Abstract
Axonemes are highly organized microtubule-based structures conserved in many eukaryotes. In an attempt to study axonemes by a proteomics approach, we selectively cloned cDNAs of axonemal proteins by immunoscreening the testis cDNA library from the ascidian Ciona intestinalis by using an antiserum against whole axonemes. We report here a 37-kDa protein of which cDNA occurred most frequently among total positive clones. This protein, named LRR37, belongs to the class of SDS22+ leucine-rich repeat (LRR) family. LRR37 is different from the LRR outer arm dynein light chain reported in Chlamydomonas and sea urchin flagella, and thus represents a novel axonemal LRR protein. Immunoelectron microscopy by using a polyclonal antibody against LRR37 showed that it is localized on the tip of the radial spoke, most likely on the spoke head. The LRR37 protein in fact seems to form a complex together with radial spoke protein 3 in a KI extract of the axonemes. These results suggest that LRR37 is a component of the radial spoke head and is involved in the interaction with other radial spoke components or proteins in the central pair projection.
INTRODUCTION
Eukaryotic cilia and flagella present in diverse types of cells perform motile, sensory, and developmental functions in organisms from protists to humans. They are centered by precisely organized, microtubule-based structures, the axonemes. The axoneme consists of two central singlet microtubules, called the central pair, and nine outer doublet microtubules. These structures are well-conserved during evolution. The outer doublet microtubules, each composed of A and B subfibers, are connected to each other by nexin links, while the central pair is held at the center of the axoneme by radial spokes. Motility in cilia and flagella is generated by sliding of outer doublet microtubules driven by inner and outer dynein arms that protrude from the A tubule (Gibbons, 1981). Approximately 250 proteins are considered to construct an axoneme; however, little information is available about the specific function of each protein and how individual components are involved in the assembly and functioning of the axoneme.
The radial spokes are T-shaped structures extending from the A-tubule of each outer doublet microtubule to the center of the axoneme. They are attached adjacent to inner arm dyneins and two or three of them are arranged within each 96-nm repeat (Warner and Satir, 1974; Witman et al., 1978; Goodenough and Heuser, 1985, 1989). Biochemical, ultrastructural, and genetic studies have suggested that spokes interact with proteins projecting from the central pair and regulate the activity of dyneins (Huang et al., 1982; Smith and Sale, 1992; Smith, 2002). Radial spokes in Chlamydomonas flagella have been shown to be composed of 17 proteins, of which five are localized at the spoke head and 12 in the spoke stalk (Huang et al., 1982). Several radial spoke proteins have been cloned and characterized (Curry et al., 1992; Curry and Rosenbaum, 1993). Radial spoke protein 3 (RSP3), for example, present at the proximal end of the spoke stalk, helps in anchoring the radial spoke to the outer doublet (Huang et al., 1981; Diener et al., 1993). Several lines of evidence suggest that radial spokes regulate the activity of inner arm dynein through protein phosphorylation and dephosphorylation (Porter and Sale, 2000).
We have recently collected cDNAs from the testis of the ascidian Ciona intestinalis (Inaba et al., 2002). Approximately 30 cDNA clones have been isolated with homology to known axonemal proteins, along with many genes apparently involved in spermatogenesis. For a comprehensive understanding of the molecular architecture of the axoneme, it would be necessary to understand the molecular nature of all the axonemal proteins. To identify and study novel axonemal proteins, polyclonal antibodies were raised against fractionated axonemes so that we could immunoscreen cDNAs of axonemal proteins. Western blot analysis revealed that >58% of the total axonemal proteins were recognized by the antiserum. By screening the testis cDNA library of C. intestinalis with this anitserum, a number of positive clones showing homology to uncharacterized proteins were obtained.
Here, we report the cloning, sequencing, and localization of a 37-kDa leusine-rich repeat (LRR) protein, LRR37. The LRR motifs are short sequences present in proteins with diverse functions and cellular locations, and widely participate in protein–protein interactions. Clones showing sequence homology to an LRR protein occurred in a higher frequency than other positive clones in this study. LRR37 is different from the LRR dynein light chain (LC) reported in Chlamydomonas (Benashski et al., 1999). We show it is a novel component of the radial spoke.
MATERIALS AND METHODS
Experimental Materials
Adult C. intestinalis were collected near the Education & Research Center of Marine Bio-Resources (Tohoku University, Miyagi, Japan). They were kept under constant light for 2–3 d for spermiation and accumulation of sperm into sperm duct.
Extraction and Fractionation of Axonemal Proteins
Sperm were obtained from sperm duct into filtered seawater (FSW) and kept on ice until an enough amount of sperm was collected. Sperm were washed once with FSW by centrifugation at 8000 × g for 10 min at 4°C. The sperm were suspended in FSW and homogenized to dissociate the heads and flagella. The heads were removed by centrifugation at 8000 × g for 5 min and the flagella in the supernatant were collected by centrifugation at 12,000 × g for 10 min. Flagella were demembranated with 0.1% Triton X-100 in buffer A (20 mM Tris-HCl, pH 8.0, 1 mM MgSO4, 0.15 M KCl, 0.5 mM EGTA) on ice for 10 min. Axonemes were obtained by centrifugation at 12,000 × g for 10 min. The axonemes were washed two to three times with buffer A. Successive extraction of the axonemes was carried out by the method of Inaba et al. (1988). The outer arm dynein was removed by suspending the axonemes in buffer A containing 0.6 M KCl and kept on ice for 30 min, followed by centrifugation at 12,000 × g for 15 min. The KCl-extracted axonemes were suspended in a low ionic strength buffer (1 mM Tris-HCl, 1 mM EDTA, pH 8.0) and dialyzed against the same buffer overnight. The suspension was centrifuged at 100,000 × g for 1 h at 4°C. Most part of the inner dynein arms, radial spokes, nexin links, and central pair apparatus are extracted in the supernatant, whereas the rest of these structures are still retained in the pellet.
Screening the Testis cDNA Library
The construction of λZAP II cDNA library of C. intestinalis testis was described previously (Padma et al., 2001). Briefly, phage particles were incubated with XL-1 Blue MRF′ host cells for 15 min at 37°C and plated at a density of 3000 phages per 10-cm dish. After incubation at 37°C for 4 h, the phage plaques were overlaid with Hybond-C extra membrane (Amersham Biosciences, Piscataway, NJ) that had been incubated in 10 mM isopropyl-1-thio-β-d-galactopyranoside solution. After incubation at 37°C for 4 h, the membranes were removed and washed in phosphate-buffered saline (PBS) with 0.05% Tween 20 (TPBS). The membranes were incubated with blocking solution (2% skim milk in TPBS) for 1 h at room temperature. The membranes were incubated with primary antibody (“anti-axoneme antiserum” containing equal volumes of anti-TD supernatant and anti-TD pellet; 1:2000 dilution) in blocking buffer for 2 h, washed with TPBS (4 × 10 min), incubated with secondary antibody (horseradish peroxidase [HRP]-conjugated rabbit IgG) in blocking buffer (1:2500 dilution) for 1 h, and washed with TPBS (3 × 10 min). The membranes were finally washed with PBS and positive plaques were visualized by HRP-catalyzed reaction by using hydrogen peroxide and diaminobenzidine. The positive plaques were picked up and suspended in suspending medium SM. The phage was plated at a density of 1000 phages per 10-cm dish and rescreened as described above. To obtain the cDNA of RSP3, the expressed sequence tag (EST) sequences of C. intestinalis testis (Inaba et al., 2002) were searched for by BLASTX, and clones showing sequence homology to RSP3 from Chlamydomonas and sea urchin were retained. Its full-length sequence was determined.
