Abstract
In the budding yeast Saccharomyces cerevisiae, an actomyosin-based contractile ring is present during cytokinesis, as occurs in animal cells. However, the precise requirement for this structure during budding yeast cytokinesis has been controversial. Here we show that deletion of MYO1, the single myosin II gene, is lethal in a commonly used strain background. The terminal phenotype of myo1Δ is interconnected chains of cells, suggestive of a cytokinesis defect. To further investigate the role of Myo1p in cytokinesis, we conditionally disrupted Myo1 function by using either a dominant negative Myo1p construct or a strain where expression of Myo1p can be shut-off. Both ways of disruption of Myo1 function result in a failure in cytokinesis. Additionally, we show that a myo1Δ strain previously reported to grow nearly as well as the wild type contains a single genetic suppressor that alleviates the severe cytokinesis defects of myo1Δ. Using fluorescence time-lapse imaging and electron microscopy techniques, we show that cytokinesis in this strain is achieved through formation of multiple aberrant septa. Taken together, these results strongly suggest that the actomyosin ring is crucial for successful cytokinesis in budding yeast, but new cytokinetic mechanisms can evolve through genetic changes when myosin II function is impaired.
INTRODUCTION
The molecular mechanism of cytokinesis represents a fundamental question in cell biology. In animal cells, cytokinesis is achieved through concerted membrane constriction and addition, involving a cortical actomyosin-based contractile ring that is thought to provide the force to drive cleavage furrow ingression (Satterwhite and Pollard, 1992; Field et al., 1999; Hales et al., 1999). Assembly of the actomyosin ring is under tight spatial and temporal controls to ensure the proper segregation of genetic material and organelles. In contrast to animal cells, cytokinesis in plant cells requires targeted vesicle fusion leading to the formation of a cell plate that divides the cell (Staehelin and Hepler, 1996). Surprisingly, despite the presence of a rigid cell wall, cytokinesis in both budding and fission yeast involves an actomyosin-based contractile ring similar to that seen in animal cells (Chang and Nurse, 1996; Bezanilla et al., 1997; Kitayama et al., 1997; May et al., 1997; Bi et al., 1998; Lippincott and Li, 1998a). The existence of an evolutionarily conserved force-generating structure suggests that the mechanism of cytokinesis may also be conserved from yeast to higher eukaryotes, making budding yeast an attractive model for studying this fundamental process.
In light of this goal, it is important to determine the extent to which cytokinesis in Saccharomyces cerevisiae is dependent on the actomyosin ring. The critical role played by myosin II in eukaryotic cell division is evidenced by the cytokinesis failure in Dictyostelium myosin II null cells grown in suspension (De Lozanne and Spudich, 1987) as well as by antibody inhibition experiments in dividing embryos (Mabuchi and Okuno, 1977). Further genetic evidence also came from disruption of myosin II genes in Drosophila and Schizosaccharomyces pombe (Karess et al., 1991; Kitayama et al., 1997; Motegi et al., 1997). However, the requirement for myosin II during cytokinesis in S. cerevisiae has been controversial. A number of studies have reported that disruption of the single myosin II heavy chain gene, MYO1, is not lethal but results in a severe cell division defect (Watts et al., 1987) and abnormal cell wall organization (Rodriguez and Paterson, 1990; Schmidt et al., 2002). However, a more recent study reported that a MYO1 null mutation did not result in a strong cell division defect (Bi et al., 1998). It also raised the possibility that the primary defect caused by the myo1Δ mutation was due to a delay in cell separation. These data raised doubts as to whether the contractile ring represents the predominant mechanism for cytokinesis in budding yeast.
A lack of clarity in the extent to which budding yeast cytokinesis is dependent on the contractile ring could stem from the fact that earlier studies were all based on analyses of myosin II null cells in different strain backgrounds, where genetic and epigenetic modifiers accumulated over time can produce inconsistent and possibly misleading phenotypes. For example, the thick unresolved septa observed in some myo1Δ cells could be a consequence of a cytokinesis defect, because abnormal build up of septal material could eventually force the closure of the mother-bud opening but result in a subsequent septation failure. In this study we have quantitatively assessed the acute effects of loss of myosin II on cytokinesis and reinvestigated a previously described myosin II null strain that was shown to be capable of efficient cytokinesis.
MATERIALS AND METHODS
Media and Genetic Manipulations
Yeast cell culture and genetic techniques were carried out by methods described in Sherman et al. (1974). Yeast extract, peptone, dextrose (YPD) contained 2% glucose, 1% yeast extract, and 2% Bactopeptone (Difco Laboratories, Detroit, MI). YPR contained 2% raffinose, 1% yeast extract, and 2% Bactopeptone. YPGR contained 2% galactose, 2% raffinose, 1% yeast extract, and 2% Bactopeptone. Synthetic complete (SC) media was prepared by the method described by Kaiser et al. (1994).
Plasmid Construction
To generate the plasmid that expresses the C-terminal 868 amino acids of Myo1p under the control of the GAL1 promoter (pNT3), pLP8 was double digested with BglII and BamHI, yielding a 2.6-kb fragment encoding the C-terminal 884 amino acids of Myo1p. This fragment was ligated into the BamHI site of pRL62, a pRS306-based vector for expression of genes under the GAL1 promoter, and the correct orientation was determined. An in-frame ATG is provided by codon 1060 of Myo1p, 48-base pairs after the GAL1 promoter sequence. A promoter-less version of Myo1 (pNT119) was created for construction of strains carrying Myo1 tagged with GFP-6myc at the chromosomal locus. Briefly, a plasmid expressing Myo1-GFP-6myc, cloned PstI-NotI in the pRS305 backbone was digested with HindIII to remove the promoter and first 654 base pairs of Myo1 and religated to create pNT119. The plasmid expressing GFP-Tub1 (pAFS125) has been described previously (Straight et al., 1997). To generate the plasmid expressing Chs2-GFP (pLP31), a DNA fragment containing a 283-base pair 5′ sequence and the entire open reading frame of CHS2 was obtained by PCR against yeast genomic DNA, using primers NC2 (5′-GCGCGAAGCTTGTCTGAAAAGAAGATAGTAGG-3′) and CC2 (5′-GCGCGGGATCCGCCCTTTTTGTGGAAAACATT-3′). This fragment was digested with HindIII (included in the 5′ primer) and BamHI (included in the 3′ primer immediately after the coding sequence for the last amino acid) and then cloned between the corresponding sites in pRL73 (a COOH-terminal green fluorescent protein [GFP]-tagging vector; Lippincott and Li, 1998a).
Strain Construction
All strains used in this study are listed in Table 1. A complete MYO1 deletion was made using a one-step, PCR-mediated technique (Longtine et al., 1998). Briefly, the kanMX6 marker from pFA6a-kanMX6 was amplified together with sequences flanking the MYO1 open reading frame, using primers NT48 (5′- CGTGGTTAGAAGATCATAACAAAGTTAGACAGGACAACAACAGCAATACGG-ATCCCCGGGTTAATTAA-3′) and NT50 (5′-GCATATTCTCATTCTGTATATACAAAACATCTCATCATTATTTTTTTAAATAAA-GGGAATTCGAGCTCGTTTAAAC-3′), and transformed into RLY323. Kanr colonies were selected using YPD plates containing 200 g/ml geneticin (Invitrogen, Carlsbad, CA), generating RLY1236. The success of the deletion was determined by PCR using a forward primer corresponding to sequences 198–177 nucleotides upstream of the MYO1 start, MNF (5′-GCGCGCTGCAGCATCATTTAGCCCAAAAGGTA-3′) and a reverse primer internal to kanMX6, NT36 (5′-GCGAGCCCATTTATACCCAT-3′). To generate strains carrying Myo1-GFP at the chromosomal locus, pNT119 was digested with BclI and integrated into wild-type (RLY261) and GAL-tail–expressing (RLY884) strains, creating RLY1450 and RLY1451, respectively. To construct the Myo1 shut-off strain, a GAL-HA-MYO1 expression plasmid (pNT28) was first generated and transformed into the heterozygous diploid myo1Δ strain RLY1236. pKT64 (GAL-MLC1; Shannon and Li, 2000) was then transformed into the resulting diploid. Sporulation and tetrad analysis were carried out to yield RLY1776 (Table 1). The myo1Δ::URA3 mutation has been described previously (Bi et al., 1998). A diploid strain heterozygous for this mutation in BF264–15Du background (RLY1400) was created by mating JMY1236 to DLY2 (congenic haploid strains were obtained from D. Lew, Duke University Medical Center, Durham, NC). Sporulation and tetrad dissection of RLY1400 produced RLY1401 (myo1Δ::URA3 (sick)) and RLY1466 and RLY1467 (both myo1Δ::URA3 (healthy)). Mating of RLY1466 and RLY1467 created a strain (RLY1468) homozygous for the myo1Δ mutation and the healthy phenotype. Mating of RLY1466 and RLY1401 created a strain (RLY1488) homozygous for the myo1Δ mutation and heterozygous for the healthy phenotype. To visualize Chs2 dynamics in both wild-type and myo1Δ (healthy) cells, DLY2 and RLY1466 were both transformed with pLP31(Chs2-GFP) and pAFS125(GFP-Tub1) to generate RLY1673 and RLY1674, respectively.
