nucleoporin Nup145p produce a rapid temperature-conditional mRNA export defect and alterations to nuclear structure. Mol Cell Biol. 1997;17:906–920. doi: 10.1128/mcb.17.2.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott DJ, Stutz F, Lescure A, Rosbash M. mRNA nuclear export. Curr Opin Genet Dev. 1994;4:305–309. doi: 10.1016/s0959-437x(05)80058-9. [DOI] [PubMed] [Google Scholar]
- Fabre E, Boelens WC, Wimmer C, Mattaj I, Hurt EC. Nup145p is required for nuclear export of mRNA and binds homopolymeric RNA in vitro via a novel conserved motif. Cell. 1994;78:275–289. doi: 10.1016/0092-8674(94)90297-6. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B, Hurt EC, Aebi U, Pante N. Molecular architecture of the yeast nuclear pore complex: localization of Nsp1p subcomplexes. J Cell Biol. 1998;143:577–588. doi: 10.1083/jcb.143.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone C, Darby MK, Gerace L. A monoclonal antibody against the nuclear pore complex inhibits nucleocytoplasmic transport of protein and RNA in vivo. J Cell Biol. 1988;107:1289–1297. doi: 10.1083/jcb.107.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay DR, Forbes DJ. Reconstitution of biochemically altered nuclear pores: transport can be eliminated and restored. Cell. 1990;60:17–29. doi: 10.1016/0092-8674(90)90712-n. [DOI] [PubMed] [Google Scholar]
- Finlay DR, Meier E, Bradley P, Horecka J, Forbes DJ. A complex of nuclear pore proteins required for pore function. J Cell Biol. 1991;114:169–183. doi: 10.1083/jcb.114.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay DR, Newmeyer DD, Hartl PM, Horecka J, Forbes DJ. Nuclear transport in vitro. J Cell Sci Suppl. 1989;11:225–242. doi: 10.1242/jcs.1989.supplement_11.17. [DOI] [PubMed] [Google Scholar]
- Finlay DR, Newmeyer DD, Price TM, Forbes DJ. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J Cell Biol. 1987;104:189–200. doi: 10.1083/jcb.104.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer WH, Karr D, Jackson B, Park M, Vale W. Microsequence analysis of proteins purified by gel electrophoresis. Methods Neurosci. 1991;6:69–84. [Google Scholar]
- Fontoura BM, Blobel G, Matunis MJ. A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J Cell Biol. 1999;144:1097–1112. doi: 10.1083/jcb.144.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MW, Wiese C, Allen TD, Wilson KL. Dimples, pores, star-rings, and thin rings on growing nuclear envelopes: evidence for structural intermediates in nuclear pore complex assembly. J Cell Sci. 1997;110:409–420. doi: 10.1242/jcs.110.4.409. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Gorsch LC, Dockendorff TC, Cole CN. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J Cell Biol. 1995;129:939–955. doi: 10.1083/jcb.129.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Dang T, Pane N, Shevchenko A, Mann M, Forbes D, Hurt E. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol Biol Cell. 1997;8:2017–2038. doi: 10.1091/mbc.8.10.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Schlaich N, Tekotte H, Hurt EC. Functional interaction of Nic96p with a core nucleoporin complex consisting of Nsp1p, Nup49p, and a novel protein Nup57p. EMBO J. 1995;14:76–87. doi: 10.1002/j.1460-2075.1995.tb06977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan T, Muller S, Klier G, Pante N, Blevitt JM, Haner M, Paschal B, Aebi U, Gerace L. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol Biol Cell. 1995;6:1591–1603. doi: 10.1091/mbc.6.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hartl P, Olson E, Dang T, Forbes DJ. Nuclear assembly with lambda DNA in fractionated Xenopusegg extracts: an unexpected role for glycogen in formation of a higher order chromatin intermediate. J Cell Biol. 1994;124:235–248. doi: 10.1083/jcb.124.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath CV, Copeland CS, Amberg DC, Del Priore V, Snyder M, Cole CN. Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiaeRAT2/NUP120 gene. J Cell Biol. 1995;131:1677–1697. doi: 10.1083/jcb.131.6.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw JE, Carragher BO, Milligan RA. Architecture and design of the nuclear pore complex. Cell. 1992;69:1133–1141. doi: 10.1016/0092-8674(92)90635-p. [DOI] [PubMed] [Google Scholar]