Isolation of Radial Spoke by KI Extraction
The radial spoke fraction is solubilized by treatment of KCl-extracted axonemes with buffer A containing 0.5 M KI on ice for 30 min (Yang et al., 2000) and centrifuged at 100,000 × g for 30 min. The supernatant was separated on a BioSilect SEC400 gel filtration column (300 × 7.8 mm; Bio-Rad, Hercules, CA) at a flow rate of 1.0 ml/min. Fractions (200 μl) were collected and proteins in each fraction were separated by SDS-PAGE with 10% polyacrylamide as the separating gel. The gel was stained with silver to check the protein pattern. For Western blot analysis, proteins were transferred to polyvinylidene difluoride membrane and immunoblotted using anti-LRR37 or anti-RSP3 antibody at a dilution of 1:5000 as described previously (Padma et al., 2001).
Sequence Analysis
The insert of cDNA in λZAP II vector was subcloned into pBluescript by in vivo excision and sequenced using a BigDye terminator sequencing kit with an ABI310 DNA sequencer. Translation of DNA sequence into amino acid sequence, calculation of molecular mass and estimation of isoelectric points were done by GENETYX-Mac software. Multiple sequence alignment was carried out by CLUSTALW. ProfileScan (http://hits.isb-sib.ch/cgi-bin/PFSCAN) was used for prediction of functional sites or domains in amino acid sequence.
Bacterial Expression and Isolation of Fusion Protein
The open reading frames in the cDNA inserts were amplified by polymerase chain reaction. Primers used were 5′-CGCGGGATCCGATCCGTTGCCTTACT-3′ (sense) and 5′-CGCGGAATTCCAA CACAAGCAAAGCA-3′ (antisense) for LRR37 and 5′-CGCGGGATCCATGGCTGCAGTGATTCCA-3′ (sense) and 5′-CGCGGAATTCGCTGTACAATACAAACGA-3′ (antisense) for RSP3. The amplified cDNA and pET 32a (+) were cleaved with EcoRI and BamHI, ligated to each other after removing the small DNA fragment by S-400 spin column (Amersham Biosciences), and transformed into the host AD494. The transformed colonies were inoculated into LB with 100 μg/ml ampicillin and incubated at 37°C until the OD600 reached ∼0.6. After addition of 2 mM isopropyl-1-thio-β-d-galactopyranoside to induce protein expression, the bacteria were further incubated overnight at 37°C. The expression of the recombinant protein was checked by SDS-PAGE with 10% polyacrylamide gel (Laemmli, 1970). The bacterial cells expressing the protein were harvested by centrifugation at 10,000 × g for 5 min at 4°C and suspended in the binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl pH 8.0, 6 M urea, and 0.5% Triton X-100). They were left at room temperature for 15 min to allow cell lysis and protein solubilization. Cell debris was removed by centrifugation at 12,000 × g for 15 min and the supernatant was loaded onto a column with Ni2+-immobilized His-Bond metal chelation resin (Novagen, Madison, WI). The column was washed with 20 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl pH 8.0, 6 M urea, and 0.5% Triton X-100 and eluted with 1 M imidazole in binding buffer. The fractions containing the protein were pooled and dialyzed against PBS. The purified protein was used as an antigen to raise an antibody.
Antibodies
Polyclonal antibodies against the recombinant proteins of LRR37 and RSP3 were raised in Balb c mice. The antigen was emulsified with Freund's complete adjuvant, and mice were given three subcutaneous injections at intervals of 10 d. A test bleed was done before collection of the antiserum.
For preparation of polyclonal antibodies against the axonemal proteins for screening the cDNA library, axonemes were suspended in a low ionic strength buffer (TD solution: 1 mM Tris-HCl, 1 mM EDTA, pH 8.0) and dialyzed overnight. The suspension was centrifuged at 100,000 × g for 1 h at 4°C to separate the extract (TD supernatant) and the pellet. The pellet was suspended in the same buffer (TD pellet). The TD supernatant and TD pellet were injected separately into two rabbits subcutaneously by mixing the antigen with Freund's complete adjuvant. Three doses were given at intervals of 10 d and finally a booster dose was given 1 wk before collection of serum.
Two-Dimensional (2D) Gel Electrophoresis
The axonemes prepared as described above were solubilized in a solution containing 8 M urea, 2 M thiourea, 10% isopropanol, 0.1% Triton X-100, 50 mM dithiothreitol, and 4% 3-[(3-cholamidopro-pyl)dimethylammonio]propanesulfonate at a protein concentration of 1.2 mg/ml. For the first dimension, ∼300 μg of axonemal proteins in the above-mentioned buffer was supplemented with 0.5% IPG buffer, incubated for 20 min at room temperature, and applied to the strip holder (for 11-cm immobilized pH gradient gel) (Amersham Biosciences). The IPG dry strips were put on the sample without trapping air bubbles and covered with silicone oil. After rehydration for 4 h at 0 V, isoelectrofocusing was started at 30 V for 10 h, followed by 200 V for 1 h, 500 V for 1 h, 1000 V for 1 h, and 8000 V for 5 h. The IPG strips were equilibrated for 30 min in a SDS-PAGE sample buffer containing 8 M urea, 50 mM dithiothreitol and subjected to second dimensional horizontal SDS-PAGE. The pI markers were purchased from Amersham Biosciences and Bio-Rad.
Western Blot Analysis
Proteins were separated by SDS-PAGE or 2D gel electrophoresis and transferred to polyvinylidene difluoride membranes. Membranes were treated with 7.5% skim milk in TPBS to prevent nonspecific protein binding. Blots were incubated with primary antibodies: anti-LRR37 at 1:5000 and anti-RSP3 at 1:5000 for 2 h at room temperature. After washing with TPBS, blots were incubated with HRP-conjugated secondary antibody at 1:5000 for 1 h at room temperature. After washing with TPBS four times, blots were developed using enhanced chemiluminescence kit, ECL-Plus (Amersham Biosciences).