Table 1.
Yeast strains
| Name | Genotype | Background | Source |
|---|---|---|---|
| RLY261 | MATa ura3-1 his3-11,15 leu2-3,112 trp1 ade2-1 bar1Δ | W303a | Elion Lab |
| RLY323 | MATa/α ura3-1/ura3-1 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ade2-1/ade2-1 bar1Δ/bar1Δ | W303a | Elion Lab |
| RLY669 | MATa ura3-1 his3-11,15 leu2-3,112 trp1-1 ade2-1 bar1Δ pMYO1-GFP (pLP8) GAL-tail∷URA3 (pNT3) | W303a | This work |
| RLY670 | MATa ura3-1 his3-11,15 leu2-3,112 trp1-1 ade2-1 bar1Δ pMYO1-GFP (pLP8) GAL-HA∷URA3 (pRL62) | W303a | This work |
| RLY884 | MATa ura3-1 his3-11,15 leu2-3,112 trp1-1 ade2-1 bar1Δ GAL-tail∷URA3 (pNT3) | W303a | This work |
| RLY1236 | MATa/α ura3-1/ura3-1 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ade2-1/ade2-1 bar1Δ/bar1Δ myo1Δ∷KAN/MYO1 | W303a | This work |
| RLY1355 | MATa ura3-1 his3-11,15 leu2-3,112 trp1-1 ade2-1 bar1Δ GAL-HA∷URA3 (pRL62) | W303a | This work |
| DLY2 | MATα ura3 his2 leu2-3,112 trp1-1 ade1 | BF264-Du | Lew lab |
| JMY1236 | MATa ura3 his2 leu2-3,112 trp1-1 ade1 myo1Δ∷URA3 | BF264-Du | Lew lab |
| RLY1400 | MATa/α ura3/ura3 his2/his2 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ade1/ade1 myo1Δ∷URA3/MYO1 | BF264-Du | This work |
| RLY1401 | MATα ura3 his2 leu2-3,112 trp1-1 ade1 myo1Δ∷URA3 (sick) | BF264-Du | This work |
| RLY1450 | MATa ura3-1 his3-11,15 leu2-3,112 trp1-1 ade2-1 bar1Δ MYO1-GFP∷LEU2 GAL-tail∷URA3 (pNT3) | W303a | This work |
| RLY1451 | MATa ura3-1 his3-11,15 leu2-3,112 trp1-1 ade2-1 bar1Δ MYO1-GFP∷LEU2 GAL-HA∷URA3 (pRL62) | W303a | This work |
| RLY1466 | MATα ura3 his2 leu2-3,112 trp1-1 ade1 myo1Δ∷URA3 (healthy) | BF264-Du | This work |
| RLY1467 | MATα ura3 his2 leu2-3,112 trp1-1 ade1 myo1Δ∷URA3 (healthy) | BF264-Du | This work |
| RLY1468 | MATa/α ura3/ura3 his2/his2 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ade1/ade1 myo1Δ∷URA3 (healthy)/myo1Δ∷URA3 (healthy) | BF264-Du | This work |
| RLY1488 | MATa/α ura3/ura3 his2/his2 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ade1/ade1 myo1Δ∷URA3 (sick)/myo1Δ∷URA3 (healthy) | BF264-Du | This work |
| RLY1673 | MATα ura3 his2 leu2-3,112 trp1-1 ade1 pCHS2-GFP (pLP31) pGFP-TUB1 (pAFS125) | BF264-Du | This work |
| RLY1674 | MATα ura3 his2 leu2-3,112 trp1-1 ade1 myo1Δ∷URA3 (healthy) pCHS2-GFP (pLP31) pGFP-TUB1 (pAFS125) | BF264-Du | This work |
| RLY1769 | MATa ura3-1 his3-11,15 leu2-3,112 trp1-1 ade2-1 bar1Δ GAL-tail∷URA3 (pNT3) CYK2-GFP:LEU2 | W303a | This work |
| RLY1771 | MATa ura3-1 his3-11,15 leu2-3,112 trp1-1 ade2-1 bar1Δ GAL-tail∷URA3 (pNT3) CDC12-GFP:LEU2 | W303a | This work |
| RLY1773 | MATa ura3-1 his3-11,15 leu2-3,112 trp1-1 ade2-1 bar1Δ cyk1Δ∷LEU2 GAL-tail∷URA3 (pNT3) CYK1-GFP:HIS3 | W303a | This work |
| RLY1776 | MATa ura3-1 his3-11,15 leu2-3,112 trp1-1 ade2-1 myo1Δ∷KAN GAL-HA-MYO1∷URA3 GAL-HA-MLC1∷LEU2 | W303a | This work |
Quantification of the Effects of GAL-Tail Expression on the Localization of Bud Neck Components
Cells were grown overnight in SC-Leu + 2% raffinose. Expression of GAL-tail was induced by the addition of galactose to 2%. At 0, 2, and 4 h after induction, the bud neck localization of GFP-tagged proteins (such as those listed in Table 2) was quantified in live cells using an Eclipse E800 microscope with a 100/1.40 oil differential-interference contrast objective (Nikon, Melville, NY). At least 100 cells were analyzed at each time point. Images were collected with a 0.5-s exposure to fluorescent light filtered through an EXHQ450/50 DM480 LP/BA465LP GFP filter set (Chroma, Brattleboro, VT) with a cooled RTE/CCD 782Y Interline camera (Princeton Instruments, Monmouth, NJ) using MetaMorph (Universal Imaging Corp., Downingtown, PA).
Table 2.
Effect of Myo1-tail overexpression on the localization of bud neck components
| Induction | % with Cdc12p-GFP ring | % with Cyk2p-GFP ring | % with Cyk1p-GFP ringa | % with Myo1p-GFP ring |
|---|---|---|---|---|
| 0-h induction | 100 | 22 ± 6 | 18 ± 2 | 85 ± 7 |
| 4-h induction | 99 ± 1 | 20 ± 4 | 19 ± 2 | 27 ± 4 |
Quantification of localization of bud neck components is described in MATERIALS AND METHODS. Strains RLY670 (Myo1p-GFP), 1769 (Cyk2p-GFP), 1771 (Cdc12p-GFP), and 1773 (Cyk1p-GFP) were used. 0- or 4-h induction refers to the time of induction of GAL-tail. The percentages are averages from two independent experiments. At least 200 total cells were counted at each time point/experiment.
Because the Cyk1p ring only exists transiently in mitotic cells, the percentages reflect the fractions of large budded cells (instead of total cells) that contained a Cyk1p ring.