- Holt GD, Snow CM, Senior A, Haltiwanger RS, Gerace L, Hart GW. Nuclear pore complex glycoproteins contain cytoplasmically disposed O-linked N-acetylglucosamine. J Cell Biol. 1987;104:1157–1164. doi: 10.1083/jcb.104.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine MK, Watkins JL, Wente SR. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J Cell Biol. 1995;131:1699–1713. doi: 10.1083/jcb.131.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Mattaj IW. RNA export. Cell. 1995;81:153–159. doi: 10.1016/0092-8674(95)90323-2. [DOI] [PubMed] [Google Scholar]
- Kessel RG. Annulate lamellae: a last frontier in cellular organelles. Int Rev Cytol. 1992;133:43–120. doi: 10.1016/s0074-7696(08)61858-6. [DOI] [PubMed] [Google Scholar]
- Macaulay C, Forbes DJ. Assembly of the nuclear pore: biochemically distinct steps revealed with NEM, GTP gamma S, and BAPTA. J Cell Biol. 1996;132:5–20. doi: 10.1083/jcb.132.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay C, Meier E, Forbes DJ. Differential mitotic phosphorylation of proteins of the nuclear pore complex. J Biol Chem. 1995;270:254–262. doi: 10.1074/jbc.270.1.254. [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Takagi M, Ban T, Miyazaki M, Yamamoto T, Kondo Y, Yoneda Y. Identification and characterization of nuclear pore subcomplexes in mitotic extract of human somatic cells. Biochem Biophys Res Commun. 1999;254:417–423. doi: 10.1006/bbrc.1998.9953. [DOI] [PubMed] [Google Scholar]
- Meier E, Miller BR, Forbes DJ. Nuclear pore complex assembly studied with a biochemical assay for annulate lamellae formation. J Cell Biol. 1995;129:1459–1472. doi: 10.1083/jcb.129.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Shaikh S, Burke B, Dreyfuss G. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 1999;18:1982–1995. doi: 10.1093/emboj/18.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U, Rout MP, Maguire S, Blobel G, Wozniak RW. The yeast nucleoporin Nup188p interacts genetically and physically with the core structures of the nuclear pore complex. J Cell Biol. 1996;133:1153–1162. doi: 10.1083/jcb.133.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M, Stutz F, Lee L, Davis LI, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- Newmeyer DD, Wilson KL. Egg extracts for nuclear import and nuclear assembly reactions. Methods Cell Biol. 1991;36:607–634. doi: 10.1016/s0091-679x(08)60299-x. [DOI] [PubMed] [Google Scholar]
- Ohno M, Fornerod M, Mattaj IW. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- Pante N, Bastos R, McMorrow I, Burke B, Aebi U. Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J Cell Biol. 1994;126:603–617. doi: 10.1083/jcb.126.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MA, Forbes DJ, Dahlberg JE, Lund E. The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J Cell Biol. 1997;136:241–250. doi: 10.1083/jcb.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MA, Macaulay C, Masiarz FR, Forbes DJ. Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J Cell Biol. 1995;128:721–736. doi: 10.1083/jcb.128.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Blobel G, Wozniak RW. Nup155 is a novel nuclear pore complex protein that contains neither repetitive sequence motifs nor reacts with WGA. J Cell Biol. 1993;121:1–9. doi: 10.1083/jcb.121.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–652. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Blobel G. Isolation of the yeast nuclear pore complex. J Cell Biol. 1993;123:771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Wente SR. Pores for thought: nuclear pore complex proteins. Trends Cell Biol. 1994;4:357–365. doi: 10.1016/0962-8924(94)90085-x. [DOI] [PubMed] [Google Scholar]