Immunofluorescence Microscopy
Immunofluorescence microscopy was performed by the method of Inaba et al. (1993) with slight modifications. Sperm from C. intestinalis were collected in artificial seawater. After appropriate dilution, they were attached on slides precoated with 1 mg/ml poly-l-lysine. After incubation for 5 min at room temperature (RT), the sperm were fixed and permeabilized by incubation in methanol at −20°C for 10 min. After quick removal of methanol by air blow, the slides were rehydrated using excess PBS. The slides were transferred to a moist chamber and incubated with blocking buffer (10% sheep serum in PBS) for 1 h at RT and then with preimmune serum or anti-LRR37 serum in the blocking buffer for 1 h at RT, followed by washes with PBS (4 × 5 min) at RT. Samples were then incubated with Texas Red-conjugated goat anti-mouse IgG (Alexa 546; Molecular Probes, Eugene, OR) at 1:1000 dilution for 1 h at RT, washed with PBS (3 × 5 min) at RT, and mounted in 50% glycerol. Slides were examined by BX50 microscope (Olympus, Tokyo, Japan) with 40 or 100× glycerol immersion objective and images were photographed on FUJI film Provia 400.
Immunogold Labeling
Immunogold labeling was done as reported by Inaba et al. (1998) with some modifications. The sperm in artificial seawater were collected by brief centrifugation and demembranated using 0.04% Triton X-100 in buffer A. After washing twice with buffer A and once with PBS, the sperm were fixed with 1% glutaraldehyde in PBS for 10 min at room temperature. After washing with PBS three times, samples were treated with 1 mg/ml NaBH4 in PBS for 10 min at room temperature to inactivate free aldehyde groups and then washed twice with PBS. Samples were incubated in a blocking buffer containing 10 mg/ml bovine serum albumin (BSA) in PBS for 2 h at 4°C with occasional agitation. The sample was then incubated with control IgG, anti-LRR37 antibody (1:100), or anti-RSP3 antibody (1:100) in blocking buffer for 12 h at 4°C with occasional agitation and washed with PBS containing 1 mg/ml BSA four times (2 h each). Samples were finally incubated at 4°C for 12 h in goat anti-mouse secondary antibody conjugated with 5-nm gold (diluted to 1:20 in blocking buffer; Biocell, Cardiff, United Kingdom). They were then washed with PBS containing 1 mg/ml BSA four times (2 h each) and fixed with 2.5% glutaraldehyde in 0.12 M phosphate buffer, pH 7.3, at room temperature for 1 h. Samples were then suspended in 1% glutaraldehyde in phosphate buffer and stored until subsequent use for electron microscopy.
Electron Microscopy
Samples fixed with 1% glutaraldehyde in phosphate buffer, as described above, were postfixed with 1% OsO4 for 1 h, dehydrated in graded ethanol series, embedded in Epon812 through propylene oxide, and thin-sectioned with an average thickness of 70 nm. Sections were stained with uranyl acetate and lead citrate and observed with a 1200EX electron microscope (JEOL, Tokyo, Japan) at 80 kV.
RESULTS
Characterization of Antiserum against Ciona Axonemal Proteins
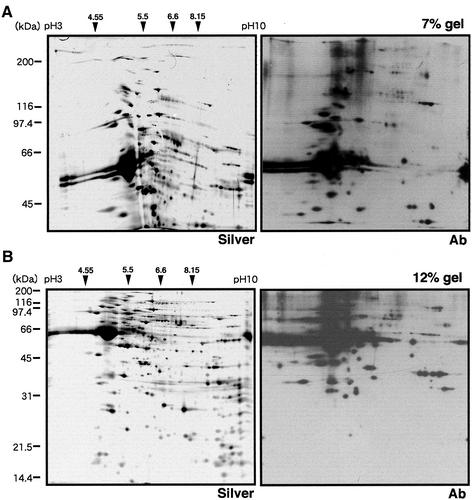
To isolate cDNAs of axonemal proteins, we first prepared antiserum that covers a number of axonemal proteins. To reduce the bias derived from the difference in antigenicity among different proteins, we used two axonemal fractions, TD supernatant and TD pellet (see MATERIALS AND METHODS). Each fraction was injected into rabbits separately and the serum collected from each rabbit was equally mixed before use (anti-axoneme antiserum). To characterize the anti-axoneme antiserum, sperm axonemal proteins were separated by 2D-gel electrophoresis and immunoblotted. Axonemal proteins of C. intestinalis were resolved into ∼400 spots. Western blot analysis showed that 230 of them were recognized by the antiserum (Figure 1), implying that the antiserum could be used for isolating cDNAs for 58% of total axonemal proteins.
Figure 1.
Two-dimensional gel electrophoresis of flagellar axonemes of C. intestinalis. Silver protein staining (left) and corresponding immunoblot with anti-axoneme antiserum (right) are shown. Axonemal proteins were first separated by isoelectric focusing (pH 3–10), followed by SDS-PAGE with 7% (A) and 12% (B) polyacrylamide for separating gels. Marker positions for isoelectric points or molecular mass (kilodaltons) are indicated.
Cloning of a Novel Axoneme LRR Protein by Immunoscreening
Screening of the C. intestinalis testis λZAPII cDNA library by using the antiserum resulted in the isolation of a number of positive cDNA clones. The average number of positive clones was 1–2 per 1000 plaques. The size of the cDNA inserts determined by polymerase chain reaction with T3 and T7 primers varied from 0.3 to 2.2 kb. At present, 76 cDNA inserts were sequenced with T3 primer and sequence homology was searched for by BLASTX (Tables 1 and 2). The 5′ sequences of 30 clones showed homology to various known axonemal proteins, such as α-tubulin, β-tubulin, tektin A1, dynein heavy chain, and dynein intermediate chains (group I). Ten clones showed homology to proteins that are known to associate with the axoneme, such as cAMP-dependent protein kinase, calcineurin B, and nucleoside diphosphate kinase (group II). Six showed homology to heat shock protein 40, which has not been identified as an axonemal protein (group III). The rest of the positive clones were divided into two groups: one (19 clones) showing homology to proteins that have been reported in the database with unknown function (group IV) and the other (11 clones) showing no sequence homology to any known proteins (group V).
Table 1.
Classification of positive cDNA clones isolated by immunoscreening with anti-axoneme antiserum
| Category | Description | Number of occurrences |
|---|---|---|
| Group I | Known axonemal proteins | 30 |
| Group II | Associated with axonemes | 10 |
| Group III | Not known as axonemal or axoneme-associated proteins | 6 |
| Group IV | Only appeared in database | 19 |
| Group V | No significant similarity | 11 |
| Total | 76 |
Table 2.