Cell Synchronization, Zymolyase Treatment, and Cell Counting
RLY884 (GAL-tail) and RLY261 (wild-type) were grown in YPR liquid media overnight at 30°C. Cells were arrested in G1, before Myo1p localization to the bud neck, using 10 ng/ml α-factor, and expression of GAL-tail was induced by the addition of galactose to 2%. After 3 h, cells were washed five times with sterile water and resuspended in YPGR. At 0, 2, and 4 h after release, aliquots of cells were fixed directly in the growth media by the addition of formaldehyde to 5% final concentration. After incubation at 25°C for 1 h with gentle rocking, fixed cells were washed twice with PBS and then once with 1 M sorbitol in 50 mM KPO4, pH 7.5. Cells were incubated with 0.2 mg/ml zymolyase 20T (Seikagaku Corporation, Tokyo, Japan) in the above sorbitol buffer containing 2 mM DTT for 10–20 min at 37°C. Typically, >90% of the treated cells lost the refractile appearance when observed under a Labophot-2 microscope (Nikon Inc.) with a Plan40 0.5 ELWD objective, indicating that cell wall removal was efficient. After zymolyase treatment, cell numbers were counted on a hemacytometer. A chain or cluster of cells that could not be separated after cell wall removal was counted as one cell.
Confocal Imaging of Chs2-GFP Dynamics
RLY1673 (Chs2-GFP, GFP-Tub1) and RLY1674 (myo1Δ (healthy), Chs2-GFP, GFP-Tub1) were cultured in SC-Leu liquid media to midlogarithmic phase and placed in a growth chamber for imaging, essentially as described (Maddox et al., 2000). Briefly, gelatin (Sigma Chemical Co., St. Louis, MO; catalogue number G-2500) was added to SC-Leu media to 25% (wt/vol) and heated to 75°C for mixing of the gelatin and the medium. Growth chambers (made fresh for each experiment) were prepared by placing 50 μl of the liquefied gelatin/medium mixture between two microscope slides and applying pressure until the gelatin had solidified. The slides were then pried apart, leaving a thin slab of gelatin on one slide. One milliliter of the log phase culture was pelleted, washed once in sterile water, and resuspended in 50 μl sterile water. Seven microliters of the concentrated cells were pipetted onto the gelatin slab and covered with an 18 × 18 mm, no. 1 cover glass. The chamber was then sealed with Valap (1:1:1 vaseline:lanolin:paraffin). Fluorescence images were collected with a Perkin Elmer-Cetus (Boston, MA) spinning disk confocal on a Nikon TE2000 inverted microscope, using a 100× 1.4 NA Plan Apo objective lens. The 488-nm line from a krypton-argon laser was selected with a Chroma (Brattleboro, VT) 488/10-nm bandpass excitation filter. A Chroma single-wavelength, 488-nm transmitting dichroic mirror and HQ550 long-pass emission filter were used. Images of z-series optical sections were acquired with a Hamamatsu (Bridgewater, NJ) ORCA ER-cooled CCD camera and a Prior (Rockland, MD) ProScan focus motor. Through-focal z-series consisting of 13–15 frames acquired at 0.2-μm intervals were collected at each time point. Z-series were collected every 30 s (RLY1674) or every 1 min (RLY1673), using an exposure time of 800 ms. Images were binned 2 × 2 to increase signal over camera noise. MetaMorph imaging software (Universal Imaging Corp.) was used to control hardware during acquisition and analyze images. Adaptive blind deconvolution was performed for 40 iterations using AutoDeblur software (AutoQuant Imaging Inc., Watervliet, NY) before image analysis. For presentation of z-series, single images were constructed by maximum-brightness projection. To measure intensity profiles, a line was drawn through the Chs2-GFP ring, and intensity values were plotted against the distance along the line, using MetaMorph. To create overlay plots for Chs2p dynamics in wild-type and myo1Δ healthy cells (see Figure 7), successive frames from time-lapse movies were thresholded, binarized, and skeletonized (using MetaMorph) and then inverted and overlaid (using Adobe Photoshop, San Jose, CA).
Figure 7.
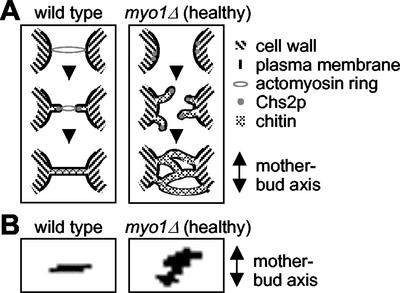
A model to explain membrane closure events during cytokinesis in the wild-type or myo1Δ (healthy) cells. (A) In wild-type cells (left panel) contraction of the actomyosin ring drives ingression of the plasma membrane, pulling Chs2p in toward the center of the bud neck. Chitin is deposited (through the action of Chs2p) outside the membrane in the deepening cleavage furrow, and this continues until a disk of chitin separates the two cells (the primary septum). In myo1Δ (healthy) cells (right panel), the lack of an actomyosin ring and the presence of a suppressor mutation may result in Chs2p no longer being concentrated in one position at the bud neck, and multiple membrane invaginations are formed in various directions. Membrane closure in myo1Δ (healthy) cells occurs in a haphazard manner that results in the enclosure of large amounts of cytoplasm between multiple septa. (B) Selected images from time-lapse movies of Chs2p-GFP in wild-type (left) and myo1Δ (healthy) (right) were overlaid, as described in MATERIALS AND METHODS. In each case, the orientation of the Chs2p-GFP ring was compared with that of the mother-bud axis.
Electron Microscopy
Cells were cultured overnight in SC-Leu media, pelleted, resuspended in YPD containing 10 μg/ml nocodazole, and arrested for 3 h at room temperature. Cells were washed three times with sterile water and resuspended in YPD, and aliquots taken at 30, 45, and 60 min after release from nocodazole arrest, to enrich for cells undergoing cytokinesis. Cells were fixed and embedded essentially as described previously (Schmidt et al., 2002). Aliquots of cells were washed in 0.1 M sodium phosphate buffer, pH 7.2 (PB) and fixed by suspension in PB containing 3% paraformaldehyde and 0.5% glutaraldehyde for 2 h at room temperature. After fixation the three time points were combined. Cells were washed twice in PB, resuspended in PB containing 1% sodium meta periodate (Pierce, Rockford, IL) for 1 h, rinsed again with PB, and quenched for 30 min in 50 mM NH4Cl in PB. After rinsing in PB, cells were dehydrated in ethanol (50% for 15 min, 70% for 15 min, 95% for 15 min, followed by 100% for 15 min twice). The cells were embedded in LR White resin (Electron Microscopy Sciences, Fort Washington, PA). Thin sections were stained in saturated uranyl acetate mixed 1:1 with acetone followed by lead citrate before examination in the electron microscope.
RESULTS
Myo1p Is Required for Cytokinesis
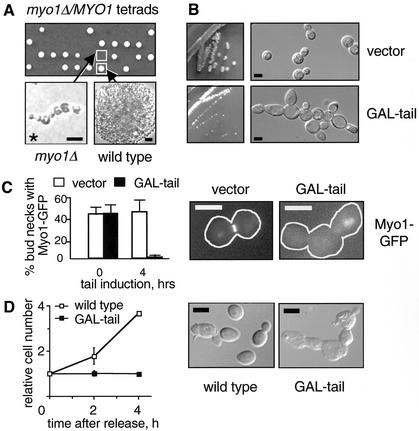
The goal of this study is to clarify the extent to which cytokinesis is dependent on the contractile ring in budding yeast. This issue has been unclear due to conflicting reports describing the phenotype of myo1Δ cells (Watts et al., 1987; Rodriguez and Paterson, 1990; Bi et al., 1998). We had previously noted that deletion of MYO1 in the W303a background was lethal, in contrast to the mild to severe growth defects described previously for myo1Δ mutants (Watts et al., 1987; Rodriguez and Paterson, 1990; Bi et al., 1998). As shown in Figure 1A, tetrad analysis of a W303a diploid strain heterozygous for myo1Δ showed 2:2 segregation for viability. The myo1Δ microcolonies exhibited a qualitatively uniform terminal morphology characterized by growth as an interconnected chain of cells, suggesting a defect in cell division.
Figure 1.