- Saitoh H, Dasso M. The RCC1 protein interacts with Ran, RanBP1, hsc70, and a 340-kDa protein in Xenopusextracts. J Biol Chem. 1995;270:10658–10663. doi: 10.1074/jbc.270.18.10658. [DOI] [PubMed] [Google Scholar]
- Shah S, Forbes DJ. Separate nuclear import pathways converge on the nucleoporin Nup153 and can be dissected with dominant-negative inhibitors. Curr Biol. 1998;8:1376–1386. doi: 10.1016/s0960-9822(98)00018-9. [DOI] [PubMed] [Google Scholar]
- Shah S, Tugendreich S, Forbes D. Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J Cell Biol. 1998;141:31–49. doi: 10.1083/jcb.141.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker DK, Vann LR, Goldberg MW, Allen TD, Wilson KL. TPEN, a Zn2+/Fe2+ chelator with low affinity for Ca2+, inhibits lamin assembly, destabilizes nuclear architecture and may independently protect nuclei from apoptosis in vitro. Cell Calcium. 1998;23:151–164. doi: 10.1016/s0143-4160(98)90114-2. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes sec13p and a sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Snow CM, Senior A, Gerace L. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J Cell Biol. 1987;104:1143–1156. doi: 10.1083/jcb.104.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderqvist H, Hallberg E. The large C-terminal region of the integral pore membrane protein, POM121, is facing the nuclear pore complex. Eur J Cell Biol. 1994;64:186–191. [PubMed] [Google Scholar]
- Stewart M, Whytock S, Mills AD. Association of gold-labeled nucleoplasmin with the centers of ring components of Xenopusoocyte nuclear pore complexes. J Mol Biol. 1990;213:575–582. doi: 10.1016/S0022-2836(05)80242-6. [DOI] [PubMed] [Google Scholar]
- Stoffler D, Fahrenkrog B, Aebi U. The nuclear pore complex: from molecular architecture to functional dynamics. Curr Opin Cell Biol. 1999;11:391–401. doi: 10.1016/S0955-0674(99)80055-6. [DOI] [PubMed] [Google Scholar]
- Sukegawa J, Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell. 1993;72:29–38. doi: 10.1016/0092-8674(93)90047-t. [DOI] [PubMed] [Google Scholar]
- Sullivan KM, Busa WB, Wilson KL. Calcium mobilization is required for nuclear vesicle fusion in vitro: implications for membrane traffic and IP3 receptor function. Cell. 1993;73:1411–1422. doi: 10.1016/0092-8674(93)90366-x. [DOI] [PubMed] [Google Scholar]
- Suprynowicz FA, Gerace L. A fractionated cell-free system for analysis of prophase nuclear disassembly. J Cell Biol. 1986;103:2073–2081. doi: 10.1083/jcb.103.6.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MT, Siniossoglou S, Podtelejnikov S, Benichou JC, Mann M, Dujon B, Hurt E, Fabre E. Two functionally distinct domains generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins. EMBO J. 1997;16:5086–5097. doi: 10.1093/emboj/16.16.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Blobel G. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic transport. J Cell Biol. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Blobel G. NUP145 encodes a novel yeast glycine-leucine-phenylalanine-glycine (GLFG) nucleoporin required for nuclear envelope structure. J Cell Biol. 1994;125:955–969. doi: 10.1083/jcb.125.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RW, Blobel G. The single transmembrane segment of gp210 is sufficient for sorting to the pore membrane domain of the nuclear envelope. J Cell Biol. 1992;119:1441–1449. doi: 10.1083/jcb.119.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- Zabel U, Doye V, Tekotte H, Wepf R, Grandi P, Hurt EC. Nic96p is required for nuclear pore formation and functionally interacts with a novel nucleoporin, Nup188p. J Cell Biol. 1996;133:1141–1152. doi: 10.1083/jcb.133.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]