Gene description and occurrence of clones isolated
| Description | Number of occurrences | Accession number |
|---|---|---|
| Group I | ||
| Dynein beta heavy chain | 1 | AB091080 |
| Dynein intermediate chain 3 of sea urchin | 1 | BP021102 |
| TNDK dynein intermediate chain | 1 | AB057786a |
| Alpha-tubulin | 25 | BP02736 |
| Beta-tubulin | 1 | BP024273 |
| Tektin A1 | 1 | AB081510 |
| Group II | ||
| cAMP-dependent protein kinase regulatory subunit | 3 | BP024535 |
| Calcineurin B subunit | 6 | AB079059a |
| Nucleoside diphosphate kinase | 1 | AB091081 |
| Group III | ||
| Heat shock protein 40 (DnaJ) | 6 | AB079057a |
| Group IV | ||
| B7 protein | 11 | AB079056a |
| D. melanogaster CG1490 | 3 | AB083180a |
| D. melanogaster CG10869 | 3 | AB091082 |
| D. melanogaster CG15373 | 2 | AB083181a |
| Group V | ||
| K. Inaba, unpublished lambda ZAPII cDNA library | 1 | AB091083 |
| K. Inaba, unpublished lambda ZAPII cDNA library | 1 | AB091084 |
| K. Inaba, unpublished lambda ZAPII cDNA library | 1 | AB091085 |
| K. Inaba, unpublished lambda ZAPII cDNA library | 1 | AB091086 |
| K. Inaba, unpublished lambda ZAPII cDNA library | 2 | AB091087 |
| K. Inaba, unpublished cDNA library | 1 | BP023928 |
| K. Inaba, unpublished cDNA library | 1 | BP019748 |
| N. Satoh, unpublished cDNA library | 1 | AV891052 |
| N. Satoh, unpublished cDNA library | 1 | AV876427 |
| N. Satoh, unpublished cDNA library | 1 | AV964166 |
Full cDNA sequences are available. Others are EST data.
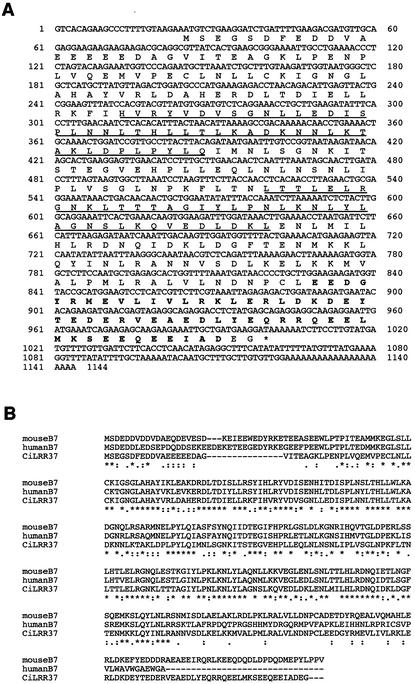
We tried to characterize each cDNA starting with a clone showing homology to mouse and human B7 protein. B7 protein was originally identified in a series of gene analysis of a gene-rich cluster at human chromosome 12p13 and its syntenic region in mouse chromosome 6 (Ansari-Lari et al., 1998), but its function has not been elucidated. Eleven clones with sequence homology to B7 protein were obtained and the one with the longest insert was sequenced. This 1.3-kb insert contained an open reading frame encoding a protein of 325 amino acid residues with a predicted molecular weight of 37,047 and a pI of 4.46 (Figure 2A). We termed this protein LRR37. LRR37 showed 51 or 41% sequence identity to mouse or human B7 protein, respectively (Figure 2B). A PROSITE search showed that LRR37 has four leucine-rich regions present in pairs at amino acid residues 77–122 and 166–207. The consensus sequence of an LRR structure is LxxLxxLxLxxNxIxxIxxLxx; where Ile and Leu can be replaced by Met, Phe, or Val (Kajava, 1998). The midregion that contains LRR regions is well conserved among human, mouse, and Ciona (Figure 2B). The LRR sequence at amino acid residues 193–262 showed high homology to the SDS22+ sequence (Ohkura and Yanagida, 1991). In the C-terminal region, a Glu-rich region is found at amino acid residues 269–324 (Figure 2A).
Figure 2.
(A) Nucleotide and deduced amino acid sequences of the full-length cDNA that encodes the leucine-rich repeat protein LRR 37 from C. intestinalis. These sequence data are available from GenBank/DDBJ/EMBL under the accession number AB079056. The regions of LRR are underlined. (B) Multiple alignment of Ciona LRR37, mouse B7, and human B7 proteins by CLUSTALW. Asterisks, colons, and dots indicate identical residues in all sequences in the alignment, conserved substitutions, and semiconserved substitutions, respectively.
Full-length sequencing of an LRR light chain of outer arm dynein was carried out to confirm that the LRR37 is different from the homolog of LRR LC1 reported in Chlamydomonas (Benashski et al., 1999). A cDNA clone from the Ciona testis cDNA EST project (Inaba et al., 2002) showing homology to Chlamydomonas LC1 was sequenced. This LRR protein has a molecular mass of 21 kDa with a pI of 5.52 (DDBJ/GenBank/EMBL accession number AB080948). This putative LRR-DLC showed no significant sequence homology to LRR37, and thus LRR37 is a novel LRR-protein in the axoneme.
A BLAST search in the Chlamydomonas EST database (http://www.kazusa.or.jp/en/plant/chlamy) revealed that an EST sequence (accession no. BE0565868) shows a significant homology to Ciona LRR37 (E = 2e-13). However, no information is available regarding its localization and function.
Western Blot Analysis of C. intestinalis Axonemal Fractions
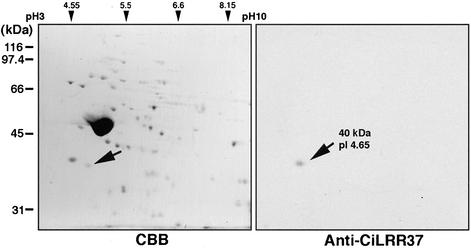
The protein-coding region of the LRR37 cDNA was subcloned into pET32 vector and an LRR37-thioredoxin fusion protein was prepared for raising polyclonal antibody. To examine the antibody specificity, axonemal proteins were separated by 2D gel electrophoresis and immunoblotted with the antibody. A protein spot with a molecular mass of 40 kDa and pI 4.65 was recognized by the antibody (Figure 3). Both values well agreed with those deduced from the cDNA sequence.
Figure 3.
Two-dimensional gel electrophoresis of flagellar axonemes of C. intestinalis. Left, Coomassie protein staining and right, corresponding immunoblot with anti-LRR37 antibody. Axonemal proteins were first separated by isoelectric focusing (pH 3–10), followed by SDS-PAGE with 10% polyacrylamide for separating gel. Marker positions for isoelectric points or molecular mass (kilodaltons) are indicated. The antibody recognized a single protein spot with molecular mass or pI of 40 kDa or 4.65, respectively.