Myo1p is required for cytokinesis in the W303a background. (A) Phenotype of myo1Δ in W303a. Tetrad analysis of a diploid strain (RLY1236) heterozygous for the myo1Δ mutation showing 2:2 segregation for viability (upper panel). The plate was photographed after 2 d growth at 30°C. The morphology of a typical myo1Δ microcolony after 20 h growth at 30°C compared with a wild-type colony at the same stage (lower panels). Asterisk indicates that the myo1Δ microcolony did not increase further in size and eventually lysed. Note that the images are not to the same scale. (B) RLY1355 (vector control) and RLY884 (GAL-tail) were struck onto a YPGR plate and grown at 25°C for 3 d (left panels). A representative colony from each strain was picked for examination of cellular morphology (right panels). (C) RLY1451 (Myo1p-GFP plus vector control) and RLY1450 (Myo1p-GFP plus GAL-tail) cells were cultured overnight in YPR at 30°C. Galactose was added to induce tail expression, and samples were taken at 0 and 4 h. The number of cells in which Myo1p-GFP was localized to the bud neck was counted and divided by the total number of budded cells counted (at least 100) to give percent bud necks with Myo1p-GFP. Representative images showing Myo1p-GFP localization at the 4-h time point are shown next to the graph, with the cell outlines drawn in for clarity. (D) RLY261 (wild-type) and RLY884 (GAL-tail) cells were cultured overnight in YPR at 30°C. Galactose and α-factor (10 ng/ml final concentration) were added and the cells were grown at 30°C for 3 h. After washing three times with water, the cells were released into YPGR, and 5-ml duplicates were fixed with formaldehyde at 0, 2, and 4 h after release. Fixed cells were treated with zymolyase and counted on a hemacytometer. The resultant cell concentration at each time point was divided by that at time 0 to give relative cell number. Representative images of zymolyase-treated cells at the 4-h time point are also shown. Scale bars: A, 10 μm; B–D, 5 μm.
To distinguish whether Myo1p is required for cytokinesis, septum formation, and/or cell separation, we wanted to examine the response of wild-type cells to an acute loss of Myo1p function. Two strategies were used to conditionally disrupt Myo1p function. First, we have found in a separate study that the C-terminal 868 amino acids of Myo1p, downstream from the motor domain and the light chain binding sites, are sufficient for localization to the bud neck (Tolliday, N. and Li, R., unpublished results), and so we reasoned that over production of this region might interfere with endogenous Myo1p function. A construct (referred to as GAL-tail) was created where the C-terminal 868 amino acids of Myo1p can be conditionally expressed under the control of the inducible GAL1 promoter. Wild-type cells carrying GAL-tail grew normally in the presence of glucose but showed a severe growth defect in the presence of galactose. After 3 d growth at 25°C on galactose-containing media, the GAL-tail–expressing strain formed much smaller colonies than a control strain expressing vector alone (Figure 1B, left panels). Examination of GAL-tail–expressing colonies using a light microscope revealed chains of connected cells (Figure 1B, right panel), consistent with the terminal phenotype of the null.
The phenotype caused by over production of Myo1-tail might be due to competition between GAL-tail and endogenous Myo1p for localization at the bud neck. To test this possibility, the GAL-tail and vector control constructs were introduced into a strain where Myo1p tagged with GFP at the C terminus was expressed at the endogenous locus. Strains were cultured overnight in selective media containing raffinose, and galactose was added to induce GAL-tail expression. Before induction, both GAL-tail and vector control strains showed Myo1p-GFP localized at ∼44% of all bud necks (Figure 1C). After 4 h growth in galactose, the percentage of bud necks showing Myo1p-GFP localization had decreased to 1% in the presence of GAL-tail, whereas this value remained at 46% in the control strain (Figure 1C). A qualitatively similar result was also obtained when Myo1-GFP was expressed on a plasmid under MYO1 promoter (Table 2). Localization of other bud neck proteins that have been implicated in cytokinesis, including Cdc12p (septin), Cyk2/Hof1p, and Cyk1/Iqg1p (Tolliday et al., 2001), were not affected by GAL-tail expression (Table 2). These results suggest that high levels of Myo1-tail specifically displace Myo1p from the bud neck, and therefore can be used to further analyze the role of Myo1p in cell division.
To dissect the role of Myo1p during cell division, parallel cultures of wild-type cells with or without GAL-tail were grown overnight in raffinose-containing media. The cells were arrested in G1 using α-factor, and galactose was added to induce GAL-tail. This arrest point is before the time of Myo1p localization to the bud neck, and therefore GAL-tail should inhibit Myo1p localization upon release. After 3 h the cells were released from G1 arrest into galactose-containing media and allowed to proceed through subsequent cell cycles. Budding and nuclear division cycle were unaffected by Myo1-tail over expression (unpublished data). However, cells expressing GAL-tail grew as chains of cell bodies. To determine whether this defect was due to a block in cytokinesis (division of the cytoplasm) or cell separation, fixed cells from each time point were treated with zymolyase and counted. After 4 h, the number of wild-type cells had increased by ∼3.6-fold, whereas the number of cells expressing GAL-tail showed no increase (Figure 1D). Additionally, the cells expressing GAL-tail remained as chains of attached cells after cell wall removal, indicating a failure in cytokinesis.
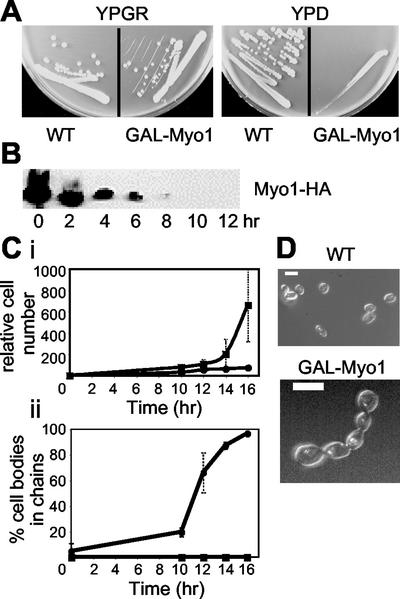
A second strategy that we used to conditionally disrupt Myo1p was to create a strain where expression of Myo1p can be turned off using the GAL1 promoter. An initial obstacle was that GAL-Myo1 itself is toxic and causes a cytokinesis defect in the wild-type background (unpublished data). We reasoned that this effect could be due to depletion of Mlc1p, a light chain for Myo1p (Boyne et al., 2000). Mlc1p level in the cell is limiting, probably due to other binding partners such as Myo2p and Cyk1/Iqg1p (Stevens and Davis, 1998; Shannon and Li, 2000). In fact, the cytokinesis defect caused by Myo1p overexpression is probably due to titration of Mlc1p away from Cyk1/Igq1p, as the interaction between the latter two proteins is essential. Thus, to be able to control Myo1p expression using the GAL1 promoter, a GAL-Mlc1 construct was cointroduced into the myo1Δ background to alleviate the toxic effect of GAL-Myo1p. GAL-Mlc1 itself has no effect on cell growth or cytokinesis (Shannon and Li, 2000). The resulting strain, RLY1776 (Table 1) grows as well as the wild type on media containing galactose but fails to grow on glucose-containing plates (Figure 2A).
Figure 2.
Cytokinesis defects caused by MYO1 shut-off. (A) Growth of wild-type (RLY261) and Myo1 shut-off (RLY1776) strains on YPGR or YPD plates, photographed after 3 d growth at 30°C. (B) RLY1776 cells were cultured overnight in YPGR at 30°C. A 10-ml sample was processed for immunoblot analysis (time 0). The rest of the cells were washed once with water and then shifted to YPD to shut off Myo1 expression. Samples were taken every 2 h after the shift and processed for immunoblotting using an anti-HA antibody to detect Myo1p level. (C) RLY261 (▪) and RLY1776 (●) strains were cultured and shifted as in B. (i) Samples were taken at 0, 10, 12, 14, and 16 h, fixed in formaldehyde, treated with zymolyase, and counted on a hemocytometer. Cell number calculation was done as described in Figure 1 legend. (ii) The percentage of cell bodies in chains was obtained by counting the number of cell bodies in chains of three or more cell bodies, divided by the total number of cell bodies in a defined field. The numbers at each time point in the plot were averages of duplicate samples. (D) Typical RLY 261 and RLY1776 cells from the 14-h time point are shown. Bar, 5 μm.