Localization of LRR37 in the Axoneme
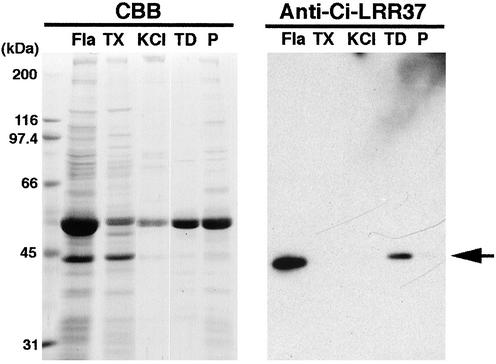
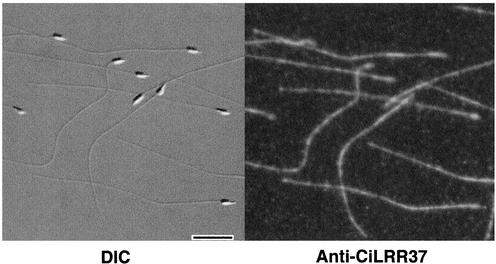
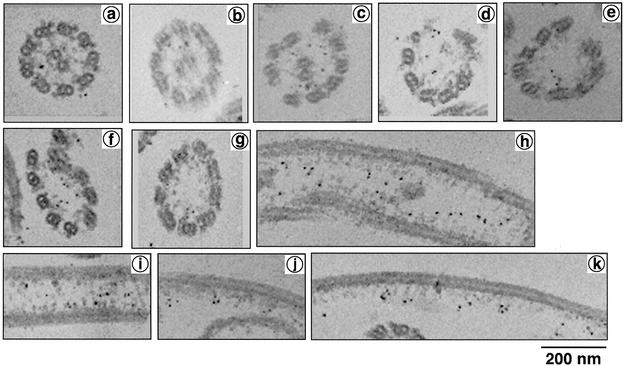
Successive extraction of axonemes gives a clue to the localization and properties of various proteins (Gibbons, 1981; Piperno et al., 1981). Western blot analysis showed that the anti-LRR37 antibody recognized a protein with molecular mass of ∼40 kDa, which was exclusively extracted into the low-ionic strength TD supernatant fraction (Figure 4). This suggests that the LRR37 protein is not a component of outer arm dynein but that of other structures, such as radial spokes, inner dynein arms, or nexin links. By immunofluorescence microscopy, it was shown that LRR37 is localized along the whole length of flagella (Figure 5), indicating that LRR 37 is a structural component of the axoneme. To specify the axonemal structure that contains LRR37, immunogold electron microscopy was performed. A small number of gold particles was observed attached to apparently intact axonemes; however, more particles were observed on the partially disrupted axoneme (Figure 6). In both cases, they were located around the tip of the radial spokes, most likely on spoke heads.
Figure 4.
Immunoblot of several fractions of axonemes by anti-LRR37 antibody. Flagella (Fla) were demembranated with Triton X-100 (TX) to obtain the axonemes. Axonemes were then successively treated with 0.6 M KCl solution (KCl), followed by the dialysis against a low ionic strength (TD, supernatant; and P, pellet after ultracentrifugation). A 40-kDa immunoreactive band was exclusively detected in TD supernatant.
Figure 5.
Immunofluorescence microscopy with anti-LRR37 antibody. Left, differential interference contrast image of Ciona sperm. Right, corresponding immunostaining with Alexa 546-conjugated secondary antibody. Bar, 10 μm.
Figure 6.
Immunogold electron microscopy with anti-LRR37 antibody. Labeling patterns in cross sections of intact (a and b) and partially disrupted axonemes (c–g), longitudinal sections of partially disrupted axonemes (h–k) are shown. Bar, 200 nm.
Comparison of Localization between LRR37 and RSP3
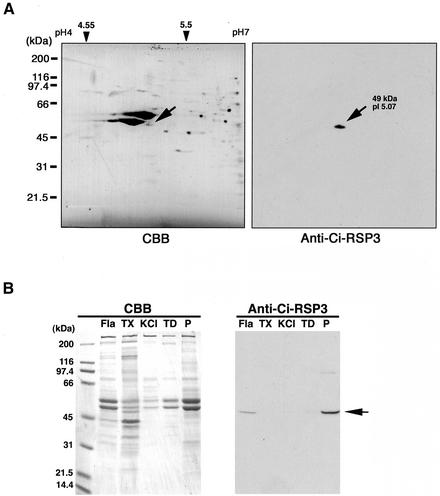
As shown above, immunoelectron microscopy showed that the LRR37 protein is probably located at the spoke head. To determine whether the LRR37 protein is a component of the radial spoke, its association with another spoke protein was examined. For this purpose, we first isolated the cDNA encoding RSP3. A cDNA insert with the 5′ EST showing homology to Chlamydomonas RSP3 (Inaba et al., 2002) was retained and full-length sequencing was carried out. The open reading frame codes for a 356 amino-acid protein with a molecular mass of 39 kDa (accession number, AB074930). Polyclonal antibody was raised against the RSP3 protein and used in experiments to elucidate the relationship between this protein and LRR37. In the 2D gel of whole axonemal proteins, the antibody recognized an axonemal protein with a molecular mass of 49 kDa and a pI of 5.07 (Figure 7A). Western blot analysis of the successive fractions of Ciona axonemes revealed that the anti-RSP3 anitibody strongly reacted with the pellet fraction after dialysis against TD low ionic strength solution (Figure 7B), in contrast with LRR37 that was soluble in the TD solution (Figure 4). In Chlamydomonas flagella, dialysis of KCl-extracted axonemes against a low ionic strength solution results in selective solubilization of the radial spoke head (Piperno et al., 1981). This further supports that LRR37 is a component of radial spoke head.
Figure 7.
(A) Two-dimensional gel electrophoresis of flagellar axonemes of C. intestinalis. Coomassie protein staining pattern (right) and corresponding immunoblot with anti-RSP3 antibody (right) are shown. Axonemal proteins were first separated by isoelectric focusing (pH 4–7), followed by SDS-PAGE with 7% polyacrylamide for separating gel. Marker positions for molecular mass (kilodaltons) and pI are indicated. The antibody recognized a single protein spot with molecular mass or pI of 49 kDa or 5.07, respectively. (B) Immunoblot of several fractions of axonemes by anti-RSP3 antibody. Flagella (Fla) were demembranated with Triton X-100 (TX) to obtain the axonemes. Axonemes were then successively treated with 0.6 M KCl solution (KCl), followed by the dialysis against a low ionic strength (TD, supernatant; and P, pellet after ultracentrifugation). A 49-kDa immunoreactive band was mostly detected in TD pellet.
Immunogold labeling of the axoneme with anti-RSP3 antibody showed that RSP3 is also localized on the radial spoke, but the gold particles were detected at loci much more proximal to the doublet microtubule (Figure 8). The full length of the radial spoke measured in the electron micrographs was 33.6 ± 2.6 nm (n = 22). Measurements of the distance from the spoke base to the gold particle clearly showed that the two proteins are localized at different positions; the average distance was 14.9 ± 4.8 (n = 45) or 25.9 ± 4.3 (n = 37) nm for RSP3 or LRR37, respectively. Taken together, the result well supports the idea that LRR37 is a component of the radial spoke head whereas RSP3 is located at the base of the radial spoke.
Figure 8.
Immunogold localization of RSP3. Labeling patterns in cross sections of the axoneme retaining 9 + 2 structure (a and b) or in partially disrupted axonemes (c–g) are shown. Bar, 200 nm.
Gel Filtration of KI Extract of the Axoneme
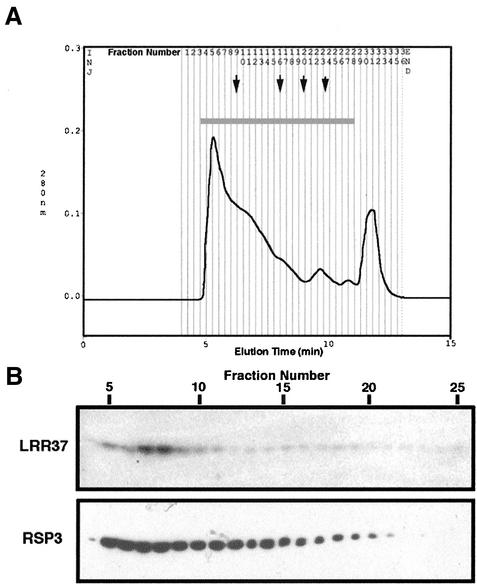
Radial spokes can be detached from axonemes with 0.5 M KI solution and isolated through sucrose density gradient as 20 S T-shaped particles in Chlamydomonas (Yang et al., 2001). The 0.5 M KI treatment of KCl-extracted Ciona axonemes solubilized a group of proteins. Both anti-RSP3 and anti-LRR37 antibodies strongly reacted with the KI extract (our unpublished data). The extract was further separated on a BioSilect SEC400 gel filtration column to isolate the radial spoke as a complex (Figure 9). Proteins in the fractions 4–25 were separated by SDS electrophoresis on a 10% gel and immunoblotted with both anti-LRR37 and anti-RSP3 antibodies. The result indicated that both proteins were eluted at the same position with an apparent molecular mass of 1300 kDa, showing that these two proteins are contained in the same complex.
Figure 9.
Gel filtration of KI extract by BioSilect SEC400 column. (A) KI extract from KCl-treated axonemes was loaded on a gel filtration column (300 × 7.8 mm) and separated at a flow rate of 1.0 ml/min. Fractions (200 μl) were collected. The numbers above the chromatogram indicate the fraction number. Arrows show the elution positions of molecular mass markers as follows: thyroglobulin (670 kDa), immunoglobulin G (150 kDa), ovalbumin (44 kDa), and myoglobin (17 kDa). (B) Proteins in the fraction 4–25 were separated by 10% SDS-PAGE and immunoblotted by anti-LRR37 and anti-RSP3 antibodies. Both proteins were coeluted at around the position with molecular mass of 1300 kDa.
DISCUSSION
In this study, we have developed a novel proteomics method for isolating cDNAs of axonemal proteins from a metazoan, C. intestinalis. We have shown that screening of a testis cDNA library with anti-axoneme antiserum results in isolation of cDNAs of distinct types of proteins. Extensive analyses of cDNAs and genomic DNA carried out recently have discovered a number of novel genes, but in many cases their functions have not been elucidated. In combination with testis cDNA EST analysis, isolation of novel axoneme-specific cDNAs would yield new insights into the molecular architecture and function of the axoneme.
The frequency of isolating cDNA of a particular protein by the present method does not always depend on its abundance in the testis. This is because the antigen used for raising antibody was a mixture of multiple axonemal proteins, and, therefore, there should be some selectivity due to the difference in antigenicity among the proteins. The isolation of a variety of cDNAs in the present study owes to the fact that the antiserum recognized at least 58% or possibly more of the total axonemal proteins, as assessed by western blot of 2D gels.
As an example showing the power of the present method, we studied a clone encoding LRR37, a protein with a homology to the B7 protein of human and mouse, and showed that it is a novel axonemal protein probably contained in the spoke head. The cDNA of this protein occurred most frequently among the initial 76 cDNA clones isolated. Sequence analysis showed that this protein possesses four LRR motifs and belongs to the SDS22+ class of the LRR protein family. The LRR motif is found in proteins with diverse intracellular and extracellular functions, and probably facilitates protein–protein interactions (Kobe and Deisenhofer, 1995; Buchanan and Gay, 1996; Kajava, 1998). Many members of the LRR family are involved in binding to components of signal transduction pathways, such as the PP1 binding protein SDS22+ (Kobe and Deisenhofer, 1994). Each repeat of 22–29 amino acids forms a short β-strand packed against an α-helix and resulting parallel β-sheets interact with the target protein. A number of proteins representing different LRR families have been reported recently. Among them is the Chlamydomonas dynein light chain 1, which belongs to the SDS22+ family (Benashski et al., 1999). Cloning and sequencing analysis revealed that the outer arm dynein from C. intestinalis also contains a 21-kDa LRR protein. Its sequence and size are different from those of LRR37. Recently, another testis-specific SDS22+-like LRR protein was identified in mouse and human (Xue and Goldberg, 2000). It is expressed in mouse pachytene spermatocyte and localized in nucleus. Although it has a Glu-rich region at C-terminal region like LRR37, it is also clearly different from LRR37 in other aspects.
The LRR37 is suggested to be localized to radial spokes. Immunogold images indicated that LRR37 is localized at more distal parts of radial spokes than RSP3. However, it should be noted that although RSP3 is known to be located at the base of the radial spoke, gold particles were observed at some distance (14.9 nm) from the base in immunolocalization of this protein. It is likely that this is due to the size of IgGs in primary and secondary antibodies. Similarly, the size of IgGs should be taken into account in considering the distance of LRR37 from the base (25.9 nm). Considering that the radial spoke is 33.6 nm in length in our measurements, we suggest that LRR37 is located at a distal part of the spoke, most likely the spoke head. This is supported by the fact that LRR37 is easily extracted by dialysis against TD low-ionic-strength solution, whereas RSP3 stays on the doublet microtubules after dialysis. Radial spokes in Chlamydomonas have been shown to be composed of 17 proteins, five located in the spoke head and at least 12 proteins in the spoke stalk (Huang et al., 1981). Although no protein homologous to LRR37 has been identified among Chlamydomonas spoke proteins thus far, it is possible that some of the five spoke head proteins are structurally or functionally related to LRR37.