To examine the immediate effects of Myo1 shut-off on cytokinesis, we first determined that it took 10–12 h growth in glucose to eliminate Myo1p in the cell (Figure 2B). Before Myo1p depletion, there was little difference in cell number increase between the wild type and the Myo1 shut-off strain (unpublished data). As Myo1p levels drop, the rate of cell number increase in the Myo1 shut-off strain slowed down significantly, in contrast to the wild-type (Figure 2Ci). A cessation in cell number increase in the Myo1 shut-off strain was accompanied by an increase in the fraction of cell bodies that existed in chains of three or more cell bodies (Figure 2, Cii and D). This result further confirms that Myo1p is required for cytokinesis.
A Single Gene Suppressor Can Alleviate myo1Δ Cytokinesis Defects
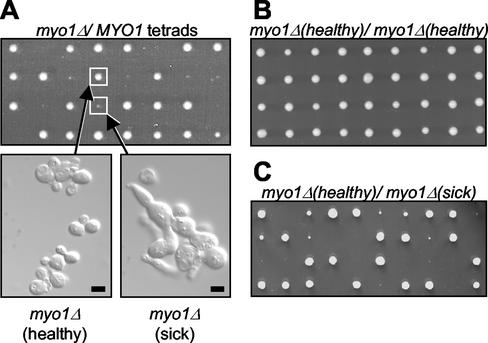
A previous study described myo1Δ growth defects that vary in severity depending on strain background (Bi et al., 1998). In particular, one strain (JMY1318) was documented to show relatively mild growth defects in comparison to wild-type cells. One possible explanation of the drastic phenotypic difference between this strain and the W303 myo1Δ strain was that this strain might have accumulated a suppressor mutation. To test this, we mated a haploid myo1Δ strain congenic with JMY1318 to the congenic wild-type strain (BF264-Du) to construct a diploid strain heterozygous for the myo1Δ mutation. Tetrad analysis showed a 2:2 segregation pattern for myo1Δ (marked with URA3) as expected (unpublished data). However, a 3:1 segregation pattern for robust growth was seen in many tetrads (Figure 3A, upper panel). Two distinct myo1Δ phenotypes were observed: one characterized by relatively normal growth (myo1Δ (healthy), referring to those with colony sizes >90% of that of the wild-type) and a second characterized by a growth defect that resulted in a severe reduction in colony size (myo1Δ (sick), referring to those with colony sizes <20% of that of the wild-type; Figure 3A, upper panel). Analysis of these phenotypes at the cellular level showed that myo1Δ (healthy) cells exhibit nearly normal morphology with the presence of only a few chains of cells, as previously reported for JMY1318 (Figure 3A, lower left panel). In contrast, examination of myo1Δ (sick) cells revealed many chains of enlarged cells that could not be separated after cell wall removal (by zymolyase treatment; Figure 3A, lower right panel), indicating a failure in cytokinesis.
Figure 3.
Phenotype of myo1Δ in BF264-Du background. (A) Tetrad analysis of a diploid strain (RLY1400) heterozygous for the myo1Δ mutation showing two distinct myo1Δ phenotypes, as represented by the boxed colonies (upper panel). The plate was photographed after 2 d growth at 30°C. An exponentially growing culture from each colony boxed above was fixed with formaldehyde and treated with zymolyase, and the cellular morphology was examined (lower panels). (B) Tetrad analysis of a diploid strain (RLY1468) created by mating two myo1Δ (healthy) strains, showing 100% robust growth. (C) Tetrad analysis of a diploid strain (RLY1488) created by mating myo1Δ (healthy) to myo1Δ (sick), showing 2:2 segregation for robust growth. Scale bar, 5 μm.
The presence of two distinct myo1Δ phenotypes together with a predominant 3:1 segregation pattern for robust growth suggest the possibility that a single suppressor mutation can alleviate the defects associated with the myo1Δ mutation. Under this hypothesis, the 3:1 and 2:2 segregation patterns for growth (Figure 3A, upper panel) would represent the tetratype and nonparental ditype patterns, respectively. Furthermore, 25% of the tetrad products from the heterozygous myo1Δ diploid strain described above would be expected to be myo1Δ (healthy), and 25% would be expected to be myo1Δ (sick). As shown in Table 3, analysis of 76 tetrad products revealed 16 myo1Δ (healthy) colonies (21%) and 22 myo1Δ (sick) colonies (29%), suggesting that the relatively normal growth of myo1Δ (healthy) is likely to be a result of a single gene suppressor mutation. To investigate this further, a diploid strain was constructed by mating two myo1Δ (healthy) strains. Tetrad analysis of this strain revealed normal growth of all tetrad products (Figure 3B). Additionally, tetrad analysis of a diploid strain created by mating myo1Δ (healthy) and myo1Δ (sick) strains showed 2:2 segregation for robust growth in 17 of 18 tetrads analyzed (Figure 3C). Taken together, these data strongly suggest that a mutation in a single gene is sufficient to suppress the growth and cytokinesis defects observed with myo1Δ in BF264-Du background.
Table 3.
Distribution of the two myo1Δ phenotypes observed in tetrad products from a strain heterozygous for myo1Δ in the BF264-Du background
| Wild type | myo1Δ (healthy) | myo1Δ (sick) | |
|---|---|---|---|
| Predicteda | 50% | 25% | 25% |
| Observed | 38/76 (50%) | 16/76 (21%) | 22/76 (29%) |
Percentages predicted if a single gene suppressor is involved.
Aberrant Septum Formation During Cell Division in Suppressed myo1Δ Cells
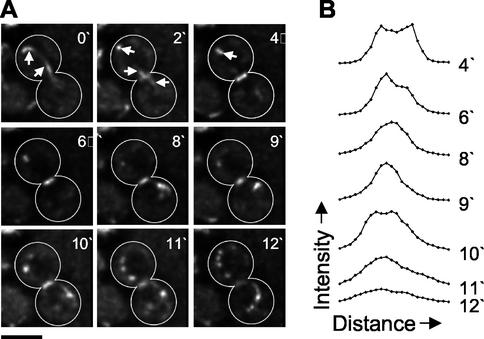
To investigate how the above suppressor could allow cytokinesis in myo1Δ cells, we characterized the events occurring during cell division in myo1Δ (healthy) cells. Chs2p, an integral membrane protein encoding Chitin Synthase II, localizes to the bud neck late in the cell cycle and is required for the formation of the primary division septum that separates the cells after cytokinesis (Shaw et al., 1991). Thus, Chs2p represents a good marker for both bud neck membrane dynamics during cytokinesis and the process of chitin deposition during septum formation. A functional plasmid-borne copy of Chs2 tagged with GFP at the C terminus was introduced into wild-type and myo1Δ (healthy) strains, in combination with GFP-tagged tubulin in order to visualize progression through the cell cycle. Time-lapse confocal microscopy was used to investigate Chs2p-GFP dynamics in living cells. Optical sections through the bud neck region of cells were acquired at regular intervals, deconvolved, and flattened to create two-dimensional projections of each time point (see MATERIALS AND METHODS). In wild-type cells, Chs2p-GFP localized as a faint ring at the bud neck, at or shortly after the time of spindle disassembly (Figure 4A, video 2′; note the distinction between the ring of Chs2p-GFP spanning the bud neck and the tubulin-GFP signal marked by arrows). The intensity of Chs2p-GFP ring fluorescence increased rapidly, and this was followed by a gradual reduction in the diameter of the Chs2p-GFP ring over the next 4–6 min to about one third of the original size (Figure 4A, 4′–9′). After this point, Chs2p-GFP then spread out again across the bud neck before fading away (Figure 4A, 10′–12′). Interestingly, discrete dots of fluorescence became visible in the cytoplasm around the time of Chs2p disappearance from the bud neck (Figure 4A, 11′ and 12′). These foci appeared to originate at and move away from the bud neck region and remained visible for at least 12 min after Chs2p-GFP ring disappearance (unpublished data).
Figure 4.