It has been well documented that protein phosphorylation controls ciliary and flagellar dynein activity. Several types of protein phosphatases and kinases responsible for regulation of phosphorylation are anchored in the axoneme (Hasagawa et al., 1987; Hamasaki et al., 1989; San Agustin and Witman, 1994; Chaudhry et al., 1995; Inaba et al., 1998, 1999; Yang et al., 2000). RSP3, located at the base of the spoke stalk helps in anchoring the spoke to the doublet microtubule (Huang et al., 1981; Diener et al., 1993). RSP3 is an A-kinase anchor protein that mediates the binding of protein kinase A, which is known to regulate the dynein activity (Howard et al., 1994), to a position near the inner dynein arms (Gaillard et al., 2001). Radial spokes also contain calmodulin (Yang et al., 2000), a cofactor of calmodulin-dependent protein kinase and type 2B protein phosphatase calcineurin. The central pair apparatus and outer doublet microtubules contain PP1 and PP2A, respectively. These phosphatases possibly control phosphorylation of an inner arm dynein IC138 (Yang et al., 2000). Like Chlamydomonas flagella, Ciona sperm flagella contain an IC that seems to be dephosphorylated upon activation of motility (Padma, Hozumi, and Inaba, unpublished data). This suggests that a similar mechanism for activation of inner arm dynein through radial spoke/central pair is present in Ciona sperm flagella.
LRR37 may participate in signal transduction by interacting with a component of the radial spoke or that of the central pair projection. In addition to the LRR motif, its C-terminal region may be also functionally important. The C-terminal region of LRR proteins has been suggested to participate in the functional modulation of LRR-mediated interaction. For example, 30 amino acid residues at the C-terminal tail of Skp2 is known to extend back toward the first LRR domain tail, resulting in possible inhibition of substrate recognition (Schulman et al., 2000). Proteins such as nucleolin (Lapeyre et al., 1987) and CENP-B (Earnshaw et al., 1987) are known to have Glu-rich regions, which are suggested to be involved in protein–protein interaction (Gingras et al., 1998). Ciona LRR37 also has a Glu-rich region at the C-terminal region. In addition, Ciona RSP3 and an IC of outer arm dynein (IC3) (Padma et al., 2001) have Glu-rich region. In both LRR37 and IC3, the regions are located at C-terminal regions. Considering that both the outer arm dynein and radial spokes are highly structured multisubunit complexes, the Glu-rich region in LRR37 may be involved in the interaction with the protein other than the one which the LRR motif interacts with. From these structural characteristics, we speculate that LRR37 might be a mediator that controls the signal transduction from radial spoke/central pair to the inner arm dynein. By analogy with the PP1-binding protein SDS22+ (Ohkura and Yanagida, 1991), it is possible that LRR37 interacts with PP1, which has been shown to be located on the central pair projection in Chlamydomonas (Yang et al., 2000). For elucidating the specific function of LRR37 in axonemal motility, a knock-down experiment is now in progress in our laboratory by introducing morpholino antisense probe to the embryo.
In conclusion, we have isolated a novel LRR protein LRR37, as a possible component of radial spoke head. It may be involved in protein–protein interaction through both LRR and Glu-rich regions. Further studies of the interaction between LRR37 and other components in the radial spoke or central pair apparatus would lead to a new insight into the structure and function of the radial spoke/central pair apparatus system in axonemal movement. Furthermore, with this work, we have started on comprehensive characterization of axonemal proteins. C. intestinalis belongs to a primitive chordate that has a basic and simple body plan similar to that of vertebrates. This may enable us to analyze the structure and function of cilia and flagella much closer to those of vertebrates than those of Chlamydomona, in a manner much simpler than in studying vertebrate cilia and flagella. Identification of a variety of axonemal proteins in Ciona will not only help understand the structure and function of the axoneme but also yield insights into axoneme-related phenomena in multicellular organisms, such as the determination of the left-right body asymmetry (Nonaka et al., 1999), male infertility (Cowan et al., 2001), and primary ciliary dyskinesia (Blouin et al., 2000; Pazour et al., 2000).
ACKNOWLEDGMENTS
We are grateful to the members of Education and Research Center of Marine Bio-Resources (Tohoku University, Miyagi, Japan) and Misaki Marine Biological Station (University of Tokyo, Tokyo, Japan) for supplying C. intestinalis and to the staff of the Electron Microscopy Facility (National Institute for Basic Biology, Okazaki, Japan) for the support in electron microscopy. We thank Dr. Yuichiro Maeda for discussion on LRR protein. P.P. is a postdoctoral fellow of the Japan Society for the Promotion of Science. This work was supported by a Grant-in-Aid for Priority Area C from the Ministry of Education, Science, Sports and Culture, Japan (to K.I.; no. 12202008).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.02–06–0089. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.02–06–0089.
REFERENCES
- Ansari-Lari MA, et al. Comparative sequence analysis of a gene-rich cluster at human chromosome 12p13 and its syntenic region in mouse chromosome 6. Genome Res. 1998;8:29–40. [PubMed] [Google Scholar]
- Benashski SE, Patel-King RS, King SM. Light chain 1 from the Chlamydomonas outer dynein arm is a leucine-rich repeat associated with the motor domain of the γ heavy chain. Biochemistry. 1999;38:7253–7264. doi: 10.1021/bi990466y. [DOI] [PubMed] [Google Scholar]
- Blouin JL, et al. Primary ciliary dyskinesia. a genome-wide linkage analysis reveals extensive locus heterogeneity. Eur J Hum Genet. 2000;8:109–118. doi: 10.1038/sj.ejhg.5200429. [DOI] [PubMed] [Google Scholar]
- Buchanan SG, Gay NJ. Structural and functional diversity in the leucine-rich repeat family of proteins. Prog Biophys Mol Biol. 1996;65:1–44. doi: 10.1016/s0079-6107(96)00003-x. [DOI] [PubMed] [Google Scholar]
- Chaudhry P, Creagh S, Yu N, Brokaw CJ. Multiple protein kinase activities required for activation of sperm flagellar motility. Cell Motil Cytoskeleton. 1995;32:65–79. doi: 10.1002/cm.970320108. [DOI] [PubMed] [Google Scholar]
- Cowan MJ, Gladwin MT, Shelhamer JH. Disorders of ciliary motility. Am J Med Sci. 2001;321:3–10. doi: 10.1097/00000441-200101000-00002. [DOI] [PubMed] [Google Scholar]
- Curry AM, Rosenbaum JL. Flagellar radial spoke: a model molecular genetic system for studying organelle assembly. Cell Motil Cytoskeleton. 1993;24:224–232. doi: 10.1002/cm.970240403. [DOI] [PubMed] [Google Scholar]