Chs2p dynamics in wild-type cells. (A) Chs2p-GFP, GFP-Tub1p expressing wild-type cells (RLY1673) were observed using three-dimensional (3D) confocal video microscopy (see online video material), as described in MATERIALS AND METHODS. A representative time-lapse series is shown, consisting of deconvolved 2D projections of each 3D image. Note that cell outlines have been drawn in for clarity. White arrows indicate signal from GFP-Tub1p, showing spindle disassembly. Scale bar, 3 μm. (B) Intensity profiles of bud neck localized Chs2p-GFP. A line was drawn through the Chs2p ring of the cell shown in A at each of the indicated time points, and pixel intensity values along the line were obtained, as described in MATERIALS AND METHODS. The measurements were then plotted against distance along the line.
To analyze Chs2p bud neck dynamics in a more quantitative manner, intensity profiles were generated, in which a line was drawn across the bud neck through the Chs2p-GFP ring, and the intensity values were plotted against distance along the line (Figure 4B). This analysis clearly shows two distinct peaks of Chs2p-GFP that move closer to each other to form a single central peak, before spreading out and fading away. This initial reduction in size of Chs2p-GFP is reminiscent of Myo1p dynamics during contraction of the actomyosin ring (Bi et al., 1998; Lippincott and Li, 1998a), suggesting that chitin deposition may be guided by contraction of the actomyosin ring.
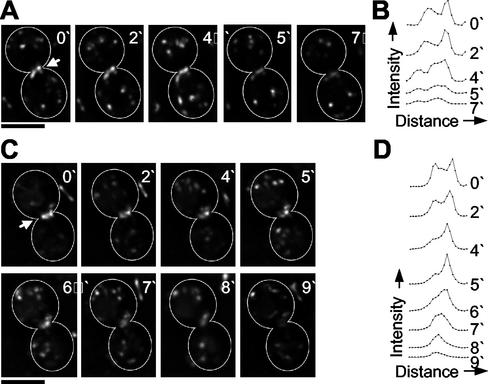
In myo1Δ (healthy) cells, Chs2p-GFP was also localized as a ring at the bud neck, although additional fluorescence was observed to extend into the cell bodies at either side of the bud neck (compare Figure 5A, video 0′ and Figure 5A, video 4′). This localization occurred ∼15–20 min after spindle disassembly, in contrast to wild-type cells (unpublished data). Two patterns of Chs2p dynamics were observed in myo1Δ (healthy) cells. In 3 of 6 movies, the diameter and intensity of the Chs2p-GFP ring was maintained for up to 4 min before the fluorescence faded away (Figure 5A, 0′–7′). Intensity profiles for Chs2p-GFP across the bud neck of these cells showed two distinct peaks that did not move closer together but remained separate before fading away (Figure 5B). In the other 3 of 6 movies, the diameter of the Chs2p-GFP ring decreased to a small dot >3–5 min before fading away (Figure 5C). However, this differed from Chs2p-GFP dynamics in wild-type cells in that only one side of the ring appeared to move inward, whereas the other faded. Analysis of intensity profiles for Chs2p-GFP at the bud neck in these cells showed two distinct peaks of fluorescence, one of which decreased in intensity rapidly, whereas the other peak moved inward, resulting in a single central peak (Figure 5D). In summary, in myo1Δ (healthy) cells Chs2p ring failed to undergo the symmetric reduction in size observed in wild-type cells, suggesting that primary septum formation in these cells may be abnormal.
Figure 5.
Chs2p dynamics in myo1Δ (healthy) cells. (A and C) Chs2p-GFP, GFP-Tub1p expressing myo1Δ (healthy) cells (RLY1674) were observed using three-dimensional (3D) confocal video microscopy, as described in MATERIALS AND METHODS. Two representative time-lapse series are shown, consisting of deconvolved two-dimensional projections of each 3D image. Note that cell outlines have been drawn in for clarity. White arrows indicate bud neck localized Chs2p-GFP. Scale bar, 3 μm. (B and D) Intensity profiles of bud neck localized Chs2p-GFP. A line was drawn through the Chs2p ring of the cells shown in A and C, respectively, at each of the indicated time points, and pixel intensity values along the line were obtained, as described in MATERIALS AND METHODS. The measurements were then plotted against distance along the line.
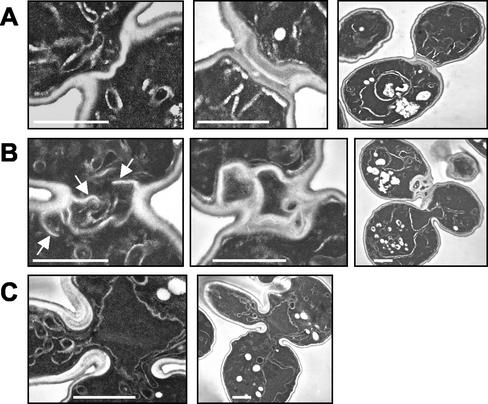
To provide further insight into the nature of the acquired suppressor mutation, we examined dividing wild-type, myo1Δ (healthy), and myo1Δ (sick) cells using electron microscopy. As shown in Figure 6A (left panel), wild-type cells assemble a primary septum 90° to the mother bud axis (Cabib et al., 1974). This is followed by deposition of secondary septa on either side of the primary septum, to form a trilaminar structure (Figure 6A, middle and right panels; Cabib et al., 1974). In contrast, multiple invaginations of the plasma membrane are observed in myo1Δ (healthy) cells (Figure 6B, right panel). These invaginations extend across the bud neck at a range of angles and result in the formation of multiple aberrant septa (Figure 6B, middle panel). This process results in enclosure of large amounts of cytoplasm between the septa (Figure 6B, middle and right panels), and at low frequencies cells with more than two cell bodies are observed (Figure 6B, right panel). This is in contrast to the gradual thickening of a wide area of cell wall at the bud neck reported for myo1Δ cells previously (Rodriguez and Paterson, 1990; Schmidt et al., 2002). The myo1Δ (sick) cells, on the other hand, did not exhibit the multisepta phenotype (Figure 6C). These data suggest that the putative suppressor mutation acquired by myo1Δ (healthy) cells results in an (indirect or direct) upregulation of membrane addition and chitin deposition in the bud neck region. This is sufficient to compensate for a lack of guidance in septum growth, resulting in formation of aberrant barriers between mother and daughter cytoplasms.
Figure 6.
Aberrant septum formation in myo1Δ (healthy) cells. Wild-type (A), myo1Δ (healthy) (B), and myo1Δ (sick) (C) cells were cultured overnight and then arrested using 10 μg/ml nocodazole. Aliquots of cells were fixed at 30, 45, and 60 min after release from arrest and pooled (to enrich for cells undergoing cytokinesis), followed by processing for electron microscopic analysis. (A) Typical wild-type cells with only the primary (left panel) or both the primary and secondary (middle and right panels, two magnifications are shown) septa. (B) Typical myo1Δ (healthy) cells in the process of septum formation (left panel) or after septa formation (middle and right panels). White arrows indicate multiple membrane invaginations observed in myo1Δ (healthy). (C) Typical two budded myo1Δ (sick) cell that had failed cytokinesis and septum formation (two magnifications are shown). Scale bars, 1 μm.
DISCUSSION
Actomyosin Ring-dependent and -independent Pathways of Cytokinesis
Experiments described above demonstrate that disruption of Myo1 function using a variety of methods results in a severe cytokinesis failure. In a separate study using synchronized yeast cell cultures, we also demonstrated that disruption of F-actin by latrunculin A blocked cytokinesis (Tolliday et al., 2002). These data strongly suggest that the actomyosin ring is crucial for cytokinesis at least in the commonly used W303a background. We hypothesize that the lack of consistency in the degree of cytokinesis defects in myo1Δ cells described in the literature was due to genetic or epigenetic modifiers that could accumulate over time. This hypothesis is supported by the identification of a suppressor mutation that masks the otherwise severe cytokinesis defects of myo1Δ cells in BF264-Du background where the healthiest myo1Δ cells were described (Bi et al., 1998). Another possible explanation for the two distinct myo1Δ phenotypes observed in the BF264-Du background is the acquisition of a synthetic lethal mutation rather than a suppressor mutation. Under this hypothesis, myo1Δ (healthy) cells would correspond to the myo1Δ mutation alone, and myo1Δ (sick) cells would correspond to myo1Δ in combination with a second mutation that drastically decreased the viability of the cells. However, if this were the case, the congenic wild-type (MYO1) strain would also have this second mutation, as indicated by the backcrossing data between the MYO1 and myo1Δ (healthy) strains. Because the myo1Δ (healthy) strain was originally derived from the MYO1 strain, it is more likely that a suppressor arose during culturing of the myo1Δ cells.
The viability of myo1Δ cells in some strain backgrounds suggests the existence of an actomyosin-independent mechanism for cytokinesis in budding yeast. It has been hypothesized that increased deposition of cell wall material at the bud neck may be sufficient to close the narrow (∼1 μm) channel that separates mother and daughter cells (Bi et al., 1998; Hales et al., 1999). This hypothesis is supported by electron microscopy studies depicting the thick abnormal septa in myo1Δ cells (Rodriguez and Paterson, 1990; Schmidt et al., 2002). However, for successful cell division, it is not merely sufficient to separate the two progeny cells; rather, cytokinesis must be coupled to the cell cycle to ensure correct segregation of nuclear and organelle materials. The accumulation of multinucleate chains of cells in the viable myo1Δ strains suggests an uncoupling of the nuclear and cell division cycles in the absence of Myo1p function. Such an uncoupling could have catastrophic effects on genetic stability, even if cytokinesis eventually occurs in some cells. Thus, the actomyosin ring is likely to be a critical target for the cell cycle machinery to coordinate cytokinesis with mitosis, in all genetic backgrounds.
Interestingly, Dictyostelium myosin II null cells, which fail to divide when grown in suspension, can undergo successful division when adhered to a substrate (De Lozanne and Spudich, 1987; Neujahr et al., 1997), suggesting that other organisms are also able to utilize an actomyosin-independent mechanism for cytokinesis. This adhesion-dependent, myosin II–independent division was initially attributed to traction-mediated cytofission, in which a giant multinucleate cell is essentially pulled apart in different directions, without coupling to the cell cycle (Spudich, 1989). However, a subsequent study showed that equatorial furrow formation in adherent myosin II null cells can occur in coordination with mitosis (Neujahr et al., 1997), suggesting that Dictyostelium can utilize at least two mechanisms for cell cycle–coupled cytokinesis. This second division mechanism is likely to involve CorA (encoding coronin, a WD-repeat containing protein) and AmiA/PiaA (a chemotaxis related gene), because cells lacking either gene show defects in adhesion-dependent cytokinesis (de Hostos et al., 1993; Chen et al., 1997; Nagasaki et al., 1998). Adherent cells lacking both myosin II and either coronin or AmiA showed significantly greater cytokinesis defects than was seen with each single mutant (Nagasaki et al., 2002), suggesting that adherent wild-type Dictyostelium are likely to utilize a combination of both mechanisms. Additionally, it has been reported that some mammalian cells can also undergo successful division when the contractile ring is disrupted (O'Connell et al., 1999, 2001). As is the case with Dictyostelium, this potential actomyosin-independent cell division is adhesion dependent.
How cells undergo cell cycle–coupled cytokinesis in the absence of a contractile ring is of considerable interest. Another process thought to be important for cytokinesis, in addition to contractile ring activity, is membrane addition (Straight and Field, 2000). A recent study showed that targeting of membrane vesicles to the cleavage furrow and contractile ring assembly are regulated separately by the cell cycle machinery (Shuster and Burgess, 2002). Thus, it is possible that the myosin II–independent pathway for cell division results from an upregulation of membrane addition events that can be subjected to correct temporal regulation. This would be consistent with our observation that in myo1Δ (healthy) cells cytokinesis is likely to result from an increased number of inward membrane/cell wall protrusions. In this strain cytokinesis seems to be coupled to the cell cycle, as evidenced by the large fraction of cells with normal morphology and nuclear content. Chitin deposited behind the membrane protrusions could serve to reinforce these inward protrusions, and the same could also be accomplished through adhesion in Dictyostelium or mammalian contractile ring–deficient cells.
Coordination of Cytokinesis and Septum Formation
During budding yeast cell division, the plasma membrane invaginates at the bud neck, and chitin is deposited in the growing invagination (Cabib et al., 1974, 2001). This process continues until a thin disk of chitin, the primary septum, separates the dividing cells. At this point, cytokinesis has been achieved. Secondary septa are then synthesized on both sides of the primary septum, forming a characteristic trilaminar structure, and cell separation is achieved through partial hydrolysis of the primary septum (reviewed in Cabib et al., 2001). A recent study has found that cells lacking MYO1 or CHS2, or both, show virtually identical cytokinesis defects, suggesting that MYO1 and CHS2 function in the same pathway to promote successful cytokinesis (Schmidt et al., 2002). We have found that Chs2p localizes to the bud neck at or around the time of spindle disassembly and undergoes a contraction-like reduction in size over ∼8 min, followed by respreading across the bud neck before fading away (Figure 4, A and B). Contraction of the actomyosin-based ring occurs over 7–9 min, concomitant with spindle disassembly (Bi et al., 1998; Lippincott and Li, 1998b). Thus our results, together with the work of Schmidt and coworkers (Schmidt et al., 2002), suggest a model in which contraction of the actomyosin ring drives invagination of the plasma membrane at the bud neck (Figure 7A). This movement may guide Chs2p to move inwardly, resulting in deposition of a ring of chitin that becomes a disk perpendicular to the mother-bud axis upon completion of cytokinesis.
In myo1Δ (healthy) cells, the guided inward movement of Chs2-GFP is no longer observed. Overlay plots of the Chs2-GFP bud neck localization (see MATERIALS AND METHODS) reveal that, in contrast to in wild-type cells, the Chs2-GFP ring is no longer oriented at 90° to the mother bud axis in myo1Δ (healthy) cells (Figure 7B). Instead, many different angles are observed at different times. This is consistent with the multiple membrane invaginations observed in dividing myo1Δ (healthy) cells by electron microscopy. The formation of multiple septa together with the expanded distribution of Chs2p around the bud neck in myo1Δ (healthy) cells suggest that the actomyosin ring not only guides the movement of Chs2p but may also be required to restrict Chs2p localization to a tight band at the division site. Interestingly, in fission yeast, the septum synthesizing enzyme Cps1p also requires the presence of an actomyosin ring for localization as a tight medial ring but not for accumulation as a diffuse band at the division site (Liu et al., 2002). Thus, the mechanism for coupling septum formation and cytokinesis may be conserved between budding and fission yeasts.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to John McMillan and Daniel Lew for providing us with the BF264 yeast strains; Jennifer Waters Shuler and the Nikon Imaging Center for assistance with confocal microscopy and image analysis; and Maria Ericsson for assistance with electron microscopy. We thank Josh Syken and Lynn Verplank for critical reading of the manuscript and John Pringle for helpful discussions. This work was supported by grant GM59964 from the National Institutes of Health to R.L.
Footnotes
Online version of this article contains video material. Online version is available at www.molbiolcell.org.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–09–0558. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–09–0558.