- Diener DR, Ang LH, Rosenbaum JL. Assembly of flagellar radial spoke proteins in Chlamydomonas: identification of the axoneme binding domain of radial spoke protein 3. J Cell Biol. 1993;123:183–190. doi: 10.1083/jcb.123.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Sullivan KF, MachLin PS, Cooke CA, Kaiser DA, Pollard TD, Rothfield NF, Cleveland DW. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J Cell Biol. 1987;104:817–829. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard AR, Diener DR, Rosenbaum JL, Sale WS. Flagellar radial spoke protein 3 is an A-kinase anchoring protein (AKAP) J Cell Biol. 2001;153:443–448. doi: 10.1083/jcb.153.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons IR. Cilia and flagella of eukaryotes. J Cell Biol. 1981;91:107s–124s. doi: 10.1083/jcb.91.3.107s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras D, White D, Garin J, Cosson J, Huitorel P, Zingg H, Cibert C, Gagnon C. Molecular cloning and characterization of a radial spoke head protein of sea urchin sperm axonemes: involvement of the protein in the regulation of sperm motility. Mol Biol Cell. 1998;9:513–522. doi: 10.1091/mbc.9.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE. Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J Cell Biol. 1985;100:2008–2018. doi: 10.1083/jcb.100.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE. Structure of the soluble and in situ ciliary dyneins visualized by quick-freeze deep-etch microscopy. In: Warner FP, editor. Cell Movement. Vol. 1. New York: Alan R. Liss; 1989. pp. 121–140. [Google Scholar]
- Hamasaki T, Murtaugh T, Satir B, Satir P. In vitro phosphorylation of Paramecium axonemes and permeabilized cells. Cell Motil Cytoskeleton. 1989;12:1–11. doi: 10.1002/cm.970120102. [DOI] [PubMed] [Google Scholar]
- Hasagawa E, Hayashi H, Asukura S, Kamiya R. Stimulation of in vitro motility of Chlamydomonas axonemes by inhibition of cAMP dependant phosphorylation. Cell Motil Cytoskeleton. 1987;8:302–311. doi: 10.1002/cm.970080403. [DOI] [PubMed] [Google Scholar]
- Howard DR, Habermacher G, Glass DB, Smith EF, Sale WS. Regulation of Chlamydomonas flagellar dynein by an axonemal protein kinase. J Cell Biol. 1994;127:1683–1692. doi: 10.1083/jcb.127.6.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Piperno G, Ramanis Z, Luck DJ. Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J Cell Biol. 1981;88:80–88. doi: 10.1083/jcb.88.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Ramanis Z, Luck DJ. Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for flagellar function. Cell. 1982;28:115–124. doi: 10.1016/0092-8674(82)90381-6. [DOI] [PubMed] [Google Scholar]
- Inaba K, Akazome Y, Morisawa M. Purification of proteasomes from salmonid fish and their localization along sperm flagella. J Cell Sci. 1993;104:907–915. doi: 10.1242/jcs.104.3.907. [DOI] [PubMed] [Google Scholar]
- Inaba K, Kagami O, Ogawa K. Tctex2-related outer arm dynein light chain is phosphorylated at activation of sperm motility. Biochem Biophys Res Commun. 1999;256:177–183. doi: 10.1006/bbrc.1999.0309. [DOI] [PubMed] [Google Scholar]
- Inaba K, Mohri T, Mohri H. B-band protein in sea urchin sperm flagella. Cell Motil Cytoskeleton. 1988;10:506–517. [Google Scholar]
- Inaba K, Morisawa S, Morisawa M. Proteasomes regulate the motility of salmonid fish sperm through modulation of cAMP-dependent phosphorylation of an outer arm dynein light chain. J Cell Sci. 1998;111:1105–1115. doi: 10.1242/jcs.111.8.1105. [DOI] [PubMed] [Google Scholar]
- Inaba K, Padma P, Satouh Y, Shin-I T, Kohara Y, Satoh N, Satou Y. EST analysis of gene expression in testis of the ascidian Ciona intestinalis. Mol Reprod Dev. 2002;62:431–445. doi: 10.1002/mrd.10131. [DOI] [PubMed] [Google Scholar]
- Kajava AV. Structural diversity of leucine-rich repeat proteins. J Mol Biol. 1998;277:519–527. doi: 10.1006/jmbi.1998.1643. [DOI] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature. 1995;374:183–185. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lapeyre B, Bourbon H, Amalric F. Nucleolin, the major nucleolar protein of growing eukaryotic cells: an unusual protein structure revealed by nucleotide sequence. Proc Natl Acad Sci USA. 1987;84:1472–1476. doi: 10.1073/pnas.84.6.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1999;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- Ohkura H, Yanagida M. S. pombe gene sds22+ essential for a midmitotic transition encodes a leucine-rich repeat protein that positively modulates protein phosphatase-1. Cell. 1991;64:149–57. doi: 10.1016/0092-8674(91)90216-l. [DOI] [PubMed] [Google Scholar]
- Padma P, Hozumi A, Ogawa K, Inaba K. Molecular cloning, and characterization of a thioredoxin/nucleoside diphosphate kinase related dynein intermediate chain from the ascidian, Ciona intestinalis. Gene. 2001;275:177–183. doi: 10.1016/s0378-1119(01)00661-8. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, Cole DG. Chlamydomonas IFT88, and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia, and flagella. J Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Huang B, Ramanis Z, Luck DJ. Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J Cell Biol. 1981;88:73–79. doi: 10.1083/jcb.88.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Sale WS. The 9 + 2 axoneme anchors multiple inner arm dyneins, and a network of kinases, and phosphatases that control motility. J Cell Biol. 2000;151:F37–F42. doi: 10.1083/jcb.151.5.f37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Agustin JT, Witman GB. Role of cAMP in the reactivation of ram sperm. Cell Motil Cytoskeleton. 1994;27:206–218. doi: 10.1002/cm.970270303. [DOI] [PubMed] [Google Scholar]
- Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- Smith EF. Regulation of flagellar dynein by the axonemal central apparatus. Cell Motil Cytoskel. 2002;52:33–42. doi: 10.1002/cm.10031. [DOI] [PubMed] [Google Scholar]
- Smith EF, Sale WS. Regulation of dynein-driven microtubule sliding by the radial spokes in flagella. Science. 1992;257:1557–1559. doi: 10.1126/science.1387971. [DOI] [PubMed] [Google Scholar]
- Warner FD, Satir P. The structural basis of ciliary bend formation: radial spoke positional changes accompanying microtubule sliding. J Cell Biol. 1974;63:35–63. doi: 10.1083/jcb.63.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman GB, Plummer J, Sander G. Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J Cell Biol. 1978;76:729–747. doi: 10.1083/jcb.76.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J-C, Goldberg E. Identification of a novel testis-specific leucine-rich proteins in humans, and mice. Biol Reprod. 2000;62:1278–1284. doi: 10.1095/biolreprod62.5.1278. [DOI] [PubMed] [Google Scholar]
- Yang P, Diener DR, Rosenbaum JL, Sale WS. Localization of calmodulin, and dynein light chain LC8 in flagellar radial spokes. J Cell Biol. 2001;153:1315–1325. doi: 10.1083/jcb.153.6.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Fox L, Colbran RJ, Sale WS. Protein phosphatases PP1, and PP2A are located in distinct positions in the Chlamydomonas flagellar axoneme. J Cell Sci. 2000;113:91–102. doi: 10.1242/jcs.113.1.91. [DOI] [PubMed] [Google Scholar]