REFERENCES
- Bezanilla M, Forsburg SL, Pollard TD. Identification of a second myosin II in Schizosaccharomyces pombe: Myp2p is conditionally required for cytokinesis. Mol Biol Cell. 1997;8:2693–2705. doi: 10.1091/mbc.8.12.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, Pringle JR. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol. 1998;142:1301–1312. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne JR, Yosuf HM, Bieganowski P, Brenner C, Price C. Yeast myosin light chain, Mlc1p, interacts with both IQGAP and class II myosin to effect cytokinesis. J Cell Sci. 2000;113(Pt 24):4533–4543. doi: 10.1242/jcs.113.24.4533. [DOI] [PubMed] [Google Scholar]
- Cabib E, Roh DH, Schmidt M, Crottie LB, Varma A. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J Biol Chem. 2001;276(23):19679–19682. doi: 10.1074/jbc.R000031200. [DOI] [PubMed] [Google Scholar]
- Cabib E, Ulane R, Bowers B. A molecular model for morphogenesis: the primary septum of yeast. Curr Top Cell Regul. 1974;8(0):1–32. doi: 10.1016/b978-0-12-152808-9.50008-0. [DOI] [PubMed] [Google Scholar]
- Chang F, Nurse P. How fission yeast fission in the middle. Cell. 1996;84:191–194. doi: 10.1016/s0092-8674(00)80973-3. [DOI] [PubMed] [Google Scholar]
- Chen MY, Long Y, Devreotes PN. A novel cytosolic regulator, Pianissimo, is required for chemoattractant receptor and G protein-mediated activation of the 12 transmembrane domain adenylyl cyclase in Dictyostelium. Genes Dev. 1997;11(23):3218–3231. doi: 10.1101/gad.11.23.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hostos EL, Rehfuess C, Bradtke B, Waddell DR, Albrecht R, Murphy J, Gerisch G. Dictyostelium mutants lacking the cytoskeletal protein coronin are defective in cytokinesis and cell motility. J Cell Biol. 1993;120:163–173. doi: 10.1083/jcb.120.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lozanne A, Spudich JA. Disruption of the Dictyostelium discoideum myosin heavy chain gene by homologous recombination. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- Field C, Li R, Oegema K. Cytokinesis in eukaryotes: a mechanistic comparison. Curr Opin Cell Biol. 1999;11:68–80. doi: 10.1016/s0955-0674(99)80009-x. [DOI] [PubMed] [Google Scholar]
- Hales KG, Bi E, Wu JQ, Adam JC, Yu IC, Pringle JR. Cytokinesis: an emerging unified theory for eukaryotes? Curr Opin Cell Biol. 1999;11(6):717–725. doi: 10.1016/s0955-0674(99)00042-3. [DOI] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Karess RE, Chang XJ, Edwards KA, Kulkarni S, Aguilera I, Kiehart DP. The regulatory light chain of nonmuscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila. Cell. 1991;65:1177–1189. doi: 10.1016/0092-8674(91)90013-o. [DOI] [PubMed] [Google Scholar]
- Kitayama C, Sugimoto A, Yamamoto M. Type II myosin heavy chain encoded by the myo2 gene composes the contractile ring during cytokinesis in Schizosaccharomyces pombe. J Cell Biol. 1997;137:1309–1319. doi: 10.1083/jcb.137.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Li R. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin, to a ring structure involved in budding yeast cytokinesis. J Cell Biol. 1998a;140:355–366. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Li R. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J Cell Biol. 1998b;143:1947–1960. doi: 10.1083/jcb.143.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Tang X, Wang H, Oliferenko S, Balasubramanian MK. The localization of the integral membrane protein Cps1p to the cell division site is dependent on the actomyosin ring and the septation-inducing network in Schizosaccharomyces pombe. Mol Biol Cell. 2002;13(3):989–1000. doi: 10.1091/mbc.01-12-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mabuchi I, Okuno M. The effect of myosin antibody on the division of starfish blastomere. J Cell Biol. 1977;74:251–263. doi: 10.1083/jcb.74.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Bloom KS, Salmon ED. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat Cell Biol. 2000;2(1):36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May KM, Watts FZ, Jones N, Hyams JS. Type II myosin involved in cytokinesis in the fission yeast. Schizosaccharomyces pombe. Cell Motil Cytoskel. 1997;38:385–396. doi: 10.1002/(SICI)1097-0169(1997)38:4<385::AID-CM8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Motegi F, Nakano K, Kitayama C, Yamamoto M, Mabuchi I. Identification of Myo3, a second type-II myosin heavy chain in the fission yeast Schizosaccharomyces pombe. FEBS Lett. 1997;420:161–166. doi: 10.1016/s0014-5793(97)01510-x. [DOI] [PubMed] [Google Scholar]
- Nagasaki A, De Hostos EL, Uyeda TQ. Genetic and morphological evidence for two parallel pathways of cell-cycle-coupled cytokinesis in Dictyostelium. J Cell Sci. 2002;115(Pt 10):2241–2251. doi: 10.1242/jcs.115.10.2241. [DOI] [PubMed] [Google Scholar]
- Nagasaki A, Sutoh K, Adachi H. A novel Dictyostelium discoideum gene required for cAMP-dependent cell aggregation. Biochem Biophys Res Commun. 1998;244(2):505–513. doi: 10.1006/bbrc.1998.8230. [DOI] [PubMed] [Google Scholar]
- Neujahr R, Heizer C, Gerisch G. Myosin II-independent processes in mitotic cells of Dictyostelium discoideum: redistribution of the nuclei, re-arrangement of the actin system and formation of the cleavage furrow. J Cell Sci. 1997;110:123–137. doi: 10.1242/jcs.110.2.123. [DOI] [PubMed] [Google Scholar]
- O'Connell CB, Warner AK, Wang Y. Distinct roles of the equatorial and polar cortices in the cleavage of adherent cells. Curr Biol. 2001;11(9):702–707. doi: 10.1016/s0960-9822(01)00181-6. [DOI] [PubMed] [Google Scholar]
- O'Connell CB, Wheatley SP, Ahmed S, Wang YL. The small GTP-binding protein Rho regulates cortical activities in cultured cells during division. J Cell Biol. 1999;144(2):305–313. doi: 10.1083/jcb.144.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JR, Paterson BM. Yeast myosin heavy chain mutant: maintenance of the cell type specific budding pattern and the normal deposition of chitin and cell wall components requires an intact myosin heavy chain gene. Cell Motil Cytoskel. 1990;17:301–308. doi: 10.1002/cm.970170405. [DOI] [PubMed] [Google Scholar]
- Satterwhite LL, Pollard TD. Cytokinesis. Curr Opin Cell Biol. 1992;4:43–52. doi: 10.1016/0955-0674(92)90057-j. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Bowers B, Varma A, Roh DH, Cabib E. In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J Cell Sci. 2002;115(Pt 2):293–302. doi: 10.1242/jcs.115.2.293. [DOI] [PubMed] [Google Scholar]
- Shannon KB, Li R. A myosin light chain mediates the localization of the budding yeast IQGAP-like protein during contractile ring formation. Curr Biol. 2000;10:727–730. doi: 10.1016/s0960-9822(00)00539-x. [DOI] [PubMed] [Google Scholar]
- Shaw JA, Mol PC, Bowers B, Silverman SJ, Valdivieso MH, Duran A, Cabib E. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1991;114(1):111–123. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink G, Lawrence C. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1974. [Google Scholar]
- Shuster CB, Burgess DR. Targeted new membrane addition in the cleavage furrow is a late, separate event in cytokinesis. Proc Natl Acad Sci USA. 2002;99(6):3633–3638. doi: 10.1073/pnas.052342699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA. In pursuit of myosin function. Cell Regul. 1989;1(1):1–11. doi: 10.1091/mbc.1.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin LA, Hepler P. Cytokinesis in higher plants. Cell. 1996;84:821–824. doi: 10.1016/s0092-8674(00)81060-0. [DOI] [PubMed] [Google Scholar]
- Stevens RC, Davis TN. Mlc1p is a light chain for the unconventional myosin Myo2p in Saccharomyces cerevisiae. J Cell Biol. 1998;142:711–722. doi: 10.1083/jcb.142.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Field CM. Microtubules, membranes and cytokinesis. Curr Biol. 2000;10(20):R760–R770. doi: 10.1016/s0960-9822(00)00746-6. [DOI] [PubMed] [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Tolliday N, Bouquin N, Li R. Assembly and regulation of the cytokinetic apparatus in budding yeast. Curr Opin Microbiol. 2001;4:690–695. doi: 10.1016/s1369-5274(01)00270-3. [DOI] [PubMed] [Google Scholar]
- Tolliday, N., VerPlank, L., and Li, R. (2002). Rho1 directs formin-mediated actin ring assembly during budding yeast cytokinesis. Curr. Biol. (in press). [DOI] [PubMed]
- Watts FZ, Shiels G, Orr E. The yeast MYO1 gene encoding a myosin-like protein required for cell division. EMBO J. 1987;6(11):3499–3505. doi: 10.1002/j.1460-2075.1987.tb02675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.